Abstract
The springs at Gypsum Hill and Colour Peak on Axel Heiberg Island in the Canadian Arctic originate from deep salt aquifers and are among the few known examples of cold springs in thick permafrost on Earth. The springs discharge cold anoxic brines (7.5 to 15.8% salts), with a mean oxidoreduction potential of −325 mV, and contain high concentrations of sulfate and sulfide. We surveyed the microbial diversity in the sediments of seven springs by denaturing gradient gel electrophoresis (DGGE) and analyzing clone libraries of 16S rRNA genes amplified with Bacteria and Archaea-specific primers. Dendrogram analysis of the DGGE banding patterns divided the springs into two clusters based on their geographic origin. Bacterial 16S rRNA clone sequences from the Gypsum Hill library (spring GH-4) were classified into seven phyla (Actinobacteria, Bacteroidetes, Firmicutes, Gemmatimonadetes, Proteobacteria, Spirochaetes, and Verrucomicrobia); Deltaproteobacteria and Gammaproteobacteria sequences represented half of the clone library. Sequences related to Proteobacteria (82%), Firmicutes (9%), and Bacteroidetes (6%) constituted 97% of the bacterial clone library from Colour Peak (spring CP-1). Most GH-4 archaeal clone sequences (79%) were related to the Crenarchaeota while half of the CP-1 sequences were related to orders Halobacteriales and Methanosarcinales of the Euryarchaeota. Sequences related to the sulfur-oxidizing bacterium Thiomicrospira psychrophila dominated both the GH-4 (19%) and CP-1 (45%) bacterial libraries, and 56 to 76% of the bacterial sequences were from potential sulfur-metabolizing bacteria. These results suggest that the utilization and cycling of sulfur compounds may play a major role in the energy production and maintenance of microbial communities in these unique, cold environments.
Relatively little is known about the diversity, abundance, and ecology of microorganisms in polar regions, where unique habitats exist, including subglacial and permanently ice-covered lakes, cryptoendoliths, polar glaciers, polar sea ice (for an overview, see references 11 and 45), permafrost and ground ice (56), and cryopegs (21). Perennial springs are extremely rare in areas underlain by deep, continuous cold permafrost because of the limited opportunity of exchange between sub- and suprapermafrost groundwater systems. In this respect, the two groups of cold perennial springs located at Expedition Fjord on west-central Axel Heiberg Island in the Canadian high Arctic (Fig. 1), at nearly 80oN, are among the few known examples of cold, nonvolcanic springs in thick permafrost on Earth (4). Other high-latitude springs have been reported in Spitsbergen and east Greenland (36, 68), and supraglacial sulfur springs arising from glacial meltwater exist on northern Ellesmere Island (24), although the microbiology has not been extensively studied.
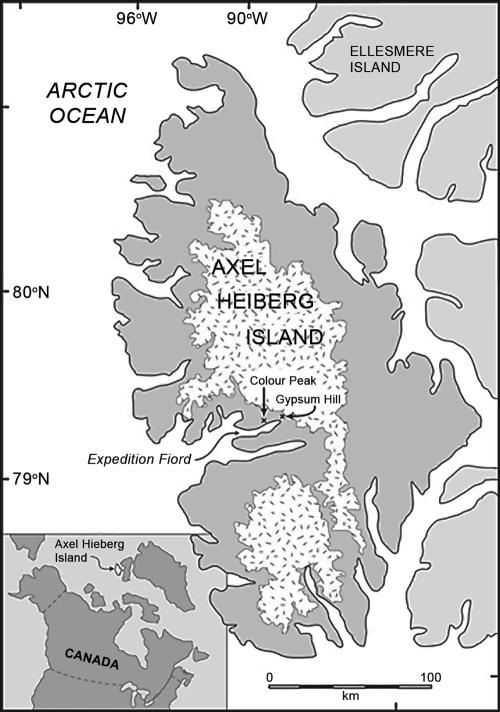
FIG. 1.
Location of the perennial springs on Axel Heiberg Island, Canada. (Reprinted from reference 4 with permission of the publisher.)
The Expedition Fjord springs are located 11 km apart at the base of Gypsum Hill and Colour Peak in a region of thick continuous permafrost over 400 to 600 m thick (60), with a mean annual air temperature of −15°C (13). Both groups of springs occur in topographically and geologically similar settings but are associated with separate piercement dome structures (44). The main springs maintain a constant temperature throughout the year despite air temperatures that drop below −40°C during the winter. The springs at both sites discharge cold brines, with high concentrations of Na+, Ca2+, SO42−, and Cl−, and a strong odor of hydrogen sulfide (H2S); if all the dissolved ions are normalized to Na+, the water displays the same basic signature as seawater (44).
It has been hypothesized that the springs originate from subpermafrost salt aquifers and rise to the surface through the permafrost (4); they are not associated with volcanic activity; the heat is provided through the local geothermal gradient. Similar low-temperature hydrosystems might occur or have occurred on Mars. Mars Global Surveyor images indicated the presence of gully-like landforms that occur primarily at high latitudes, providing evidence of recent fluvial activity (38). These features appear to be geologically young enough to have been formed under the present climatic conditions that include mean surface temperatures of −60°C and extensive permafrost. Given the absence of any association between these flow features and obvious geothermal heat sources (e.g., volcanic features), eutectic brines present in the Martian subsurface have been suggested as the likely fluid that formed these features (26). These two sets of Arctic springs represent useful terrestrial analogues with which to study the requisites that would have enabled life to develop and be maintained in Martian hydrosystems.
Previous studies have explored the microbiota of sulfur springs, but most of them focused on hot springs (6, 27) and deep-sea hydrothermal vents (25, 58). The cold Ancaster sulfur spring in Ontario (14) and the mesophilic spring of Zodletone Mountain in Oklahoma (16) were shown to host rich and complex microbial communities with abundant microbial mats at the spring sources and at their channels. In these systems, sulfide supports a diversity of phototrophic microorganisms (cyanobacteria, purple and green sulfur bacteria, and Chloroflexi spp.). The microbial characterization of cold sulfidic springs in Germany revealed a novel string-of-pearls community comprised of novel Archaea organisms in close association with a sulfide-oxidizing bacteria related to the genus Thiothrix (40).
In 1999, a preliminary investigation of the microbial composition of a biofilm formed on a glass slide placed in a channel within the spring flow at one Colour Peak discharge site detected sulfur-metabolizing phylotypes (3). In this study, molecular phylogenetic approaches (denaturing gradient gel electrophoresis [DGGE] and 16S rRNA clone library analyses) were used to examine the bacterial and archaeal composition of the microbial communities in the sediments of seven springs at Gypsum Hill and Colour Peak.
MATERIALS AND METHODS
Site description.
The Gypsum Hill site is located at 79°24′30"N, 90°43′05"W and is situated on the northwest side of Expedition River, approximately 2.5 km downstream from the terminus of the White and Thompson glaciers and 7 km upstream from the head of Expedition Fjord. Forty springs and seeps discharge along a band nearly 300 m long and 30 m wide, between 10 and 20 m above sea level (asl) at the base of a steep southeast-facing slope formed by the Expedition Diapir (Gypsum Hill) (44). The Colour Peak springs are located at 79°22′48"N, 91°16′24"W on the north side of Expedition Fjord, roughly 3 km from the head of Expedition Fjord. At least 20 springs discharge into a series of deep gullies located 30 to 40 m asl near the base of a south-facing slope of Colour Peak (44). In this study, four representative springs from Gypsum Hill (springs GH-1, GH-2, GH-3, and GH-4) and three representative springs from Colour Peak (springs CP-1, CP-2, and CP-3) were chosen for all or for selected analyses, as indicated below.
