Abstract
Achieving efficient heterologous protein production and secretion by Bacillus subtilis is an attractive prospect, although often disappointingly low yields are reached. The expression of detoxified Clostridium perfringens β-toxin (β-toxoid) is exemplary for this. Although β-toxin can be efficiently expressed and secreted by Bacillus subtilis, the genetically detoxified, and industrially interesting, β-toxoid variant is difficult to obtain in high amounts. To optimize the expression of this putative vaccine component, we studied the differences in the global gene regulation responses of B. subtilis to overproduction of either β-toxin or β-toxoid by transcriptomics. A clear difference was the upregulation of the CssRS regulon, known to be induced upon secretion stress, when β-toxoid is produced. YkoJ, a protein of unknown function, was also upregulated, and we show that its expression is dependent on cssS. We then focused on the heterologous protein itself and found that the major secretion bottleneck can be traced back to a single amino acid substitution between the β-toxin and the β-toxoid, which results in the rapid degradation of β-toxoid following secretion across the cytoplasmic membrane. In contrast to β-toxin, β-toxoid protein is more prone to degradation directly after secretion, most likely due to poor folding characteristics introduced with point mutations. Our results show that although the host can be adapted in many ways, the intrinsic properties of a heterologous protein can play a decisive role when optimizing heterologous protein production.
Bacillus subtilis is widely used for protein production and secretion (12). Nevertheless, the secretion of heterologous proteins is often problematic, and many attempts have been made to overcome the poor production of secreted heterologous proteins, with varying success. A major problem encountered when using Bacillus subtilis as a production platform is protein degradation by extracellular proteases secreted by this organism. By using extracellular protease-deficient strains, this problem can be reduced to some extent, but such strains are more prone to lysis and have a reduced growth rate (34).
Another bottleneck is the rapid degradation of the secreted protein by a quality control mechanism in the cell wall environment (3, 14, 27, 30). Typically, this degradation takes place before the protein can fold into its native and usually protease-resistant conformation. B. subtilis responds to the overexpression of secreted proteins with the so-called secretion-stress response. CssRS controls this stress response and regulates the expression of HtrA and HtrB, two serine proteases that also can act as chaperones (7). Secretion stress is thought to be triggered by unfolded proteins at the trans side of the membrane due to problems that occur in late stages of protein secretion (19), presumably as a consequence of slow folding at the membrane cell wall interface (5, 13).
To promote correct and rapid folding of the secreted heterologous proteins, several measures can be taken. Expression of chaperones and proteases can be altered (33), the charge of the cell wall or the secreted protein can be adapted (28), and the availability of divalent metal ions can accelerate folding of the proteins (27). However, such measures usually improve secretion by only a factor of 1.5 to 3.
In this work, we report on important factors influencing the secretion of β-toxoid, a genetically inactive form of Clostridium perfringens β-toxin. This protein is of industrial interest since it is a major component in vaccine preparations protecting against C. perfringens type B and C infections. The wild-type (WT) C. perfringens β-toxin is a potent toxin that requires chemical deactivation before it can be used as a safe vaccine component. Point mutations have been introduced that render this toxin no longer toxic but still immunogenic. However, this altered β-toxoid is very poorly secreted by B. subtilis (21).
In an attempt to identify the bottleneck in secretion of β-toxoid, we compared levels of global gene expression during the overproduction of β-toxin and β-toxoid. We tested whether altering the expression of the strongest upregulated gene could improve secretion yield. Unfortunately this did not yield the desired results.
