Abstract
ISXax1 is a novel insertion sequence belonging to the IS256 and Mutator families. Dot blot, Southern blot, and PCR analyses revealed that ISXax1 is restricted to Xanthomonas axonopodis pv. phaseoli (variants fuscans and non-fuscans) and X. axonopodis pv. vesicatoria strains. Directed AFLP also showed that a high degree of polymorphism is associated with ISXax1 insertion in these strains.
Insertion sequences (ISs) are small, mobile, genetic elements that are ubiquitously distributed within bacterial genomes (6, 19). For prokaryotes, IS elements have been classified into about 20 families based on (i) open reading frame organization, (ii) length and similarity of terminal inverted repeat (IR) sequences, (iii) length of target site duplication (direct target repeat [DR]), and (iv) features of the DDE motif, a conserved amino acid motif called the catalytic triad of bacterial transposases (Tpases) (http://www-is.biotoul.fr/is.html) (19, 29). ISs mediate various DNA rearrangements which can lead to diverse phenotypic effects (11). The association between ISs and pathogenic or virulence functions has been frequently observed in plant as well as animal pathogens (9, 13, 14, 25, 36). ISs play an important role in evolution by facilitating horizontal gene transfer between bacterial populations. They contribute significantly to the diversity of bacteria, enhancing the organisms' adaptive and evolutionary capacities (9, 25, 36). ISs can also be used as markers for bacterial species typing and for epidemiological investigations (2, 15, 30, 34).
Here, we report ISXax1, a newly identified insertion sequence in Xanthomonas axonopodis pv. phaseoli and its fuscans variant, the causative agents of common bacterial blight of bean (4). Both pathogens have the same host range and similar biochemical phenotypes except that the fuscans variant can produce a melanin-like pigment in culture (8). We also show that ISXax1 is carried by X. axonopodis pv. vesicatoria but not by Xanthomonas vesicatoria, the two causal agents of the bacterial spot disease of tomato and pepper (26, 32, 33).
Isolation and characterization of ISXax1.
A subtractive hybridization performed between the X. axonopodis pv. phaseoli variant fuscans CFBP4834 strain (data not shown) and four strains from closely related species (Xanthomonas campestris pv. campestris CFBP2350, Xanthomonas hortorum pv. pelargonii CFBP2533, and X. vesicatoria CFBP1941 and CFBP2537) led to the isolation of a 166-bp DNA fragment (data not shown). A BLAST search (http://www.ncbi.nih.gov/BLAST/) (1) revealed that this subtracted DNA fragment has high similarities with IS-like elements. We then performed inverse PCR (24) and directed-AFLP experiments to isolate the flanking sequences of this DNA fragment. To perform directed-AFLP experiments, we digested genomic DNA by EcoRI; the restricted DNA fragments were then ligated to Ecoadapt (an EcoRI-specific, double-stranded adapter resulting from hybridization between Ecoadapt 1 and 2 oligonucleotides [Table 1 ]) by using T4 DNA ligase and amplified with an either Xpha5-Ecocore or Xpha6-Ecocore pair of primers (Table 1). Both methods allowed us to determine the complete nucleotide sequence of this putative transposase gene. This novel IS element was named ISXax1 according to the IS Finder database (http://www-is.biotoul.fr/is.html).
TABLE 1.
Oligonucleotides used as primers and adapters to amplify ISXax1 sequences and ISXax1 flanking regions
| Oligonucleotide | Sequence (5′-3′) | Position on the ISXax1 sequence (5′-3′) |
|---|---|---|
| Xpha1 | ACC CGC TGG GCC GGC TTC | 382-399 |
| Xpha2 | CCT GCC ACG CCT TGA CCT C | 547-529 |
| Xpha5 | GTG TAG TTG ACC AAC AGG CT | 128-109 |
| Xpha6 | TGG CTG CAA GTC GTG ACG GA | 718-736 |
| Xpha10 | GGC CAC AGG CAG GAG ACT GCC | 28-48 |
| Xpha11 | CCT CAG CGG TAA CCC AAA CCA | 1302-1282 |
| Ecocore | GAC TGC GTA CCA ATT C | |
| Ecoadapt1 | CTC GTA GAC TGC GTA CC | |
| Ecoadapt2 | P-AAT TGG TAC GCA GTCa |
Ecoadapt2 is phosphorylated (P) at the 5′ end.