Physicochemical analyses of spring waters and sediments.
Various physicochemical parameters were recorded in July of 2004 and 2005. Temperatures were measured with a digital thermometer (Fisher Scientific Ltd., Nepean, Ontario, Canada). Total soluble sulfide and dissolved oxygen measurements were conducted with colorimetric assays (CHEMetrics, Calverton, VA). Oxidoreduction potential and pH were measured with a Hanna HI 9025 portable meter (Hanna Instruments, Laval, Quebec, Canada). Salinity, conductivity, and total dissolved solids were measured with a sensION5 meter (Hach, Loveland, CO) after a 1:1 dilution of spring waters in distilled water for the Gypsum Hill springs and a 1:2 dilution for the Colour Peak springs. Major cations, anions, total Kjeldahl nitrogen, and carbon content from the GH-4 and CP-2 sediments were determined at Maxxam Analytique, Inc. (Lachine, Quebec, Canada).
Sampling and DNA extraction.
In July 2004, 50 ml of composite spring sediments (top 10 cm) were aseptically collected from the seven springs for molecular analyses. Samples were processed at the McGill High Arctic Research Station within 12 h to minimize changes in microbial populations. Total community DNA was extracted from 5 g of each sediment sample with an UltraClean soil DNA isolation kit (Mo Bio Laboratories, Solana Beach, CA). The bead beating time (performed with a Mo Bio vortex adapter) was optimized to obtain DNA of suitable quality for phylogenetic studies and to avoid chimera production during PCR, as follows: bead beating times ranging from 30 s to 10 min were tested, and the quantity and quality of the total DNA recovered were checked by electrophoresis on precast E-Gels (0.8% agarose) using an E-Gel PowerBase (Invitrogen Canada, Burlington, ON) with Lambda/HindIII as the molecular weight DNA ladder. Two-minute vortexing gave an intense band of high-molecular-weight DNA (≥23 kb) with no visible shearing; this vortexing time was chosen for subsequent extractions. The DNA was eluted in TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) instead of the 10 mM Tris provided with the kit and kept at 4°C.
PCR amplification of the 16S rRNA gene.
The 16S rRNA gene analyses were performed in order to assess the prokaryotic phylogenetic composition of the spring sediments. A ∼590-bp fragment of the 16S rRNA gene, corresponding to variable regions V3, V4, and V5, of the Escherichia coli 16S rRNA gene, was amplified by PCR with community DNA as template, and the resulting amplicons were used for DGGE analysis and the construction of clone libraries. The combinations of the Bacteria-specific forward primer E341F (5′-CCTACGGGIGGCIGCA-3′) and universal reverse primer U926 (5′-CCGTCAATTCCTTTRAGTTT-3′) and of the Archaea-specific forward primer A344F (5′-ACGGGGTGCAGCAGGCGCGA-3′) and reverse primer A934R (5′-GTGCTCCCCCGCCAATTCCT-3′) were used to amplify the 16S rRNA genes of Bacteria and Archaea, respectively. The forward primers used for DGGE possessed a GC clamp (5′-GCGGGCGGGGCGGGGGCACGGGGGGCGCGGCGGGCGGGGCGGGGG-3′) at the 5′ end. For additional information on the primers used in this study, refer to Baker et al. (5). Each 50-μl PCR mixture contained ∼5 ng of template DNA, 25 pmol of each of the forward and reverse primers, 200 μM of each deoxynucleoside triphosphate (dNTP), 1 mM MgCl2, 1× PCR buffer, and 2.5 units of DNA polymerase. The DNA polymerases used for PCR were the rTaq polymerase (Amersham Biosciences, Baie d'Urfe, Qc, Canada) used to generate amplicons for DGGE and Easy-A cloning enzyme (Stratagene, La Jolla, CA) for cloning. PCR negative controls were prepared by replacing the template DNA with sterile water. After the initial denaturation (95°C for 5 min), DNA polymerase was added to the reaction mixture at a temperature of 80°C.
DGGE analyses.
The 16S rRNA gene amplicons from six to eight PCRs were combined for each sediment sample and concentrated by ethanol precipitation for DGGE analysis. To increase the specificity of the amplification and to reduce the formation of spurious by-products, a touchdown PCR (12) was performed as follows: the annealing temperature was set to 60°C (for archaeal PCR) or 65°C (for bacterial PCR) and decreased by 1°C at every cycle for 10 cycles, and then 20 additional cycles were performed. Denaturation was carried out at 94°C for 1 min, the annealing time was 1 min, and the primer extension was 72°C for 3 min. A final extension at 72°C for 30 min was added to avoid the generation of double bands on the DGGE gel (28). For each sample, 350 ng of archaeal amplicons and 600 ng of bacterial amplicons were applied to an 8% (wt/vol) acrylamide gel containing a 40 to 70% (archaeal) or 40 to 60% (bacterial) denaturing gradient: the 100% denaturant consisted of 7 M urea and 40% formamide. Gels were run at 60°C for 16 h at 80 V in 1× Tris-acetate-EDTA (TAE) buffer using a Bio-Rad Dcode universal mutation detection system (Bio-Rad Laboratories, Mississauga, ON, Canada). Gels were stained for 30 min in 1× TAE containing a 1:10,000 dilution of Vistra Green (Amersham Biosciences), destained for 30 min in 1× TAE, and visualized with a FluorImager System model 595 (Molecular Dynamics, Sunnyvale, CA). Selected DGGE bands were excised from the gels and eluted in 60 μl of water at 37°C overnight. One microliter of DNA was reamplified with the appropriate corresponding Bacteria or Archaea primers without the GC clamps as follows: an initial denaturation of 5 min at 95°C, followed by 25 to 28 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s. Sequencing and phylogenetic analysis were performed as described below.
Clone libraries of 16S rRNA genes and restriction fragment length polymorphism.
A total of four 16S rRNA gene clone libraries were constructed, one bacterial and one archaeal from two spring sites. For each library, three PCRs were combined to minimize bias. PCR conditions were as follows: 25 cycles of 94°C for 1 min, 55°C (archaeal) or 45°C (bacterial) for 1 min, and 72°C for 2 min, and a final extension at 72°C for 10 min. The PCR products were purified with a MinElute PCR purification kit (QIAGEN, Mississauga, ON, Canada) and cloned using a QIAGEN PCR cloning kit at an insert:vector ratio of 3:1. The ligations were transformed by electroporation into Escherichia coli strain DH10B (Invitrogen). Transformants were selected on Luria-Bertani medium supplemented with ampicillin (100 mg liter−1), 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-Gal; 80 mg liter−1), and isopropyl-beta-d-thiogalactopyranoside (IPTG; 50 μM). Between 155 and 174 randomly selected colonies from each library were screened by restriction fragment length polymorphism (RFLP) analysis with MspI and HhaI restriction endonucleases (New England Biolabs, Ipswich, MA) as described previously (29). Clones with identical patterns were visually regrouped, and one to five representatives of each RFLP pattern were selected for sequencing of both strands. Sequencing was performed at the McGill University Genome Quebec Innovation Centre using a model 3730XL DNA analyzer system (Applied Biosystems, Foster City, CA).
Phylogenetic and cluster analyses.