Strikingly, the wild-type β-toxin protein can be efficiently secreted by B. subtilis. We therefore focused on the protein itself and analyzed the specific effects of the amino acid substitutions that differ between β-toxin and β-toxoid. Surprisingly, this revealed that only a single amino acid residue dictates the difference between high and very poor secretion yields.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Lactococcus lactis strains were grown at 30°C in M17 broth with 0.5% glucose (GM17) (29). B. subtilis strains were grown at 37°C under vigorous agitation in TY (1% tryptone, 0.5% yeast extract, 1.0% NaCl) or minimal medium (25). For the selection of transformants, appropriate antibiotics were added to the growth media at the following concentrations: chloramphenicol, 5 μg/ml; spectinomycin, 100 μg/ml; hygromycin, 125 μg/ml; and erythromycin, 5 μg/ml.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Genotype and description | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis 168 | trpC2 | 1 |
| B. subtilis NZ8900 | trpC2 amyE::spaRK | 4 |
| B. subtilis WB800 | nprE aprE epr bpr mpr::ble nprB::bsr Δvpr wprA::hyg | 34 |
| B. subtilis NZ8900-ΔykoJ | trpC2 amyE::spaRK ykoJ::spcR | This study |
| B. subtilis HT100A | NZ8900 containing Phtra-GFP fusion integrated in the chromosome | This study |
| B. subtilis PykoJ-GFP strain | NZ8900 containing PykoJ-GFP fusion integrated in the chromosome | This study |
| L. lactis MG1363 | Plasmid-free strain | 10 |
| L. lactis NZ9000 | penN::NisRK | 16 |
| Plasmids | ||
| pSG1151 | bla cat gfp | 20 |
| pPykoJ-GFP | pSG1151::PykoJ-gfp | This study |
| pPhtrA-GFP | pSG1151::PhtrA-gfp | This study |
| pDG1726 | Vector containing spectinomycin resistance cassette | 11 |
| pXB10 | pUB110 containing β-toxin-coding region | 25 |
| pNZ8048 | Nisin vector, Emr | 9 |
| pNZbtoxoid (pNZbtox) | pNZ8048::β-toxoid | 21 |
| pNZbtoxin | pNZ8048::β-toxin | This study |
| pNZ8901 | Cloning vector containing subtilin-inducible promoter, Cmr | 4 |
| pNZ8903 | Cloning vector containing WT subtilin-inducible promoter, Emr | 4 |
| pNRS-ykoJ | pNZ8901 containing ykoJ gene | This study |
| pNRS-βtoxoid (AAA) | pNZ8903 containing β-toxoid gene | This study |
| pNRS-βtoxin (DDK) | pNZ8903 containing β-toxin gene | This study |
| pNRS-βtox-DAA | pNZ8903 containing β-tox-DAA gene | This study |
| pNRS-βtox-ADA | pNZ8903 containing β-tox-ADA gene | This study |
| pNRS-βtox-AAK | pNZ8903 containing β-tox-AAK gene | This study |
| pNRS-βtox-DDA | pNZ8903 containing β-tox-DDA gene | This study |
| pNRS-βtox-DAK | pNZ8903 containing β-tox-DAK gene | This study |
| pNRS-βtox-ADK | pNZ8903 containing β-tox-ADK gene | This study |
| pRNΔykoJ_Sp | pUC18 containing up- and downstream regions of ykoJ flanking a spectinomycin resistance cassette | This study |
Strain constructions and transformation.
The cloning and transformation procedures were performed according to established techniques (6, 23) and suppliers' manuals. Restriction enzymes, DNA polymerases, deoxynucleotides, and T4 DNA ligase were obtained from Roche Diagnostics (Mannheim, Germany) and Fermentas Life Sciences (Vilnius, Lithuania) and used as specified by the suppliers. Table 2 lists the nucleotide sequences of primers used for PCR.
TABLE 2.