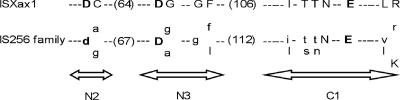
Based on data from the IS Finder database, several features (size, IRs, and DRs) (Fig. 1) suggest that ISXax1 is a new member of the IS256 family (Table 2). BLAST searches also revealed that ISXax1 shares high similarities with putative Tpase genes classified within the IS256 family. Interestingly, 100% identity was found with a putative Tpase gene (locus XCVc0009; accession no. NC_007506) from X. axonopodis pv. vesicatoria strain 85-10 (31). In X. axonopodis pv. vesicatoria strain 85-10, this ISXax1 copy is unique and resides in plasmid pXCV38 (31). Additionally, the putative ISXax1 Tpase harbors a DDE motif at its C-terminal region (Fig. 2), matching the consensus sequence determined for the IS256 family (19). Finally, ISXax1 can also be classified within the Mutator family, consisting of Tpases from eukaryotes and prokaryotes. Indeed, by analyzing the PROSITE database (http://www.expasy.org/prosite/), the signature of the Mutator family of Tpases (accession no. PS01007) was found within the ISXax1 Tpase sequence (Fig. 3).
FIG. 1.
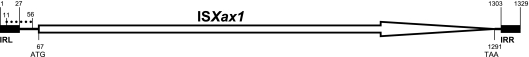
Schematic representation of the main features of the ISXax1 nucleotide sequences in X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria strains. The orientation of the ISXax1 gene is shown by an open arrow with the location of the ATG start codon (at position 67) and the TAA stop codon (at position 1291). The dotted line of the ISXax1 sequence (nucleotides 11 to 56) indicates the location of a putative promoter. The black boxes represent the positions of the two imperfect, 27-bp, terminal inverted repeats (the left IR [IRL], nucleotides 1 to 27, and the right IR [IRR], nucleotides 1303 to 1329) with five mismatches.
TABLE 2.
Main ISXax1 features compared to those of IS256 family membersa
| Feature | Value for:
|
|
|---|---|---|
| ISXax1 | IS256 family | |
| Size | 1,329 bp | 1,300 to 1,500 bp |
| No. of ORFs | 1 (408 aa) | 1 |
| Size of inverted terminal repeats (IRs) | 27 bp | 24 to 41 bp |
| Size of direct target repeats (DRs) | 8 bp | 8 or 9 bp |
| Conserved terminal base pair 5′ to 3′b | GG | G(g/a) |
Data are derived from IS Finder database (http://www-is.biotoul.fr/is.html).
Uppercase letters refer to mostly (and often) conserved bases. Lowercase letters separated by slashes indicate alternative conservation at that position.
FIG. 2.
Alignment of the ISXax1 DDE motif and the corresponding consensus sequence of the IS256 family (IS Finder database [http://www-is.biotoul.fr/is.html]). Highly conserved amino acids are shown in bold letters. For the IS256 family, uppercase letters indicate conservation within the family and lowercase letters indicate predominant amino acids. The numbers of amino acids between the motif regions are shown in parentheses. The N2, N3, and C1 domains containing the conserved DDE amino acids are also shown (19). The spacing between the three amino acids forming the DDE catalytic triad is variable depending upon IS256 family members (19).
FIG. 3.
Alignment of protein sequences of 10 IS elements (belonging to the IS256 family according to the IS Finder database [http://www-is.biotoul.fr/is.html]) which shows the signature of the Mutator family of transposases (PROSITE database [http://www.expasy.ch/cgi-bin/nicesite.pl?PS01007]). The bacterial origin and the accession number for each insertion sequence are as follows: for ISXax1 (Xanthomonas axonopodis pv. phaseoli and variant fuscans), AY935340; for IS6120 (Mycobacterium smegmatis), P35883; for IS1201 (Lactobacillus helveticus), P35880; for IS1081 (Mycobacterium bovis), P60231; for IST2 (Acidithiobacillus ferrooxidans), P35884; for ISRm5 (Sinorhizobium meliloti), Q52873; for IS905 (Lactococcus lactis), P35881; for ISRm3 (Sinorhizobium meliloti), P80011; for IS406 (Burkholderia cepacia), P24575; and for IS256 (Staphylococcus aureus), P19775. The location of the signature within each protein sequence is shown in parentheses. Black and gray backgrounds indicate identical and similar amino acids, respectively. The consensus protein sequence is shown below the alignment. This alignment was made by BOXSHADE software, version 1.6.6 (http://www.ch.embnet.org/software/BOX_form.html).