The 16S rRNA gene sequences were submitted for comparison to the GenBank databases using the BLAST algorithm (2). Sequences having ≥98% similarity and matching the same GenBank sequence were assigned to the same phylotype. The sequences were checked against the contaminant sequences commonly found in 16S rRNA gene clone libraries (59). The occurrence of chimeric sequences was determined manually and with the CHECK_CHIMERA function from the Ribosomal Database Project-II release 8.1 (http://wdcm.nig.ac.jp/RDP/cgis/chimera). The remaining sequences were then aligned with their closest relatives using ClustalW. Phylogenetic trees (neighbor-joining algorithm with Jukes-Cantor corrections) were constructed using a MacVector 7.2 software package (Accelrys). The robustness of inferred topologies was tested by 1,000 bootstrap resamplings of the neighbor-joining data. In addition, phylogenetic classification was inferred by submitting the sequences to the RDP Classifier from the Ribosomal Database Project-II release 9(http://rdp.cme.msu.edu/classifier). Comparisons of the microbial community compositions from the seven springs sampled were performed by cluster analysis of the DGGE banding patterns using Dendron 2.4 software (Solltech Inc., Oakdale, LA). Dendrograms were constructed by the unweighted pair group method with arithmetic mean (UPGMA) groupings with a similarity coefficient (SAB) matrix. The stability of the dendrograms was evaluated by randomizing the sample order 100 times and recalculating the dendrograms with 95% background noise.
Diversity indices and statistical analysis.
Rarefaction analysis was performed and diversity indices were calculated to characterize the bacterial and archaeal diversity of the spring sediment samples. The rarefaction curves were constructed using Analytic Rarefaction 1.3(http://www.uga.edu/∼strata/software/index.html). The coverage of the libraries was calculated as defined by Good (23), with the following formula: C = (1 − n1/N) × 100, where n1 is the number of phylotypes appearing only once in a library and N is the library size. The Shannon index (H′) of diversity, the reciprocal of Simpson's index (1/D) of dominance, and the Chao1 estimator of total species richness (9) were determined with EstimateS 7.5 (http://viceroy.eeb.uconn.edu/estimateS) (10). Evenness (the relative abundance of each phylotype) was calculated with the formula E = eH′/N, where H′ is the Shannon index of diversity and N is the total number of phylotypes (35). The phylotype compositions of the clone libraries were compared using the Sorensen index, S = 2 × c/(a + b), where c is the number of phylotypes found in both sample A and sample B and a is the number of phylotypes in sample A and b is the number of phylotypes in sample B (37). The LIBSHUFF program (50) (http://libshuff.mib.uga.edu) was used to evaluate the significance of differences between the clone libraries. The sequences of each clone, deduced by their RFLP patterns, were aligned using ClustalX (63), and the DNADIST program of PHYLIP (version 3.65) software (http://evolution.genetics.washington.edu/phylip.html) was used (with the Jukes-Cantor model) to generate the distance matrix submitted to LIBSHUFF.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database under accession numbers DQ521089 to DQ521211.
RESULTS
Physicochemical parameters of spring waters and sediments.
The physicochemical parameters of the spring discharge waters are summarized in Table 1 and are in close agreement with similar analyses (temperature, pH, and conductivity) performed with GH-1, CP-1, and CP-2 in 1997 and 1998 by Omelon et al. (42). Oxidoreduction potential values ranged from −283 to −375 mV, indicating that the spring outlets are highly reducing environments, although not anaerobic as low dissolved oxygen concentrations (0.05 to 0.2 ppm) were detected in the water layer just overlying the sediments. Most of the parameters measured were similar for springs at the same site, but some clear differences were observed between the Gypsum Hill and Colour Peak sites, especially an approximately twofold greater salinity measured at the CP springs (∼15.6%) than at the GH springs (∼7.6%). The spring waters were shown to be rich in sulfur compounds, containing 25 to 100 ppm of sulfide (measured in this study) and 3,724 mg/liter (at GH-1) and 2,300 mg/liter (at CP-1 and CP-2) of SO42− as measured previously (42). Spring sediment samples contained high concentrations of salts and SO42−. The CP-2 sediment contained 20 to 24 g/kg of Na+, Cl−, Ca2+, and total Fe. The GH-4 sediment also had high concentrations of Ca2+ and total Fe (∼16 g/kg) but contained three to four times less Na+ (5.4 g/kg) and Cl− (6.6 g/kg) than the CP-2 sediment. While the SO42− concentration was higher in the GH water, its concentration was approximately three times higher in the CP-2 sediment (6.7 g/kg) than in the GH-4 sediment (1.9 g/kg). The total Kjeldahl nitrogen concentrations in the sediments were 210 and 350 mg/kg, respectively, in GH-4 and CP-2. Previous studies measured dissolved inorganic carbon at 13.1 to 17.2 mg/liter (42) and found undetectable dissolved organic carbon (3). In the GH-4 sediment, total organic carbon was 3,700 mg/kg and total inorganic carbon was 3,900 mg/kg, while in the CP-2 sediment, total organic carbon was 8,300 mg/kg and total inorganic carbon was 4,500 mg/kg.
TABLE 1.
Field measurements of physicochemical parameters of the spring waters from Gypsum Hill and Colour Peak
| Spring | Temp (oC) | pH | ORP (mV)a | Salinity (%) | Conductivity (mS/cm)b | TDS (g/liter)c | Sulfide (ppm) | O2 (ppm) |
|---|---|---|---|---|---|---|---|---|
| GH-1 | 6.3 | 7.42 | −324 | 7.9 | 126 | 63.3 | 25 | 0.05 |
| GH-2 | −0.5 | 7.47 | −283 | 7.5 | 120 | 60.0 | 25 | 0.2 |
| GH-3 | 5.8 | 7.42 | −316 | 7.6 | 122 | 61.2 | 25 | 0.05 |
| GH-4 | 6.9 | 7.43 | −287 | 7.5 | 121 | 60.6 | 50 | 0.2 |
| CP-1 | 5.7 | 6.92 | −375 | 15.5 | 231 | 120.0 | 100 | 0.2 |
| CP-2 | 5.5 | 6.91 | −348 | 15.8 | 244 | 122.0 | 100 | 0.2 |
| CP-3 | 3.1 | 6.91 | −345 | 15.5 | 241 | 121.0 | >100 | 0.2 |
ORP, oxidoreduction potential.
S, siemens.
TDS, total dissolved solids.
DGGE analysis.
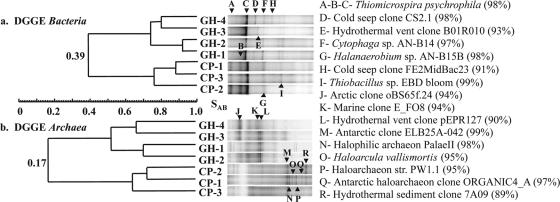
DGGE analyses of bacterial and archaeal 16S rRNA genes were performed to compare the microbial composition of the seven springs, and the DGGE banding patterns were used to construct dendrograms (Fig. 2). DGGE banding patterns divided in two clusters based on their geographic origin, i.e., Gypsum Hill and Colour Peak. Cluster analysis demonstrated low SAB between the two sets of springs (archaeal SAB [SAB arc], 0.17; bacterial SAB [SAB bac], 0.39). Specific phylogenetic information was determined by sequencing of the individual DGGE bands. All bands that migrated to the same position in a gel were identified by the same letter and gave identical sequencing results. Banding patterns from the bacterial DGGE gel showed a limited diversity, with five to seven bands for both the GH and the CP samples (Fig. 2a). DNA sequences obtained from bands A and C, found in all seven springs, were 100% identical despite the different migration and were 98% identical to the sulfur oxidizer Thiomicrospira psychrophila (33); bands B and I in CP were also related to sulfur-oxidizing bacteria. Bands F (GH) and G (CP) were closely affiliated with Cytophaga sp. strain AN-B14 and Halanaerobium sp. strain AN-B15B, respectively, isolated from a hypersaline brine/seawater interface. The archaeal DGGE gel displayed a higher number of bands than the bacterial DGGE gel (Fig. 2b), with the CP samples having approximately twofold the number of bands (14-16) as the GH samples (7-9). A group of six bands (M to R) from CP migrated close together, and all bands except R were related to haloarchaea. The four nonhaloarchaeal sequences were only distantly related to archaeal 16S rRNA clones from marine, hydrothermal vent, and Arctic Ocean environments.