Primers used in this study
| Primer name | Descriptiona | Sequence (5′ to 3′) |
|---|---|---|
| Btoxoid-RN2-fw | Forward primer for β-toxoid cloning in pNZ8048 | CGTTGCCATGGAGAAAAAATTTATTTCATTAGTTATAG |
| Btoxoid-RN2-rv | Reverse primer for β-toxoid cloning in pNZ8048 | CGCTCTAGATTAAATAGCTGTTACTTTGTGAG |
| Btox-fw_BstEII | Forward primer for Btoxoid_SS, introduces BstEII and NdeI sites | TCGGTGACCCATATGAAGAAAAAATTTATTTCATTAG |
| Btoxoid-RN2-XhoI | Reverse primer for β-toxoid, introduces XhoI site | CGCCTCGAGTTAAATAGCTGTTACTTTGTGAG |
| RNlacZ-fw | Universal primer on pUC18-derived MCS | GTGAGCGGATAACAATTTCACACAGG |
| RNlacZ-rv | Universal primer on pUC18-derived MCS | GGTTTTCCCAGTCACGACGTTGTAA |
| PhtrA_fw-KpnI | Forward primer for promotor region htrA, introduces KpnI site | CGTGAGGTACCGGCTTCTGTTTCTGCC |
| PhtrA_rv | Reverse primer for promotor region htrA, introduces HindIII site | CATCACGAAGCTTATCCATCATGTTCACTCCG |
| PykoJ-fw1 | Forward primer for amplification of ykoJ promoter, introduces KpnI site | CTGGTACCGCAGTGAATCCATCTGCCATGAC |
| PykoJ-rv1 | Reverse primer for amplification of ykoJ promoter, introduces HindIII site | ACTAAGCTTGAGCATTTGTGAGCCCTCCTTTGT |
| ykoJ-fw1 | Forward primer to amplify ykoJ for overexpression, introduces BstEII site | TGTGGTGACCCAAATGCTCAAGAAAAAATGGATGGTCGGTCTTTTAG |
| ykoJ-rv1 | Reverse primer to amplify ykoJ for overexpression, introduces XhoI site | ACACTCGAGTTAGTCATCTATCTCCTGTTTGATAATG |
| up_ykoJ-fw | Forward primer to amplify upstream region of ykoJ, internal pstI site is amplified | CTTTTGCTGCAGCAGCCATTTTAG |
| up_ykoJ-rv | Reverse primer to amplify upstream region of ykoJ, introduces HindIII site | TCTTAAGCTTTTGTGAGCCCTCCTTTGTTT |
| down_ykoJ-fw | Forward primer to amplify downstream region of ykoJ, introduces HindIII site | TCTAAGCTTCAGGAGATAGATGACTAATCAA |
| down_ykoJ-rv | Reverse primer to amplify downstream region of ykoJ, introduces XbaI site | CGATCTAGAAGCATCCAGCTGCATTA |
| btox-change_FW | Universal forward primer for mutagenesis of β-toxin, introduces Eco31I site | CGGTCTCATATTCATCTGAAATGACAACTTTAATAAACTTAAC |
| RV-btox_DAA | Reverse primer, changes DDK → DAA | CGGTCTCAAATAAGCAGCATCGATAAATCTAGCATCTATAGATGCAGTAA |
| RV-btox_ADA | Reverse primer, changes DDK → ADA | CGGTCTCAAATAAGCATCAGCGATAAATCTAGCATCTATAGATGCAGTAA |
| RV-btox_AAK | Reverse primer, changes DDK → AAK | CGGTCTCAAATATTTAGCAGCGATAAATCTAGCATCTATAGATGCAGTAA |
| RV-btox_DDA | Reverse primer, changes DDK → DDA | CGGTCTCAAATAAGCATCATCGATAAATCTAGCATCTATAGATGCAGTAA |
| RV-btox_DAK | Reverse primer, changes DDK → DAK | CGGTCTCAAATATTTAGCATCGATAAATCTAGCATCTATAGATGCAGTAA |
| RV-btox_ADK | Reverse primer, changes DDK → ADK | CGGTCTCAAATATTTATCAGCGATAAATCTAGCATCTATAGATGCAGTAA |
MCS, multiple cloning site.
Inducible β-toxin and ykoJ plasmids.
