Cells often rely on precise temporal coordination and can go through great troubles to time events in response to internal signals and environmental cues, as demonstrated by circadian clocks, cell-cycle control, and morphogenesis. But for any given set of signals and cues there can also be substantial random variability from cell to cell. The origin of such variation is understood in general terms—chemical reactions involve random collisions between diffusing molecules—but its extent cannot be inferred from first principles. Most single-cell studies have also focused on fluctuations in molecule numbers (Elowitz et al, 2002; Ozbudak et al, 2002), and there are rather few quantitative measurements (Bean et al, 2006) of intracellular timing even in the best-studied model systems. In this issue of Molecular Systems Biology, Stavans and co-workers (Amir et al, 2007) address this problem in an insightful study of how temporal fluctuations propagate along the lytic cascade of bacteriophage λ.
When λ phage infects a bacterium, it chooses between two paths: it either hijacks cellular resources, overproduces phage particles, and busts the cell open (lysis), or it integrates into the host chromosome and protects the cell from further phage infection (lysogeny). Lysogenic phage then quietly replicates with the chromosome until DNA damage activates the RecA protein, which degrades the λ repressor CI and derepresses phage promoters pL and pR. Promoter pL drives the expression of an anti-terminator for pR expression, allowing read-through to lytic downstream genes. A drop in CI thus only triggers lysis if it lasts long enough for pR to remain derepressed by the time the anti-terminator reaches a high enough concentration to allow read-through. This gives the phage a way of jumping ship if the host is in trouble, and gives the cell a grace period in which to repair DNA without a phage mutiny. It also filters out any fast spontaneous stochastic fluctuations in CI concentration that otherwise could trigger lysis without DNA damage.
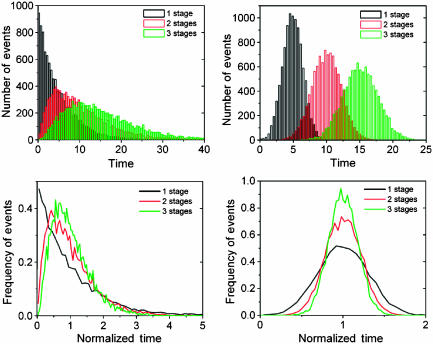
To study this process, Amir et al exposed λ-infected Escherichia coli cells to UV light and used fluorescent reporters to measure single-cell response times at different steps in the cascade that leads to lysis. Exposure to high UV led to lysis in all cells, whereas intermediate exposure caused some cells to lyse and others to merely delay division after a slight increase in pR activity. Interestingly, the UV dosage had no effect on the time between exposure and lysis, suggesting a binary switch that either launches or fails to launch the lysis cascade. When measuring the distribution of waiting times between events, they found a coefficient of variation (CV, the standard deviation divided by the mean) of around 15% for the lysis time in individual cells. They also found that the CV in the accumulated time decreased along the cascade, which was supported by a second observation that the waiting times in the early versus late stages were nearly uncorrelated. Amir et al emphasize that although each stage of the process may have evolved to provide a specific biochemical function, the mere fact that the signal is transmitted through successive steps could reduce variability in timing (Figure 1; Box 1). They also point out that the lack of correlations between different stages indicates that the factors responsible for the variability affect the timing of individual stages separately rather than affecting the cell as a whole. That does not necessarily rule out global factors like RNA polymerase or ribosomes as the main noise source: as the different parts of the cascade are far from identical and do not sense the environment at the same time, large and intermediately fast global fluctuations could randomize the timing of each stage without producing significant correlations between stages. Thus, the relative contribution of intrinsic and extrinsic noise in protein numbers to the timing of different stages remains to be examined in future studies.
Figure 1.
Histograms of number of events and normalized frequencies at each stage of a three-step cascade. The steps are independent and each lasts 5 time units on average. (Left) Exponentially distributed individual step times and gamma-distributed accumulated times. (Right) Normally distributed individual step times and normally distributed accumulated times.