FIG. 2.
Clusteranalysis of DGGE banding patterns based on position of bands using unweighted pair groupings of an SAB matrix. (a) Dendrogram for DGGE Bacteria; (b) dendrogram for DGGE Archaea. Phylogenetic affiliations of the sequenced DGGE bands are shown on the right, with the percentage of similarity.
Clone libraries.
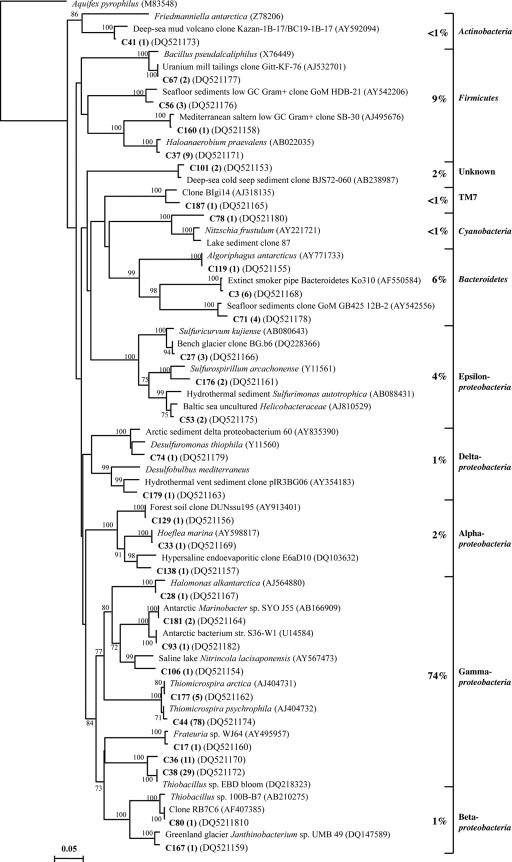
Since the DGGE analysis revealed similar banding patterns for the springs at the same site but distinct differences between the two sites, we selected one spring at Gypsum Hill and one at Colour Peak for a more thorough characterization. Bacterial and archaeal 16S rRNA gene clone libraries were constructed for the GH-4 and CP-1 spring sediments. A total of 649 clones, 311 from the GH-4 sample and 338 from the CP-1 sample, were screened by RFLP and grouped into identical restriction patterns (phylotypes). Clones from each phylotype were sequenced, and the ∼590-bp sequences were analyzed to determine their phylogenetic affiliations. Phylogenetic trees illustrating affiliations and the occurrence of each phylotype are presented in Fig. 3 through Fig. 6. Grouping of the bacterial sequences into different phyla using the RDP Classifier was generally in agreement with the phylogenetic tree branching.
FIG. 3.
Phylogenetic relationships of the 46 bacterial 16S rRNA gene sequences obtained from the GH-4 clone library. The tree was inferred by neighbor-joining analysis of 558 homologous positions of sequence from each organism or clone. Aquifex pyrophilus was used as the outgroup. Numbers on the nodes are the bootstrap values (percentages) based on 1,000 replicates. The scale bar indicates the estimated number of base changes per nucleotide sequence position. Bold type indicates GH-4 clones, with their prevalence in the clone library.
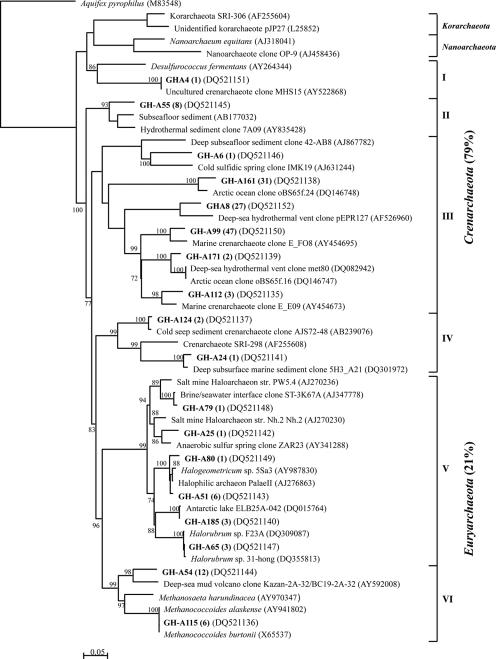
FIG. 6.
Phylogenetic relationships of the 29 archaeal 16S rRNA gene sequences obtained from the CP-1 clone library. The tree was inferred by neighbor-joining analysis of 402 homologous positions of sequence from each organism or clone. Aquifex pyrophilus was used as the outgroup. Numbers on the nodes are the bootstrap values (percent) based on 1,000 replicates. Scale bar indicates the estimated number of base changes per nucleotide sequence position. Bold type indicates CP-1 clones, with their prevalence in the clone library.
Bacterial clone libraries.
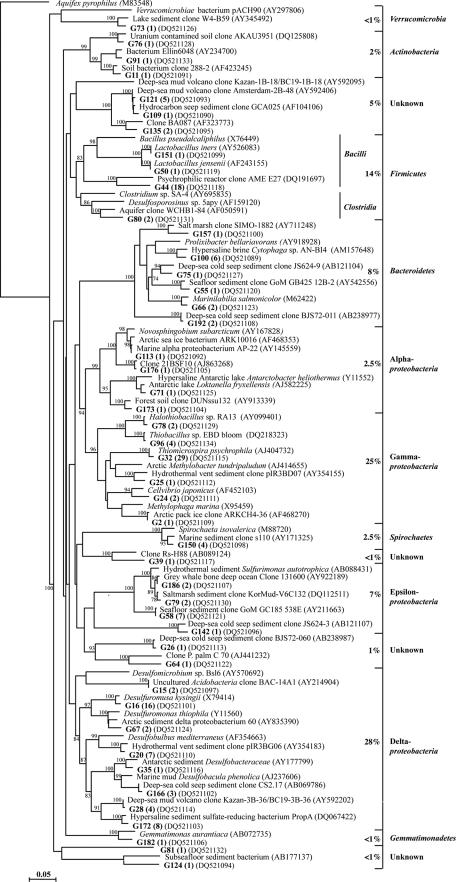
The GH-4 bacterial library was composed of 155 clones that grouped into 46 phylotypes (Fig. 3). The bacterial phylotypes could be divided into seven phyla, as follows: Proteobacteria (63% of the total clones), Firmicutes (14%), Bacteroidetes (8%), Spirochaetes (2.5%), Actinobacteria (2%), Gemmatimonadetes (<1%), and Verrucomicrobia (<1%). Eight phylotypes could not be classified into known phylogenetic groups by the RDP Classifier or by phylogenetic tree branching. Deltaproteobacteria and Gammaproteobacteria phylotypes represented half of the clone library. The Deltaproteobacteria phylotypes were classified into either the family Desulfobacteriaceae or Desulfuromonaceae by the RDP Classifier (95% confidence threshold). The Gammaproteobacteria were represented by six phylotypes; three phylotypes were related to characterized sulfur-oxidizing bacteria (Thiobacillus sp. strain EBD bloom, Halothiobacillus sp. strain RA13, and Thiomicrospira psychrophila). Phylotype G32, 98% identical to Thiomicrospira psychrophila, represented 30% of all Proteobacteria and 19% of the total clones.