To construct the B. subtilis subtilin-inducible plasmids pNRS-βtoxoid and pNRS-βtoxin, the β-toxoid gene was amplified by PCR from the pBtox-1 plasmid, and the β-toxin gene was amplified from plasmid pXB10, respectively, using primers Btox-fw_BstEII and Btoxoid-RV2-XhoI. These PCR products were digested with BstEII and AvaI and ligated into the likewise digested replicative vector pNZ8903 containing the unmodified subtilin-inducible spaS promoter (4). To construct pNRS-ykoJ, the ykoJ gene was amplified by PCR from B. subtilis 168 chromosomal DNA using primers ykoJ-fw1 and ykoJ-rv1, digested with BstEII and AvaI, and ligated into the likewise digested replicative vector pNZ8901 containing the subtilin-inducible spaS promoter (4).
To construct the L. lactis plasmids pNZ-βtoxoid and pNZ-βtoxin, the β-toxoid gene was amplified by PCR from the pBtox-1 plasmid and the β-toxin gene was amplified by PCR from plasmid pXB10 using primers Btoxoid-RN2-fw and Btoxoid-RN2-rv. This product was digested with NcoI and AvaI and ligated into the likewise digested replicative vector pNZ8048 containing the nisin-inducible promoter (9).
Ligation mixtures were transferred to electrocompetent L. lactis MG1363 culture or L. lactis NZ9000 culture using a Gene Pulser (Bio-Rad Laboratories, Hercules, CA), as described previously (18). Colonies were selected on solid medium for the erythromycin resistance. Isolated plasmids were checked for correct ligation by AvaI-AvaII digestion and DNA sequencing.
B. subtilis NZ8900 was transformed with the constructed replicative plasmids isolated from L. lactis and selected on solid medium for appropriate resistance.
ykoJ deletion construct.
Upstream and downstream regions of the ykoJ gene were amplified by PCR using primers up_ykoJ-fw1 and up_ykoJ-rv1 and down_ykoJ-fw1 and down_ykoJ-rv1, respectively, and the resulting PCR products were digested with PstI plus HindIII and HindIII plus XbaI, respectively. These products were ligated into a four-point ligation to both sides of a HindIII-digested spectinomycin resistance cassette, obtained by PCR using pDG1726 as a template and primers RNlacZ-fw and RNlacZ-rv, and a PstI-plus-XbaI-digested pUC18 plasmid. The resulting plasmid, pRNΔykoJ_Sp, was amplified with Escherichia coli and transformed to B. subtilis NZ8900 to create NZ8900-ΔykoJ. Colonies were checked for integration of the spectinomycin resistance cassette via double crossover at the locus of ykoJ by PCR.
Intermediate β-toxin mutants.
All β-toxin variants were constructed by PCR on the template plasmids pNRS-βtoxin (to create pNRS-βtox-A54DK, -A54DA, and -A54AK) and pNRS-βtoxoid (to create pNRS-βtox-D54AK, -D54DA, and -D54AA). The plasmids were amplified using a forward primer annealing next to the mutagenesis target site and containing an Eco31I recognition site and a specific reverse primer containing the desired point mutation and an Eco31I site (Table 2). After amplifying the whole plasmid, the PCR product was digested with Eco31I and circularized by self-ligation. The resulting plasmid was electroporated to L. lactis MG1363. Mutants were checked for the appearance/disappearance of the ClaI site contained within the first codon of the DDK region and subsequently checked by DNA sequencing (Baseclear, Leiden, The Netherlands). Correctly constructed plasmids were transformed to B. subtilis NZ8900 and selected for erythromycin resistance.
Protein expression, protein isolation, gel electrophoresis, and Western blotting.
B. subtilis cultures were diluted from an overnight culture to a starting optical density at 600 nm (OD600) of 0.1. β-Toxin or β-toxoid expression was induced when the culture reached an OD600 of ∼0.5 by the addition of 0.75% of subtilin containing supernatant of strain ATCC 6633 prepared as described previously (4). Two hours after induction, cells were separated from the supernatant by centrifugation for 1 min at 14,000 rpm. Supernatant proteins were concentrated 20-fold following trichloroacetic acid precipitation and prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (17). Cell fractions were prepared for SDS-PAGE as described previously (32). Proteins were separated by SDS-PAGE and either stained with Coomassie brilliant blue directly or transferred to a polyvinylidene difluoride membrane (Molecular Probes, Inc., Eugene, OR). β-Toxoid protein was visualized using a monoclonal anti-β-toxoid antibody (Intervet Int., Boxmeer, The Netherlands) and a secondary horseradish peroxidase-conjugated goat anti-mouse antibody (Amersham Biosciences, Buckinghamshire, United Kingdom). Protein sizes and concentrations were determined with a prestained protein marker (Fermentas, Vilnius, Lithuania), and Quantity One software (Bio-Rad, Hercules, CA).