The IS256 family is widely distributed in bacteria since members of this family have been disclosed in Actinobacteria, Firmicutes (Clostridia, Bacillales, and Lactobacillales), and Proteobacteria (Alpha-, Beta-, and Gammaproteobacteria) (29). However, the description of ISXax1 is original as it appears that this IS element is the first characterized member of the IS256 family in Xanthomonas strains. Do genomes of X. axonopodis pv. phaseoli and its variant fuscans harbor other IS elements belonging to the IS256 family? Strikingly, among the plant-pathogenic bacteria whose genomes have been sequenced completely, Pseudomonas savastanoi pv. phaseolicola, a pathogen of bean, as are X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans, is the unique bacterium harboring many IS elements belonging to the IS256 family (12). The forthcoming sequencing of the genome of our model strain, XapfCFBP4834, will help us to speculate on the role played by this IS family in the evolution of bacteria and, in particular, of bean pathogens.
ISXax1 distribution in X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans strains.
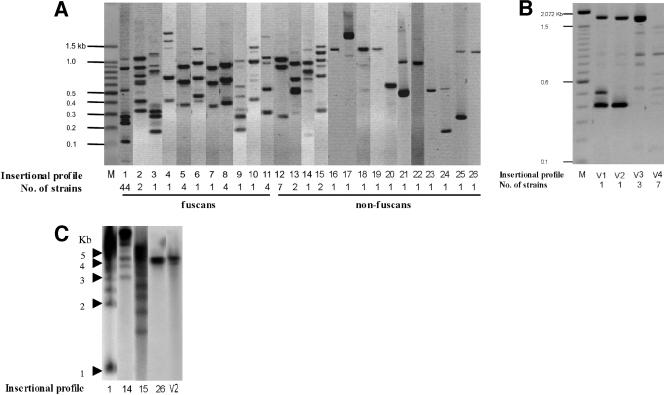
We studied the distribution of ISXax1 within a worldwide collection of both X. axonopodis pv. phaseoli (23 strains) and X. axonopodis pv. phaseoli variant fuscans (64 strains) by directed AFLP. ISXax1 was present in all tested strains (Fig. 4; data not shown), suggesting that ISXax1 has evolved in genomes of both X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans for a long time. This result is supported by the fact that the G+C content of ISXax1 (62.3 mol%) is similar to the average value of total DNA for X. axonopodis pv. phaseoli (∼65 mol%) (32) and that the codon usage in ISXax1 and its hosts (X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans) does not differ (our unpublished data). A high polymorphism was revealed since the copy number ranged from 1 to 7 (confirmed by Southern blot hybridization) (Fig. 4), and 26 different ISXax1 insertional profiles were observed. No DNA fragment was common to all tested X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans strains, and bands shared among different patterns were rare. This high polymorphism of insertion might reflect the ability of ISXax1 to insert into many different sites in genomes of both X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans. Fifteen ISXax1 insertion profiles were generated by X. axonopodis pv. phaseoli strains, whereas only 11 profiles were obtained with X. axonopodis pv. phaseoli variant fuscans strains. This result confirms that the non-fuscans variant is more heterogeneous than the fuscans variant is (3, 21). Moreover, as identical ISXax1 profiles can result from strains isolated in distant countries, our approach confirms that the diversity observed in both X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans was not geographically structured for either of the two pathogens (20, 21). This observation is not surprising since X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans are both seed-borne pathogens, and their dissemination through contaminated seeds is well known (27, 35). The lack of geographical differentiation has important practical implications, as available host resistance genes are likely to be effective for managing the disease in diverse geographical areas (20). Interestingly, no common ISXax1 insertion profile was shared by both pathogens. Thus, our study provides further data to show that X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans are genetically distinct. Indeed, our results are in complete agreement with the following previously used methods: DNA-DNA hybridization (10), random amplified polymorphic DNA (3, 18), repetitive-element PCR (18, 20, 21), pulsed-field gel electrophoresis (5), restriction fragment length polymorphism analyses using hrp and pectate lyase genes as probes (5), and PCR-restriction fragment length polymorphism of the ribosomal genes (20).