The CP-1 bacterial library was composed of 174 clones that grouped into 30 phylotypes (Fig. 4). The phylotypes could be divided into six phyla (Proteobacteria [82%], Firmicutes [9%], Bacteroidetes [6%], Actinobacteria [<1%], and Cyanobacteria [<1%] and the candidate division TM7 [<1%]). The Proteobacteria were highly dominant, comprising 19 different phylotypes with representatives from all five subclasses (Alpha-, Beta-, Delta-, Epsilon-, and Gammaproteobacteria). The Gammaproteobacteria dominance (74%) was largely due to the high proportions of phylotype C44 (98% identical to Thiomicrospira psychrophila), which alone represented 45% of the clone library, and of phylotypes C36/C38 (related to Thiobacillus sp. strain EBD bloom) (23%). The two deltaproteobacterial phylotypes were related to the sulfur-reducing bacterium Desulfuromonas thiophila and the sulfate-reducing bacterium Desulfobulbus mediterraneus.
FIG. 4.
Phylogenetic relationships of the 30 bacterial 16S rRNA gene sequences obtained from the CP-1 clone library. The tree was inferred by neighbor-joining analysis of 501 homologous positions of sequence from each organism or clone. Aquifex pyrophilus was used as the outgroup. Numbers on the nodes are the bootstrap values (percentages) based on 1,000 replicates. Scale bar indicates the estimated number of base changes per nucleotide sequence position. Bold type indicates CP-1 clones, with their prevalence in the clone library.
Archaeal clone libraries.
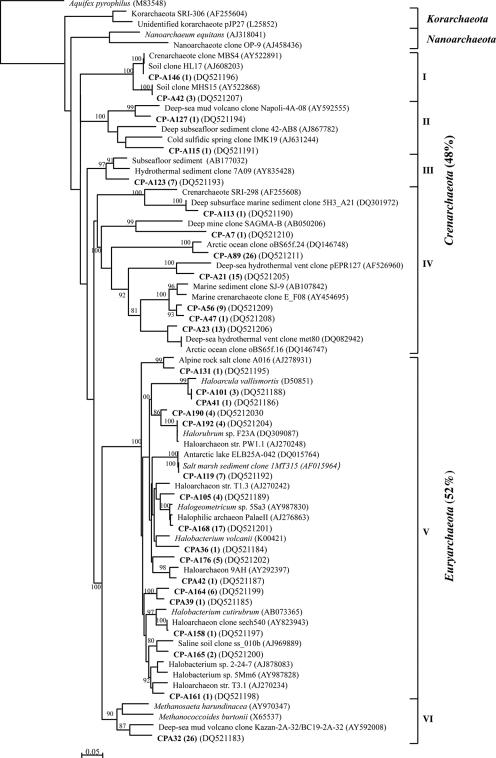
The 156 GH-4 archaeal clones represented 18 archaeal phylotypes that fell into six clusters in the phylogenetic tree (Fig. 5): four clusters of Crenarchaeota (I to IV) and two of Euryarchaeota (V to VI). Crenarchaeota phylotypes accounted for 79% of the GH-4 archaeal clone library, with the three most abundant phylotypes (GH-A99, GH-A161, and GHA8; 90% of the Crenarchaeota clones) grouping with cluster III. Cluster III phylotypes had their best BLAST matches (84 to 95%) with archaeal clones from diverse marine environments. Euryarchaeota clusters V and VI were composed of sequences related to the orders Halobacteriales and Methanosarcinales, respectively. One phylotype was 99% identical to the psychrotolerant methanogens Methanococcoides burtonii (20) and M. alaskense (49). The most abundant Euryarchaeota phylotype (GH-A54) grouped with the methanogens but was only distantly related (90%) to an uncultured archaeon from a methane-rich deep-sea mud volcano.
FIG. 5.
Phylogenetic relationships of the 18 archaeal 16S rRNA gene sequences obtained from the GH-4 clone library. The tree was inferred by neighbor-joining analysis of 402 homologous positions of sequence from each organism or clone. Aquifex pyrophilus was used as the outgroup. Numbers on the nodes are the bootstrap values (percentages) based on 1,000 replicates. Scale bar indicates the estimated number of base changes per nucleotide sequence position. Bold type indicates GH-4 clones, with their prevalence in the clone library.
Twenty-nine archaeal phylotypes were defined from 164 clones screened in the CP-1 archaeal clone library. As was observed for GH-4, the CP-1 phylotypes divided into four Crenarchaeota clusters (I to IV; 12 phylotypes; 48% of the clones) and two Euryarchaeota clusters (V to VI; 17 phylotypes; 52% of the clones) (Fig. 6). Crenarchaeota cluster I phylotypes were related to clones retrieved from soil, while cluster II and III phylotypes grouped with clones from deep-sea environments or from the surface waters of a cold sulfidic spring in Germany (48). The seven phylotypes from cluster IV (40% of the clones) were related to clones from deep subsurfaces or marine environments. The CP-1 Euryarchaeota were comprised of 16 phylotypes that grouped with Halobacteriales sequences in cluster V and one phylotype, CPA32 (16% of the clones), that was associated with the methanogen cluster (VI). CPA32 was only distantly related to 16S rRNA sequences in the public databases but was identical to phylotype GH-A54 from the GH-4 library.
Comparison and statistical analysis of the clone libraries.
The number of clones, phylotypes, and biodiversity indices calculated for the four clone libraries are summarized in Table 2. The coverage of the clone libraries was high, ranging from 84 to 96%, suggesting that the major part of the microbial diversity was identified in this study. The high coverage values are corroborated by the rarefaction curves that reached a near plateau, except for the GH-4 bacterial libraries (data not shown). The rarefaction curves, supported by the Shannon index and Chao1, indicated that the GH-4 bacterial population was the most diverse, while the GH-4 archaeal population was the least diverse. The diversity of the CP-1 bacterial and archaeal libraries was similar in numbers of phylotypes, but the Shannon index, Chao1, and Evenness (E) estimated that the archaeal diversity was higher in both species richness and evenness.
TABLE 2.