DNA microarray experiments.
DNA microarray procedures were performed as described by Lulko et al. (20a). In short, RNA was isolated from three independently grown cultures of B. subtilis NZ8900 containing either pNRS-βtoxin or pNRS-βtoxoid. β-Toxin or β-toxoid expression was induced as described above, and samples for RNA isolation were taken 1.5 h after induction with subtilin. Single-strand reverse transcription (amplification) and indirect labeling of total isolated RNA with either Cy3 or Cy5 dye were performed, and labeled cDNA samples were hybridized overnight (O/N) at 48°C on in-house-printed microarray slides containing 70-meric oligonucleotides covering all B. subtilis open reading frames. After hybridization, slides were washed and scanned. Slide data were processed and normalized as described previously (8), yielding average ratios of gene expression levels of the strain expressing the β-toxoid compared to those of the strain expressing the WT β-toxin. Expression of a gene was considered to be significantly altered when its expression ratio was >1.75 or <0.57 and had a CyberT Bayesian P value of <0.001. All DNA microarray data, including the slide images and raw data, obtained in this study are available online (http://molgen.biol.rug.nl/publication/btox_data/).
PhtrA-GFP and PykoJ-GFP analysis.
The htrA and ykoJ promoter regions were amplified by PCR using primers PhtrA-fw-kpnI and PhtrA-rv and PykoJ-fw and PykoJ-rv, respectively. The PCR products were digested with HindIII and KpnI and ligated into the likewise digested plasmid pDG1151. The plasmids were transferred to E. coli, and correct clones were checked by PCR and DNA sequencing. The pPhtrA-GFP plasmid was integrated via single crossover in the chromosomal DNA of B. subtilis strain NZ8900 at the locus of the htrA promoter, creating B. subtilis strain HT100A, and the pPykoJ-GFP plasmid was likewise integrated at the locus of the ykoJ promoter, creating the B. subtilis YkoJ-GFP strain. Green fluorescent protein (GFP) production was measured using a Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Mijndrecht, The Netherlands). The average fluorescence of 20,000 gated cells was determined using WinMDI 2.8 (http://facs.scripps.edu/software.html) software.
Assay of β-toxin and β-toxoid stability.
An O/N culture of L. lactis NZ9000 containing either pNZβtox or pNZβtoxin was diluted to an OD600 of 0.1 and grown for 2.5 h until an OD600 of 0.5 was achieved. Nisaplin (stock 50 mg/ml) in a final dilution of 1 × 10−7 was added to induce the nisin-inducible promoter, and 2 h after induction, total supernatant was harvested by centrifugation and subsequent filtration over a 0.2-μm syringe filter (Schleicher and Schuell Microscience, Dassel, Germany).
To collect spent supernatants of B. subtilis strains 168 and WB800, the strains were grown in TY medium, and supernatant samples were taken 2 h into the stationary growth phase. Supernatant was separated by centrifugation and subsequently passed through a 0.2-μm filter. The β-toxin and β-toxoid samples harvested from L. lactis were mixed 1:1 with the spent B. subtilis supernatant and incubated for 10 min and 1 h, respectively, at 37°C. As a control, fresh TY medium was used. After incubation, total protein was concentrated 10-fold upon trichloroacetic acid precipitation as described before and analyzed using SDS-PAGE. The concentrations of β-toxin and β-toxoid were determined by Coomassie brilliant blue staining followed by densitometric scanning (Bio-Rad GS-800 scanner) and analysis with Quantity One software (Bio-Rad, Hercules, CA).