Temporal fluctuations in a sequential process.
The relative variation in the time it takes to complete a sequential process can be formulated in terms of the properties of the individual steps. If these are statistically independent, the average and variance in the total times are simply sums over the individual steps. Keeping with the notation of Amir et al, the accumulated average and variance at the ith step of the cascade follow
where subscript denotes accumulated values and parentheses denote individual values. If all individual substeps also have the same averages and variances, Tind and Vind, this simplifies to
The coefficient of variation at step i thus decreases with the inverse square root of the number of steps, and adding more steps always decreases relative fluctuations. If the steps are independent but not identical, which approximately applies to the study of Stavans and co-workers, this is not necessarily true: adding another step could either increase or decrease the total relative variation. To have a decreasing accumulated CV at step i
then, each step must add more to the total average than to the total variance. If each step is exponentially distributed, so that  , this in turn requires that
, this in turn requires that
The last step must thus be fast compared to previous steps. If the individual times are not independent but rather positively correlated, the noise reduction is diminished. For negative correlations, one step of the cascade can compensate for another, leading to more efficient noise reduction.
Is the observed variation surprising? A reasonable naïve guess is that waiting times are exponentially distributed (CV=100%), as for ‘memoryless' elementary chemical reactions, which occur with a constant probability per time unit. However, because each stage of the cascade involves a large number of elementary steps, it is equally reasonable to expect that the total waiting time is normally distributed, with a variance that decreases with the number of steps. The problem is that the apparent complexity of a system is a poor indicator of its effective dynamic complexity: simple bimolecular reactions could give non-exponential behavior whereas extraordinarily complex systems can be perfectly memoryless. Everything is an approximation of something else and every step can be divided into further sub-steps. As Amir et al point out, this in turn means that the variation in the accumulated time does not necessarily decrease along the cascade. For example, if a slow exponential step is added to a cascade composed of many fast exponential steps, the precision is lost and the total time will approximately follow the exponential of the rate-limiting step. The observed decrease in the total variation along the process is thus non-trivial, and raises the issue of whether the individual steps have evolved statistical properties to increase the total precision. This is an open question and it is not clear how the phage would benefit from precision or variability in lysis times.
Variability in molecule numbers can drastically affect the timing of events, but the results of Amir et al also demonstrate the importance of the inverse question: How does the timing of events affect the variability in molecule numbers? Most models to date assume that complicated processes—like transcription, translation, or RNA degradation—can be approximated as single exponentially distributed transitions despite the large number of sub-steps involved. Many cases are well approximated by exponentials, but because the randomness in the timing is a main determinant of fluctuations in molecule numbers, this is an issue that deserves more attention. We are still far from a general mathematical or experimental understanding of the interdependency of the variation in the timing of events and the variation in molecule numbers. On the mathematical side, we need more general theory, perhaps based on queuing or renewal theory—the mathematical study of random waiting lines—rather than on the standard Chemical Master Equations that are now used. On the experimental side, we need more studies like the one of Stavans and co-workers to measure directly the distribution of waiting times between events, as well as more detailed measurements following single cells over real time with single-molecule resolution. Such experimental techniques are now starting to become available (Golding et al, 2005; Cai et al, 2006) and we believe the field is poised for a major conceptual advance.
References
- Amir A, Kobiler O, Rokney A, Oppenheim AB, Stavans J (2007) Noise in timing and precision of gene activities in a genetic cascade. Mol Syst Biol 3: 71 17299413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR (2006) Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell 21: 3–14 [DOI] [PubMed] [Google Scholar]
- Cai L, Friedman N, Xie XS (2006) Stochastic protein expression in individual cells at the single molecule level. Nature 440: 358–362 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC (2005) Real-time kinetics of gene activity in individual bacteria. Cell 123: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A (2002) Regulation of noise in the expression of a single gene. Nat Genet 31: 69–73 [DOI] [PubMed] [Google Scholar]