Numbers of clones and phylotypes analyzed for the four 16S rRNA gene clone libraries and their diversity indices
| Clone library | Total no. of clones | No. of phylotypes | Coverage (%) | Shannon index (H′) | Simpson's index (1/D) | Evenness (E) | Chao1 | Sorensen similarity index |
|---|---|---|---|---|---|---|---|---|
| Bacterial libraries | 0.16 | |||||||
| GH-4 | 155 | 46 | 84 | 3.17 | 14.82 | 0.52 | 71 | |
| CP-1 | 174 | 30 | 91 | 2.16 | 4.25 | 0.33 | 50 | |
| Archaeal libraries | 0.51 | |||||||
| GH-4 | 156 | 18 | 96 | 2.12 | 5.94 | 0.47 | 23 | |
| CP-1 | 164 | 29 | 92 | 2.77 | 12.18 | 0.61 | 68 |
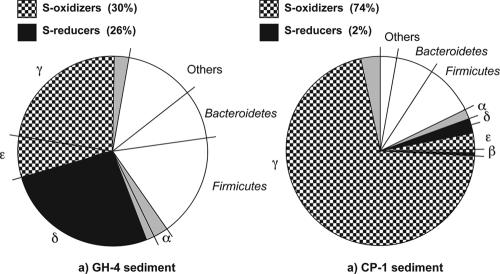
The microbial compositions of the clone libraries were compared by calculating the Sorensen similarity index. Phylotype comparisons between the CP-1 and GH-4 bacterial libraries revealed that six phylotypes gave the same best-BLAST matches, for a low similarity value of 0.16. Even though the two bacterial libraries were dominated by sequences from potentially sulfur-metabolizing bacteria, there was a significant disparity in the distribution of the S-oxidizing and the S-reducing populations (Fig. 7). The proportions of S oxidizers and S reducers, representing 56% of the clone library, were similar in the GH-4 sample. In the CP-1 sample, the S oxidizers were highly dominant (74%) and the S reducers represented only 2% of the clones. The similarity value (0.51) of the archaeal libraries was substantially higher than that determined for the bacterial libraries; 8/10 of GH-4 Crenarchaeota phylotypes and 4/8 of GH-4 Euryarchaeota phylotypes (67% of the total GH-4 phylotypes) had the same best-BLAST matches as the CP-1 library. The CP-1 archaeal library had more sequences related to haloarchaea, which is consistent with its higher salinity. Comparison of the GH-4 and CP-1 clone libraries using LIBSHUFF analysis indicated that both bacterial and archaeal libraries were significantly different (P < 0.05). The uniqueness of the spring microbial communities was estimated by comparing the degree of relatedness of the spring microbial sequences with their closest matches in GenBank. Overall, 31% of the total phylotypes were related to known sequences at <95% identity, which suggests that these sequences represent novel microbial genera not yet retrieved from any other environment.
FIG. 7.
Distribution of the putative sulfur-metabolizing Proteobacteria of the spring sediments.
DISCUSSION
The present study provides the first characterization of the microbial diversity present in the cold saline springs found at Expedition Fjord in the Canadian high Arctic. Physical and chemical analyses from this study and previous investigations (3, 42) indicate that the spring water and sediment environments are permanently cold, moderately saline, and highly reducing and probably contain anoxic and microaerophilic zones. A variety of potential electron donors (sulfides, organic C, CH4, and Fe) and electron acceptors (O2, SO42−, CO2, and Fe) have been identified in these environments, potentially supporting diverse heterotrophic and autotrophic modes of microbial metabolism, and representative microorganisms of these two major metabolic groups were detected in the clone libraries. The ability to metabolize sulfur compounds appears to be widespread among microorganisms of widely different phylogenetic and physiological types, and the microbial communities of sulfur-rich habitats are therefore influenced by the environmental conditions, i.e., pH, temperature, sulfide, sulfur, or sulfate concentrations, redox conditions, the presence of other electron acceptors, light availability, and organic carbon content (16). In this respect, the majority of 16S rRNA gene sequences obtained from the springs were most closely related to sequences retrieved from the sediments of marine environments (including diverse deep-sea habitats), subsurfaces (subseafloor and mines), and hypersaline habitats (hypersaline lakes, Dead Sea) and from both polar regions. These environments and the springs share some environmental properties that could have acted as selective factors for the establishment of the spring microbial communities, namely, the presence of sulfur compounds, high salinity, a cold temperature, and a deep subsurface origin. Molecular studies of the microbial communities of other cold saline environments such as Antarctic hypersaline lake sediments (7, 31) and Antarctic and Arctic marine sediments (8, 46, 47) also revealed highly diverse bacterial populations. Deltaproteobacteria sulfate reducer and Gammaproteobacteria sulfur oxidizer phylotypes were found to dominate in Arctic marine sediments (47). Euryarchaeota sequences related to extreme haloarchaea and methanogens dominated clone libraries from hypersaline and freshwater Antarctic lakes, respectively (7, 31). Heterotrophic Proteobacteria phylotypes, including Pseudomonas, Polaromonas, Burkholderia, and Marinobacter, the last of which was also observed in our study, were detected in supraglacial sulfur springs on Ellesmere Island (24).
Since there are known biases associated with DNA extraction and PCR amplification (39, 66), the abundance of a phylotype in a clone library does not necessarily reflect its abundance in the sample, and the corresponding ecological function cannot be inferred with certitude from the phylogenetic affiliation (1). Such assumptions should be made only when there is a high degree of sequence similarity between the phylotypes and known cultivated species. In this study, a number of phylotypes from nearly every phylogenetic group had sequence homology with cultivated microorganisms at the species or genus levels, allowing some prediction of ecological function within the spring sediments. Many phylotypes (56 to 76%) were from putative sulfur-metabolizing bacteria, suggesting that the utilization and cycling of sulfur compounds may play a major role in energy production and maintenance of microbial communities in these permanently cold saline environments.
A major metabolic process in both springs appeared to be the oxidation of reduced sulfur compounds. Sequences related to sulfur oxidizers were the most abundant and grouped into three subclasses of the Proteobacteria (Beta-, Epsilon-, and Gammaproteobacteria); the majority of these phylotypes were related to Thiomicrospira, especially T. psychrophila, and Thiobacillus, two genera frequently isolated from marine environments. T. psychrophila is a psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium that was initially isolated from marine Arctic sediments (33). The Epsilonproteobacteria group of sulfur oxidizers was related to the Sulfuricurvum and Sulfurimonas genera. Reduction of oxidized sulfur compounds, as part of the spring microbial metabolism, was exemplified by sequences of Deltaproteobacteria that were comprised of phylotypes closely related to diverse genera (Desulfuromusa, Desulfuromonas, Desulfobulbus, and Desulfobacula) of sulfur- and sulfate-reducing bacteria. A phylotype of Epsilonproteobacteria from CP-1 was related to the sulfur-reducing bacterium Sulfurospirillum arcachonense (19). Two additional Epsilonproteobacteria phylotypes were not related to any cultivated bacteria. While recent reports (41, 57) have demonstrated the metabolic diversity of cultivated Epsilonproteobacteria, their role in the S cycle as either reducing elemental sulfur to sulfide or oxidizing sulfide to sulfur has long been established (61, 67), so it is likely that these phylotypes participate in the cycling of S compounds in the GH and CP spring systems. Other detected phylotypes may be involved in the oxidoreduction of sulfur compounds. For example, a GH-4 phylotype was related to Loktanella fryxellensis, an Alphaproteobacteria sp. within the Rhodobacteraceae family (65). Some species of Rhodobacteraceae are capable of oxidizing reduced sulfur compounds under both oxic and anoxic conditions (32, 62). Phylotypes related to Marinobacter, Halomonas, and Cytophaga spp. were also detected in the spring sediments; representatives from these genera are capable of heterotrophic sulfur oxidation (22, 43, 53).
The spring archaeal populations are also likely to participate in the spring sulfur metabolism. Some haloarchaea can slowly reduce elemental sulfur (15, 64) and oxidize thiosulfate to tetrathionate (54). Elshahed et al. (15) retrieved haloarchaeal clones and isolated sulfur-reducing haloarchaea from an anoxic mesophilic sulfide- and sulfur-rich spring, suggesting that these microorganisms play a role in sulfur metabolism in sulfur-rich anaerobic ecosystems. Only one low-temperature crenarchaeote has been cultivated to date (34), but many hyperthermophilic crenarchaeotes isolated from sulfur-rich high-temperature environments are able to use oxidized or reduced sulfur compounds in their metabolic energy-yielding reactions (55). Given the similarities of some of the spring archaeal phylotypes with sequences from sulfur-rich environments (cold sulfidic spring and hydrothermal vents), at least some of the Crenarchaeota detected in the GH and CP spring sediments may rely on sulfur metabolism for their energy production.