RESULTS
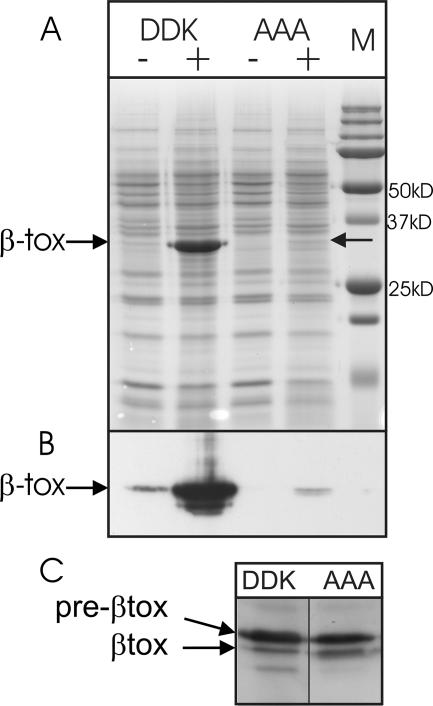
Recently, we have described the difficulties in secretion of Clostridium perfringens β-toxoid by Bacillus subtilis (21). Yet, it has been reported that wild-type β-toxin protein can be efficiently expressed in B. subtilis (26). To directly compare the secretion efficiencies of both proteins, we cloned them in the subtilin-inducible SURE vector pNZ8903. After induction for 1.5 h, cells were separated from the growth medium, and β-toxin and β-toxoid levels of production and secretion were assayed by SDS-PAGE and Western blotting using monoclonal anti-β-toxin antibody (Fig. 1). Clearly, the β-toxin protein was secreted in high amounts, up to 50% of total extracellular protein, whereas the β-toxoid variant never yielded more than 4%. The intracellular levels of β-toxin and β-toxoid were similar, and both induced cultures showed no significantly decreased growth rate compared to that of the uninduced cultures. Furthermore, no significant growth differences between the β-toxin and β-toxoid production strains were observed (data not shown).
FIG. 1.
Production and secretion of β-toxin and β-toxoid. (A) Coomassie brilliant blue-stained 12% SDS-polyacrylamide gel containing 10× concentrated supernatant of B. subtilis strain NZ8900 1.5 h after induction of the inducible β-toxin/β-toxoid plasmids. (B) Detection of secreted β-toxin and β-toxoid in the samples shown in panel A, examined by Western blotting using monoclonal antibodies against β-toxin. (C) Detection of intracellular β-toxin and β-toxoid examined by Western blotting using monoclonal antibodies against β-toxin. Pre-βtox and βtox are indicated by the arrows. DDK, WT β-toxin (D54DK); AAA, β-toxoid (A54AA); M, protein marker; +, induction with subtilin; −, no subtilin added.
Global gene expression differences between β-toxin and β-toxoid overproduction.
As shown in Fig. 1, it is apparent that there is a large difference in the secretion yields of β-toxin and detoxified β-toxoid. To identify possible bottlenecks in the secretion of β-toxoid, we compared the two production strains using DNA microarrays. Either β-toxin or β-toxoid expression was induced, and samples for RNA isolation were taken 1.5 h after induction. Following statistical analysis of the obtained data, several genes showed a clear difference in expression levels between the two strains. An overview of the results of the DNA microarray is presented in Table 3.
TABLE 3.