FIG. 4.
Representation of the different insertional profiles obtained by directed AFLP (A and B) or by Southern blot hybridization (C) (28). (A and B) Genomic DNA was first digested by EcoRI, which does not cut within ISXax1, and the restricted fragments were then ligated to Ecoadapt (an EcoRI-specific, double-stranded adapter resulting from hybridization between Ecoadapt 1 and 2 oligonucleotides [Table 1]) by using T4 DNA ligase and amplified with the Xpha-Ecocore pair of primers (Table 1). (C) Genomic DNA was digested by EcoRI, which does not cut within ISXax1, and the restricted fragments were hybridized with the ISXax1 probe (generated by using Xpha10 and Xpha11 primers [Table 1]). DNAs are from (A) 64 X. axonopodis pv. phaseoli variant fuscans strains and 23 X. axonopodis pv. phaseoli strains and from (B) 12 X. axonopodis pv. vesicatoria strains. Profiles 1 to 11 belong to the fuscans strains of X. axonopodis pv. phaseoli, and those from 12 to 26 belong to the non-fuscans strains. Profiles V1 to V4 were generated by X. axonopodis pv. vesicatoria strains. The number of strains sharing the same insertional profile is indicated below each profile number. M corresponds to the molecular mass marker.
ISXax1 is restricted to X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria strains.
To investigate whether ISXax1 is widespread in bacteria, further dot blot and Southern blot hybridizations (28) were performed on a large collection of strains belonging to X. axonopodis pathovars and other bacterial species (data not shown). No fragments homologous to ISXax1 were detected in any of the strains tested except X. axonopodis pv. vesicatoria strains (Fig. 4; data not shown). This result was confirmed by PCR with primer pairs Xpha1-Xpha2 and Xpha10-Xpha11 (Table 1), allowing the amplification of internal ISXax1 DNA fragments from all 13 X. axonopodis pv. vesicatoria strains tested, originating from different countries (data not shown). A polymorphism of the ISXax1 insertion within X. axonopodis pv. vesicatoria strains was shown by directed AFLP (Fig. 4).
Finally, the fact that ISXax1 is carried by only three taxa (X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria) belonging to the same species (X. axonopodis) may suggest that ISXax1 was already present in X. axonopodis strains before host specialization occurred. However, ISXax1 is not present in X. axonopodis pathovars, such as X. axonopodis pv. aurantifolii, X. axonopodis pv. citrumelo, X. axonopodis pv. dieffenbachiae, or X. axonopodis pv. manihotis, which belong to the same DNA-DNA homology, AFLP, or repetitive-element PCR groups (26, 33). So, ISXax1 would have been lost in certain X. axonopodis pathovars during evolution. Another explanation could be that X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria acquired ISXax1 from an unrelated bacterial species through a horizontal gene transfer event, and then ISXax1 was transmitted vertically within X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria. This last hypothesis is supported by recent observations in Gammaproteobacteria (including Xanthomonas strains). Analyses of Xanthomonas genomes clearly show that these bacteria have been subjected to numerous horizontal gene transfers during evolution, sometimes from phylogenetically distant organisms (7, 17). Moreover, gene acquisition is considered to be a major factor contributing to the genomic diversity of these bacteria but, paradoxically, once acquired, these genes are rarely transferred among lineages (16).