Two lines of evidence suggest that methanogenesis may occur in the Expedition Fjord spring sediments. First, we detected low concentrations of methane in the spring waters at both the Gypsum Hill and Colour Peak sites (data not published). Second, the most abundant Euryarchaeota phylotype from both spring libraries was associated with the Methanosarcinales cluster in the phylogenetic trees, and one GH-4 phylotype was 99% identical to psychrotolerant methanogens (M. burtonii and M. alaskense) that can use methylamines for growth (20, 49). As sulfate reducers outcompete methanogens for most energy sources, the persistence of methanogens in saline environments where sulfate is not limiting is associated with the utilization of noncompetitive substrates such as methylamines.
Sulfide-rich springs, from all ranges of temperature, are commonly sustained by the activity of phototrophic bacteria (14, 16, 51) that often form abundant photosynthetic microbial mats. However, photosynthetic prokaryotes do not seem to play an important role as the primary producers in the GH and CP springs as we did not visually or microscopically observe evidence of photosynthetic microorganisms, prokaryotic or eukaryotic, in any of the spring outlets, and only one phototrophic clone (cyanobacteria-related) was detected in CP-1. Moreover, chlorophyll was not detected over the surface of the carbonates from ∼100 spring locations using a pulse amplitude modulation fluorometer (3). Considering the high sulfide concentrations and the 24-h light illumination during the sampling period, anoxygenic phototrophs that use sulfide or other reduced sulfur compounds as electron donors in photosynthesis were unexpectedly not detected. The high salinity of the springs is not likely to be the reason for this absence as anoxygenic phototrophs were found in a hypersaline endoevaporitic microbial mat collected from a pond with 20% salinity (52). Based on these observations, the sulfide emerging from the springs may support populations of chemolithoautotrophic sulfur oxidizers that act as primary producers in the spring systems. Sulfur-based chemolithotrophy, mainly performed by Epsilonproteobacteria, can sustain microbial ecosystems devoid of light such as hydrothermal vents (30) and aphotic (cave) sulfidic springs (17, 18). This nonphotosynthesis-based primary production could hypothetically sustain the Axel Heiberg springs' microbial communities during the ∼3 months of total darkness that occur seasonally at high latitudes. This would be of significant interest in astrobiology, particularly for the search for life in subsurface waters which may exist on Mars (4, 42).
The microbial phylotypes retrieved in this study are currently being used as guides to develop appropriate culturing methodologies for isolating novel indigenous bacteria for further characterization and to develop activity assays to identify microbial communities active under in situ conditions.
Acknowledgments
We thank Gavin Whissell and Blaire Steven for assistance in the field in 2004 and 2005. Logistic support was provided by the Canadian Polar Continental Shelf Project (PCSP project numbers 666-05, 635-04, and 633-03) and by McGill University's High Arctic Research Station.
This work was supported by grants from NASA's Exobiology program (NAG5-12395) and the Natural Sciences and Engineering Research Council of Canada (NSERC). Additional funding for student research was provided by the Department of Indian and Northern Affairs—Northern Scientific Training Program and the Fonds Québécois de la Recherche sur la Nature et les Technologies (FQRNT).
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Achenbach, L. A., and J. Coates. 2000. Disparity between bacterial phylogeny and physiology. ASM News 66:714-715. [Google Scholar]
- 2.Altschul, S., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, D. 2004. Perennial springs in the Canadian high Arctic: analogues of hydrothermal systems on Mars. Ph.D. thesis. McGill University, Montreal, Canada.
- 4.Andersen, D. T., W. H. Pollard, C. P. McKay, and J. Heldmann. 2002. Cold springs in permafrost on Earth and Mars. J. Geophys. Res. 107:1-7. [Google Scholar]
- 5.Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. [DOI] [PubMed] [Google Scholar]
- 6.Barns, S., R. Fundyga, M. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman, J. P., S. A. McCammon, S. M. Rea, and T. A. McMeekin. 2000. The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol. Lett. 183:81-88. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Statistics 11:265-270. [Google Scholar]
- 10.Colwell, R. 2005. EstimateS: statistical estimation of species richness and shared species from samples. Version 7.5. User's guide and application. http://viceroy.eeb.uconn.edu/EstimateS.
- 11.Deming, J. 2002. Psychrophiles and polar regions. Curr. Opin. Microbiol. 5:301-309. [DOI] [PubMed] [Google Scholar]
- 12.Don, R., P. Cox, B. Wainwright, K. Baker, and J. Mattick. 1991. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Aids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doran, P. T., C. P. Mckay, W. P. Adams, M. C. English, R. A. Wharton, and M. A. Meyer. 1996. Climate forcing and thermal feedback of residual ice covers in the high Arctic. Limnol. Oceanogr. 41:839-848. [Google Scholar]
- 14.Douglas, S., and D. Douglas. 2001. Structural and geomicrobiological characteristics of a microbial community from a cold sulfide spring. Geomicrobiol. J. 18:401-422. [Google Scholar]
- 15.Elshahed, M. S., F. Z. Najar, B. A. Roe, A. Oren, T. A. Dewers, and L. R. Krumholz. 2004. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide- and sulfur-rich spring. Appl. Environ. Microbiol. 70:2230-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elshahed, M. S., J. S. Senko, F. Z. Najar, S. M. Kenton, B. A. Roe, T. A. Dewers, J. R. Spear, and L. R. Krumholz. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engel, A. S., N. Lee, M. L. Porter, L. A. Stern, P. C. Bennett, and M. Wagner. 2003. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engel, A. S., M. L. Porter, L. A. Stern, S. Quinlan, and P. C. Bennett. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria.” FEMS Microbiol. Ecol. 51:31-53. [DOI] [PubMed] [Google Scholar]
- 19.Finster, K., W. Liesack, and B. Tindall. 1997. Sulfurospirillum arcachonense sp. nov., a new microaerophilic sulfur-reducing bacterium. Int. J. Syst. Bacteriol. 47:1212-1217. [DOI] [PubMed] [Google Scholar]
- 20.Franzmann, P. D., N. Springer, W. Ludwig, E. C. Conway de Macario, and M. Rohde. 1992. A methanogenic Archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst. Appl. Microbiol. 15:573-581. [Google Scholar]
- 21.Gilichinsky, D., E. Rivkina, V. Shcherbakova, K. Laurinavichuis, and J. Tiedje. 2003. Supercooled water brines within permafrost: an unknown ecological niche for microorganisms: a model for astrobiology. Astrobiology 3:331-341. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez, J., R. Kiene, S. Joye, D. Sorokin, and M. Moran. 2002. Oxidation of organic and inorganic sulfur compounds by aerobic heterotrophic marine bacteria, p. 291-310. In V. P. Singh and R. D. Stapleton, Jr. (ed.), Biotransformations: bioremediation technology for health and environmental protection. Elsevier Science B.V., Amsterdam, The Netherlands.