DNA microarray results
| Gene | Ratioc | Bayesian P value | Function (putative)d |
|---|---|---|---|
| Upregulated genesa | |||
| ykoJ | 6.87 | 2.25E−14 | Unknown |
| yvtA | 2.66 | 1.47E−10 | htrB, serine protease, secretion stress |
| htrA | 2.39 | 6.06E−10 | Serine protease, secretion stress |
| spollR | 2.33 | 0.000902 | Sporulation |
| ykzD | 2.28 | 5.95E−05 | Unknown, directly downstream of ykoJ |
| yvqH | 2.09 | 3.12E−06 | lial, part of liaRS regulon |
| cggR | 1.97 | 0.000437 | Transcriptional repressor of gapA |
| yvql | 1.95 | 2.15E−05 | liaH, part of liaRS regulon |
| ldh | 1.92 | 0.000111 | l-Lactate dehydrogenase |
| mtlD | 1.92 | 6.48E−06 | PTS, mannitol-specific enzyme |
| mtlA | 1.88 | 2.57E−06 | PTS, mannitol-specific enzyme |
| cssR | 1.76 | 0.000847 | Two-component regulator, secretion stress |
| Downregulated genesb | |||
| purN | −2.61 | 6.84E−11 | Purine biosynthesis |
| purM | −2.55 | 1.06E−05 | Purine biosynthesis |
| purH | −2.47 | 1.49E−08 | Purine biosynthesis |
| rocR | −2.12 | 1.89E−08 | Regulator, binding box upstream of ykoJ |
| yomM | −1.94 | 0.00081 | Unknown |
| yddE | −1.9 | 3.65E−05 | Unknown, operon on transposon region |
| yddB | −1.86 | 3.72E−05 | Unknown, operon on transposon region |
| yecA | −1.85 | 0.000455 | Transport/binding proteins and lipoproteins |
| ydcP | −1.8 | 0.000163 | Unknown, operon on transposon region |
| yosP | −1.79 | 0.000742 | Unknown, operon on transposon region |
| yddF | −1.77 | 0.0003 | Unknown, operon on transposon region |
Upregulated genes, higher expression when β-toxoid expression is compared to β-toxin expression.
Downregulated genes, lower expression when β-toxoid expression is compared to β-toxin expression.
Ratio, expression ratio comparing expression level for strain NZ8900 overproducing β-toxoid (target strain) to that for NZ8900 overexpressing β-toxin (control).
PTS, phosphotransferase system.
The most strongly upregulated gene in the β-toxoid-expressing strain was ykoJ, a gene of unknown function. Furthermore, htrA and htrB were upregulated, as was cssR, part of the two-component system known to regulate htrA and htrB expression (7). An upregulation of htrA upon expression of β-toxoid, as monitored by a PhtrA-lacZ fusion, was previously described (21). Clearly, this upregulation is lower for the wild-type β-toxin, since a difference in htrA expression is found in the array comparison. An overview of the other differentially expressed genes and their (putative) functions is given in Table 3.
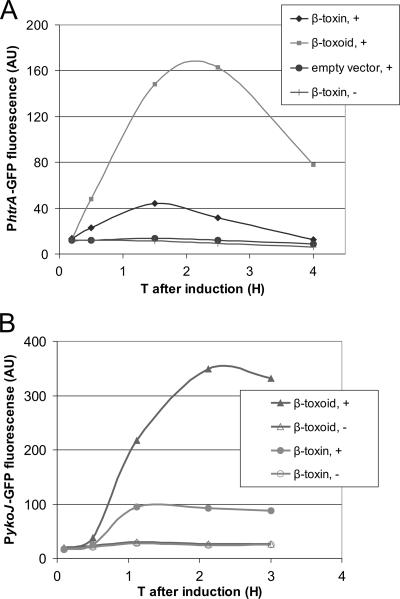
To validate the DNA microarrays, we looked specifically at the expression of ykoJ and htrA by using promoter GFP reporter fusions. We measured the average GFP expression per cell using flow cytometry. As shown in Fig. 2A, htrA expression in a strain that expressed β-toxoid was three times higher than that in a strain that expressed β-toxin. It should be noted that without induction, the PhtrA-GFP levels are much lower, indicating that expression and secretion of β-toxin also causes secretion stress. As shown in Fig. 2B, the results obtained with a PykoJ-GFP fusion confirmed the transcriptome result as well. Like htrA, PykoJ was moderately upregulated when β-toxin was induced and strongly upregulated when β-toxoid was induced.
FIG. 2.