What could be the contribution of ISXax1 in X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria genomes? We tried to answer this question by analyzing the flanking sequences of eight independent ISXax1 insertions in strains XapfCFBP4834, XapCFBP6991, and XapCFBP6994. We showed that ISXax1 is inserted into noncoding regions (except for one copy in strain XapfCFBP4834 inserted into the aceK homolog, encoding an isocitrate dehydrogenase kinase/phosphatase, X. axonopodis pv. vesicatoria [accession no. CAJ25689]). We also observed that the locations of these ISXax1 insertions seem variable from strain to strain (Fig. 4; data not shown). These results suggest that this IS element is likely not involved in the host specificity or pathogenicity of X. axonopodis pv. phaseoli and X. axonopodis pv. phaseoli variant fuscans. It has been shown that Xanthomonas genomes were invaded by numerous IS elements since more than 100 transposase genes were disclosed in each genome except those in X. axonopodis pv. vesicatoria (31). An interesting feature is that these genomes carry distinctive sets of transposable elements; some of these IS elements are shared by all Xanthomonas genomes, whereas others exhibit more restricted distributions (22, 23). Therefore, we suggest that ISXax1 might be, as are other IS elements in Xanthomonas (22, 23), an important driver of the differentiations and evolutions of X. axonopodis pv. phaseoli, X. axonopodis pv. phaseoli variant fuscans, and X. axonopodis pv. vesicatoria genomes.
Nucleotide sequence accession number.
The sequence for the novel IS element was deposited in GenBank under accession no. AY935340.
Acknowledgments
We are grateful to M.-A. Jacques (INRA) for her critical reading of the manuscript. We thank M.-A. Jacques (INRA), O. Pruvost (CIRAD), and M. Guénard (SNES-GEVES) for kindly providing strains.
We acknowledge the financial support received from Région Pays de la Loire. S. M. Alavi is supported by a grant from NIGEB, Tehran, Iran.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch, P. R. J., L. J. Hyman, R. Taylor, A. F. Opio, C. Bragard, and I. K. Toth. 1997. RAPD PCR-based differentiation of Xanthomonas campestris pv. phaseoli from Xanthomonas campestris pv. phaseoli var. fuscans. Eur. J. Plant Pathol. 103:809-814. [Google Scholar]
- 4.Broughton, W. J., G. Hernandez, M. Blair, S. Beebe, P. Gepts, and J. Vanderleyen. 2003. Beans (Phaseolus spp.)—model food legumes. Plant Soil 252:55-128. [Google Scholar]
- 5.Chan, J. W. Y. F., and P. H. Goodwin. 1999. Differentiation of Xanthomonas campestris pv. phaseoli from Xanthomonas campestris pv. phaseoli var. fuscans by PFGE and RFLP. Eur. J. Plant Pathol. 105:867-878. [Google Scholar]
- 6.Chandler, M., and J. Mahillon. 2002. Insertion sequences revisited, p. 305-366. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 7.Comas, I., A. Moya, R. K. Azad, J. G. Lawrence, and F. Gonzalez-Candelas. 2006. The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol. Biol. Evol. 23:2049-2057. [DOI] [PubMed] [Google Scholar]
- 8.Goodwin, P. H., and C. R. Sopher. 1994. Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentisic acid. Can. J. Microbiol. 40:28-34. [Google Scholar]
- 9.Hammerschmidt, S., R. Hilse, J. P. van Putten, R. Gerardy-Schahn, A. Unkmeir, and M. Frosch. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192-198. [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrand, D. C., N. J. Palleroni, and M. N. Schroth. 1990. Deoxyribonucleic acid relatedness of 24 xanthomonad strains representing 23 Xanthomonas campestris pathovars and Xanthomonas fragariae. J. Appl. Bacteriol. 68:263-269. [Google Scholar]
- 11.Hübner, A., and W. A. Hendrickson. 1997. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2,4,5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J. Bacteriol. 179:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson, L. M. Brinkac, S. C. Daugherty, R. DeBoy, A. S. Durkin, M. G. Giglio, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. Sullivan, J. Crabtree, T. Creasy, T. Davidsen, D. H. Haft, N. Zafar, L. Zhou, R. Halpin, T. Holley, H. Khouri, T. Feldblyum, O. White, C. M. Fraser, A. K. Chatterjee, S. Cartinhour, D. J. Schneider, J. Mansfield, A. Collmer, and C. R. Buell. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearney, B., and B. Staskawicz. 1990. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J. Bacteriol. 172:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J. F., A. O. Charkowski, J. R. Alfano, A. Collmer, and S. V. Beer. 1998. Sequences related to transposable elements and bacteriophages flank avirulence genes of Pseudomonas syringae. Mol. Plant-Microbe Interact. 11:1247-1252. [Google Scholar]