- 23.Good, I. 1953. The population frequencies of species and the estimation of population of parameters. Biometrika 40:237-264. [Google Scholar]
- 24.Grasby, S. E., C. C. Allen, T. G. Longazo, J. T. Lisle, D. W. Griffin, and B. Beauchamp. 2003. Supraglacial sulfur springs and associated biological activity in the Canadian high arctic—signs of life beneath the ice. Astrobiology 3:583-596. [DOI] [PubMed] [Google Scholar]
- 25.Hoek, J., A. Banta, H. Forrest, and A.-L. Reysenbach. 2003. Microbial diversity of a sulphide spire located in the Edmond deep-sea hydrothermal vent field in the Central Indian Ridge. Geobiology 1:119-127. [Google Scholar]
- 26.Hoffman, N., P. L. Knauth, S. Klonowski, D. Burt, S. R. Saunders, R. W. Zurek, P. T. Doran, and S. L. Forman. 2000. Ideas about the surface runoff features on Mars. Science 290:711-714. [DOI] [PubMed] [Google Scholar]
- 27.Hugenholtz, P., C. Pitulle, K. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janse, I., J. Bok, and G. Zwart. 2004. A simple remedy against artifactual double bands in denaturing gradient gel electrophoresis. J. Microbiol. Methods 57:279-281. [DOI] [PubMed] [Google Scholar]
- 29.Juck, D., B. T. Driscoll, T. C. Charles, and C. W. Greer. 2003. Effect of experimental contamination with the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine on soil bacterial communities. FEMS Microbiol. Ecol. 43:255-262. [DOI] [PubMed] [Google Scholar]
- 30.Karl, D., C. Wirsen, and H. Jannasch. 1980. Deep-sea primary production at the Galapagos hydrothermal vents. Science 207:1345-1347. [Google Scholar]
- 31.Karr, E. A., J. M. Ng, S. M. Belchik, M. W. Sattley, M. T. Madigan, and L. A. Achenbach. 2006. Biodiversity of methanogenic and other Archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 72:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly, D., J. Shergill, W. Lu, and A. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 33.Knittel, K., J. Kuever, A. Meyerdierks, R. Meinke, R. Amann, and T. Brinkhoff. 2005. Thiomicrospira arctica sp. nov. and Thiomicrospira psychrophila sp. nov., psychrophilic, obligately chemolithoautotrophic, sulfur-oxidizing bacteria isolated from marine Arctic sediments. Int. J. Syst. Evol. Microbiol. 55:781-786. [DOI] [PubMed] [Google Scholar]
- 34.Könneke, M., A. Bernhard, J. de la Torre, C. Walker, J. Waterbury, and D. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 35.Krebs, C. 1989. Ecological methodology. Harper and Row, New York, NY.
- 36.Lauritzen, S., and S. Bottrell. 1994. Microbial activity in thermoglacial karst springs, south Spitsbergen. Geomicrobiol. J. 12:161-173. [Google Scholar]
- 37.Magurran, A. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 38.Malin, M. C., and K. S. Edgett. 2000. Evidence for recent groundwater seepage and surface runoff on Mars. Science 228:2330-2335. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Laurent, F., L. Philippot, S. Hallet, R. Chaussod, J. Germon, G. Soulas, and G. Catroux. 2001. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moissl, C., C. Rudolph, and R. Huber. 2002. Natural communities of novel archaea and bacteria with a string-of-pearls-like morphology: molecular analysis of the bacterial partners. Appl. Environ. Microbiol. 68:933-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa, S., K. Takai, F. Inagaki, H. Hirayama, T. Nunoura, and K. Horikoshi. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 42.Omelon, C. R., W. H. Pollard, and D. T. Andersen. 2006. A geochemical evaluation of perennial spring activity and associated mineral precipitates at Expedition Fjord, Axel Heiberg Island, Canadian high Arctic. Appl. Geochem. 21:1-15. [Google Scholar]
- 43.Podgorsek, L., R. Petri, and J. Imhoff. 2004. Cultured and genetic diversity, and activities of sulfur-oxidizing bacteria in low-temperature hydrothermal fluids of the North Fiji basin. Mar. Ecol. Prog. Ser. 266:65-76. [Google Scholar]
- 44.Pollard, W., C. Omelon, D. Andersen, and C. McKay. 1999. Perennial spring occurrence in the Expedition Fiord area of western Axel Heiberg Island, Canadian High Arctic. Can. J. Earth Sci. 36:105-120. [Google Scholar]
- 45.Priscu, J., and B. Christner. 2004. Earth's icy biosphere, p. 130-145. In A. T. Bull. (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, DC.
- 46.Ravenschlag, K., K. Sahm, and R. Amann. 2001. Quantitative molecular analysis of the microbial community in marine arctic sediments (Svalbard). Appl. Environ. Microbiol. 67:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rudolph, C., C. Moissl, R. Henneberger, and R. Huber. 2004. Ecology and microbial structures of archaeal/bacterial strings-of-pearls communities and archaeal relatives thriving in cold sulfidic springs. FEMS Microbiol. Ecol. 50:1-11. [DOI] [PubMed] [Google Scholar]
- 49.Singh, N., M. M. Kendall, Y. Liu, and D. R. Boone. 2005. Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica. Int. J. Syst. Evol. Microbiol. 55:2531-2538. [DOI] [PubMed] [Google Scholar]
- 50.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjörleifsdottir, V. T. Marteinsson, S. K. Petursdottir, O. Holst, and J. K. Kristjansson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sørensen, K. B., D. E. Canfield, A. P. Teske, and A. Oren. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71:7352-7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorokin, D. 2003. Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72:641-653. [PubMed] [Google Scholar]
- 54.Sorokin, D., T. Tourova, and G. Muyzer. 2005. Oxidation of thiosulfate to tetrathionate by an haloarchaeon isolated from hypersaline habitat. Extremophiles 9:501-504. [DOI] [PubMed] [Google Scholar]
- 55.Stetter, K. O. 2001. Microorganisms in high-temperature sulfur environments. In Encyclopedia of life sciences. John Wiley and Sons, Ltd., Chichester, United Kingdom. http://www.els.net/. doi: 10.1038/npg.els.0006101. [DOI]
- 56.Steven, B., R. Léveillé, W. H. Pollard, and L. G. Whyte. Microbial ecology and biodiversity in permafrost. Extremophiles 10:259-267. [DOI] [PubMed]
- 57.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, and Y. Sako. 2003. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 58.Takai, K., F. Komatsu, F. Inagaki, and K. Horikoshi. 2001. Distribution of archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67:3618-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor, A., and A. Judge. 1976. Canadian geothermal data collection. In E. P. Branch (ed.), Geothermal series, number 6. EMR, Ottawa.
- 61.Taylor, C. D., C. O. Wirsen, and F. Gaill. 1999. Rapid microbial production of filamentous sulfur mats at hydrothermal vents. Appl. Environ. Microbiol. 65:2253-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin, and D. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tindall, B., and H. Trüper. 1986. Ecophysiology of the aerobic halophilic archaebacteria. Syst. Appl. Microbiol. 7:202-212. [Google Scholar]
- 65.Van Trappen, S., J. Mergaert, and J. Swings. 2004. Loktanella salsilacus gen. nov., sp. nov., Loktanella fryxellensis sp. nov., and Loktanella vestfoldensis sp. nov., new members of the Rhodobacter group, isolated from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 54:1263-1269. [DOI] [PubMed] [Google Scholar]
- 66.von Wintzingerode, F., U. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 67.Wirsen, C., H. Jannasch, and S. Molyneaux. 1993. Chemosynthetic microbial activity at Mid-Atlantic Ridge hydrothermal vent sites. J. Geophys. Res. 98:9693-9703. [Google Scholar]
- 68.Worsley, P., and S. Gurney. 1996. Geomorphology and hydrogeological significance of the Holocene pingos in the Karup Valley, Traill Island, northern east Greenland. J. Quat. Sci. 11:249-262. [Google Scholar]