Multi-drug combinations are vital in modern medicine (Keith et al, 2005; Fitzgerald et al, 2006). Such drug combinations can also be used to probe the relationships between proteins in a network, and progress towards using drug interactions to infer network connectivity has been made in recent years. A current study by Lehár et al (2007) takes this effort a large step further by developing tools to use the entire data in a drug–drug interaction dose–response surface to give useful information on the networks in which the drug targets are embedded.
Classically, combinations of perturbations—drugs or mutations—have been categorized into one of three interaction types: additive, synergistic, or antagonistic (Bliss, 1939; Loewe, 1953; Hartman et al, 2001). The expected null interaction is called additive, although exactly how this should be defined has been a subject of some controversy (Bliss, 1939; Loewe, 1953; Greco et al, 1995). Synergy occurs when the combination of two perturbations has an effect greater than expected from the individual effects of the single perturbations. Antagonism describes a combination with less than expected effect. These classifications have proved powerful in dissecting the modularity and connectivity of the underlying biological networks (Tong et al, 2001; Schuldiner et al, 2005; Segre et al, 2005; Yeh et al, 2006). There are some intuitive expectations for combined effects of two drugs. Let us say, for example, that drugs A and B block two alternative metabolic pathways, of which at least one is needed. In this case, each drug may have very little effect, but the combination will be strongly synergistic. Similarly, antagonistic interaction may result from drugs acting on two parallel pathways that are both needed: inhibition of one pathway can make its product a limiting factor, thereby neutralizing the effect of inhibition of other pathways. This simple intuition can be elaborated and applied at the system level to analyze a complex interaction network composed of additive, synergistic, and antagonistic links (Tong et al, 2001; Schuldiner et al, 2005; Segre et al, 2005; Yeh et al, 2006).
But the three classical interaction types represent a radical simplification for drug combinations. Many drug combinations exhibit different types of interactions ‘within a drug pair' depending on dose. Drugs may be additive at one set of combined concentrations and synergistic or antagonistic in another. ‘Response surfaces' (also represented by ‘isobolograms'), which show combined drug effects over a 2-D gradient of concentrations, can exhibit a rich set of patterns. Can we use the abundant information in these response surfaces to infer more specific and detailed information regarding the underlying biological network?
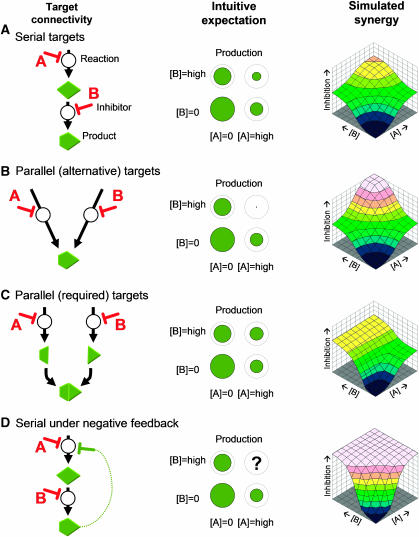
Lehár et al (2007) used simulated Michaelis–Menten metabolic pathways to ask whether drug–response surfaces can yield information regarding the underlying connectivity of molecular targets (Figure 1). They experimentally tested the results of their simulations in the well-studied sterol metabolism pathway. Indeed, in both simulation and experiments, different drug pairs showed different complex response surfaces. Knowing the underlying connectivity of the system, it was then possible to establish links between various local network topologies and the different response surfaces.
Figure 1.
The relationship between target connectivity and synergy for paired inhibitors. The underlying connectivity for two inhibitors A and B is shown schematically along with intuitive expectations for their combined effect and the simulated dose-dependent response surfaces. (A) If their targets are serial in a pathway, A and B should help each other reduce production. If they inhibit parallel pathways (B, C), the combination effect ought to reflect the rate-limiting reaction for required (‘AND') junctions but should be very synergistic for alternative (‘OR') pathways. (D) More complex network topologies without an intuitive expectation can still be simulated. Lehár et al (2007) sought to categorize and link the set of possible response surfaces to underlying network connectivity (Figure courtesy of J Lehár).