- 15.Kivi, M., X. Liu, S. Raychaudhuri, R. B. Altman, and P. M. Small. 2002. Determining the genomic locations of repetitive DNA sequences with a whole-genome microarray: IS6110 in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:2192-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerat, E., V. Daubin, H. Ochman, and N. A. Moran. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima, W. C., M. A. Van Sluys, and C. F. Menck. 2005. Non-gamma-proteobacteria gene islands contribute to Xanthomonas genome. OMICS 9:160-172. [DOI] [PubMed] [Google Scholar]
- 18.López, R., C. Asensio, and R. L. Gilbertson. 2006. Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204-1213. [DOI] [PubMed] [Google Scholar]
- 19.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahuku, G. S., C. Jara, M. A. Henriquez, G. Castellanos, and J. Cuasquer. 2006. Genotypic characterization of the common bean bacterial blight pathogens, Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans by rep-PCR and PCR-RFLP of the ribosomal genes. J. Phytopathol. 154:35-44. [Google Scholar]
- 21.Mkandawire, A. B. C., R. B. Mabagala, P. Guzman, P. Gepts, and R. L. Gilbertson. 2004. Genetic diversity and pathogenic variation of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) suggests pathogen coevolution with the common bean. Phytopathology 94:593-603. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro-Vitorello, C. B., M. C. De Oliveira, M. M. Zerillo, A. M. Varani, E. Civerolo, and M. A. Van Sluys. 2005. Xylella and Xanthomonas mobil'omics. OMICS 9:146-159. [DOI] [PubMed] [Google Scholar]
- 23.Ochiai, H., Y. Inoue, M. Takeya, A. Sasaki, and H. Kaku. 2005. Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39:275-287. [Google Scholar]
- 24.Ochman, H., A. S. Gerber, and D. L. Harlt. 1988. Genetic application of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poussier, S., P. Thoquet, D. Trigalet-Demery, S. Barthet, D. Meyer, M. Arlat, and A. Trigalet. 2003. Host plant-dependent phenotypic reversion of Ralstonia solanacearum from non-pathogenic to pathogenic forms via alterations in the phcA gene. Mol. Microbiology 49:991-1003. [DOI] [PubMed] [Google Scholar]
- 26.Rademaker, J. L. W., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. Appl. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 27.Saettler, A. W. 1991. Common bacterial blight, p. 29-30. In R. Hall (ed.), Compendium of bean diseases. The American Phytopathological Society, St. Paul, MN.
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Siguier, P., J. Filée, and M. Chandler. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9:526-531. [DOI] [PubMed] [Google Scholar]
- 30.Stanley, J., N. Baquar, and E. J. Threlfall. 1993. Genotypes and phylogenetic relationships of Salmonella typhimurium are defined by molecular fingerprinting of IS200 and 16S rrn loci. J. Gen. Microbiol. 139:1133-1140. [DOI] [PubMed] [Google Scholar]
- 31.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klösgen, T. Patschkowski, C. Rückert, O. Rupp, S. Schneiker, S. C. Schuster, F.-J. Vorhölter, E. Weber, A. Pühler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bact. 45:472-489. [Google Scholar]
- 33.Vauterin, L., J. Rademaker, and J. Swings. 2000. Synopsis on the taxonomy of the genus Xanthomonas. Phytopathology 90:677-682. [DOI] [PubMed] [Google Scholar]
- 34.Vera Cruz, C. M., E. Y. Ardales, D. Z. Skinner, J. Talag, R. J. Nelson, F. J. Louws, H. Leung, T. W. Mew, and J. E. Leach. 1996. Measurement of haplotypic variation in Xanthomonas oryzae pv. oryzae within a single field by rep-PCR and RFLP analyses. Phytopathology 86:1352-1359. [Google Scholar]
- 35.Vidaver, A. K. 1993. Xanthomonas campestris pv. phaseoli: cause of common bacterial blight of bean, p. 40-44. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman and Hall, London, United Kingdom.
- 36.Ziebuhr, W., V. Krimmer, S. Rachid, I. Löβner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]