Lehár et al (2007) produced a reference set of four response surface models to which experimental data could be classified. They found in their simulations that particular target connectivities produced distinct responses in terms of these shape models. The first reference model, based on the standard Loewe dose additivity (Loewe, 1953), is a best fit for combinations of drugs that affect the same target. A second shape model, ‘Bliss Boost', an extension of the statistical Bliss independence (Bliss, 1939), provided the best fits for separate targets in an unregulated pathway. A third model, ‘Highest Single Agent', assumes that the inhibition for the combined drugs equal the highest (the most limiting) single drug inhibition (Yeh et al, 2006), and was found to predominate for cross-pathway combinations. A fourth model of ‘Potentiation', which allows an inherent asymmetry in the response surface where the presence of a certain drug increases or decreases the effective concentration of another, was found to be a better fit when cellular targets of drugs are in pathways regulated by negative feedback.
This paper highlights the utility of models and simulations for a task as critical as that of rationalizing and modeling the effect of drug combinations. Indeed, quantitative approaches to drug interactions have had impact in pharmacology for more than a century. In the mid-nineteenth century, Fraser (1872) published a landmark paper that showed how two drugs, atropia and physostigma, can have hyperantagonistic effects that resulted in one drug reversing the effects of the other (Fraser, 1872). Fraser termed this ‘physiological antidote'. Such suppressive interactions also exist in antimicrobial agents (Yeh et al, 2006). It would be interesting to explore whether the Lehár et al's (2007) approach can be extended to link such hyperantagonistic suppressive interactions to possible underlying connectivities of the biological network.
This study offers a ‘proof of principle' that we can link the rich functional information encoded in complete drug–drug dose–response surfaces to the connectivity between biological targets. It will be interesting to see how far this kind of analysis takes us in the real world: how much will ‘off-target' effects confound the analysis? To what extent is the mapping between network topologies and response surfaces a one-to-one function? Further, it would be important to extend this idea to three or more drug components. Such multi-drug treatments are already being used, but there is little understanding of how their response surface (or, more accurately in this case, ‘response spaces') should behave. In the case of three-drug combinations, the question of how to define additivity is even more complicated than for two-drug combinations, as the null expectation for additivity must be based not only on the effect of the single drugs but also on pre-existing knowledge of all their pairwise interactions. Lehár et al's (2007) surface responses and shape models offer an excellent starting point for conceptual and experimental explorations of such combinations of three or more drugs.
References
- Bliss CI (1939) The toxicity of poisons applied jointly. Ann Appl Biol 26: 585–615 [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK (2006) Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2: 458–466 [DOI] [PubMed] [Google Scholar]
- Fraser TR (1872) The antagonism between the actions of active substances. 2: 485–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco WR, Bravo G, Parsons JC (1995) The search for synergy—a critical-review from a response-surface perspective. Pharmacol Rev 47: 331–385 [PubMed] [Google Scholar]
- Hartman JL, Garvik B, Hartwell L (2001) Cell biology—principles for the buffering of genetic variation. Science 291: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent therapeutics for networked systems. Nat Rev Drug Disc 4: 71–78 [DOI] [PubMed] [Google Scholar]
- Lehár J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF III, Giusti LC, Nolan GP, Magid OA, Lee MS, Borisy AA, Stockwell BR, Curtis T, Keith CT (2007) Chemical combination effects predict connectivity in biological systems. Mol Syst Biol 3: 80 17332758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe S (1953) The problem of synergism and antagonism of combined drugs. Arzneimittel-Forsc-Drug Res 3: 285–290 [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ (2005) Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519 [DOI] [PubMed] [Google Scholar]
- Segre D, DeLuna A, Church GM, Kishony R (2005) Modular epistasis in yeast metabolism. Nat Genet 37: 77–83 [DOI] [PubMed] [Google Scholar]
- Tong AHY, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CWV, Bussey H, Andrews B, Tyers M, Boone C (2001) Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Yeh P, Tschumi AI, Kishony R (2006) Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38: 489–494 [DOI] [PubMed] [Google Scholar]