(A)PhtrA-GFP upon overexpression of β-toxin and β-toxoid, shown by internal GFP fluorescence over time (T) of B. subtilis HT100A containing either β-toxin of β-toxoid on an inducible plasmid. After induction at T0 of β-toxin/β-toxoid, the response of the htrA promoter was measured by quantifying the average GFP fluorescence per cell using a flow cytometer. (B) PykoJ-GFP upon overexpression of β-toxin and β-toxoid, shown by internal GFP fluorescence over time of the B. subtilis PykoJ-GFP strain in its chromosome and either β-toxin or β-toxoid on an inducible plasmid. After induction at T0 of β-toxin/β-toxoid, the response of the ykoJ promoter was measured by quantifying the average GFP fluorescence per cell using a flow cytometer. Cultures not induced were also measured. +, induced; −, not induced.
Since the expression of ykoJ resembled the expression pattern of htrA, we tested whether the expression of ykoJ was controlled by the CssRS two-component system. For this test, a cssS disruption (13) was introduced into the PykoJ-GFP reporter strain. As shown in Fig. 3, no response of the ykoJ promoter occurred upon induction of β-toxin or β-toxoid when cssS was mutated. Together, the microarray analysis data show that the induction of the secretion stress regulon governed by CssRS is most apparent upon induction of β-toxoid.
FIG. 3.
Effect of cssS disruption on responses of PykoJ-GFP and PhtrA-GFP strains. Internal fluorescence of B. subtilis HT100A or the PykoJ-GFP strain was measured. Response was determined with or without a cssS disruption in the strain. In all strains, β-toxin (D54DK) or β-toxoid (A54AA) was induced by the addition of subtilin at T0.
YkoJ deletion does not improve β-toxoid secretion.
The transcriptome data suggest that the CssRS regulon could be a target when improving secretion of β-toxoid. In a previous study we already showed that the mutation of the CssRS two-component system does not positively effect production of β-toxoid (21). Since a mutation of CssRS will effectively prevent the induction of HtrA and HtrB (7), deletion of these two induced proteases separately is unlikely to improve secretion of β-toxoid. In studies performed by Vitikainen et al. (33), it was also observed that downregulation or mutation of HtrA and/or HtrB proteases does not improve secretion but instead induces severe stress in the cells, resulting in poor growth and generally lower secretion yields. However, Since ykoJ was strongly overexpressed in our study, we tested whether this protein itself influences the efficiency of secretion of β-toxoid. YkoJ contains two PepSY domains that suggest a peptidase-inhibiting action (36), but the specific function of YkoJ is still unknown. A deletion of ykoJ showed no noteworthy improvement of β-toxoid secretion. Also, the secretion level of β-toxin in this strain did not differ from that of the wild-type strain (data not shown). We constructed a YkoJ overproduction strain using a SURE expression system. Unfortunately, upon mild induction, the cultures stopped growing and started lysing, indicating that the overexpression of ykoJ is lethal to B. subtilis (data not shown). These results indicate that YkoJ alone is not directly involved in the large difference in secretion level between β-toxin and β-toxoid.
Amino acid differences between β-toxin and β-toxoid.
Since altering the expression of host genes did not improve the yield of secreted β-toxoid, we focused on the nature of the protein itself. The differences between β-toxin and β-toxoid are three consecutive mutations at the N-terminal side of the mature protein (D54A, D55A, and K56A). Based on a homology model of the mature β-toxin protein available at the MODBASE protein model database (22), we have looked at the positions of these residues in the folded protein. According to the model, β-toxin consists largely of β sheets. However, the residues 54, 55, and 56 (as counted from the first residue of the mature protein) are situated in a loop at the surface of the protein and consist of two negatively charged aspartic acids and a positively charged lysine. In β-toxoid, these residues are replaced with alanines. It is likely that this change in charge distribution affects the folding characteristics of the protein.
Since the amino acid substitutions in the β-toxin mutants might influence folding and stability of the protein, we tested its susceptibility to proteases. β-Toxoid and β-toxin produced by L. lactis were incubated with spent supernatant of a stationary-phase B. subtilis culture. This culture supernatant contains many proteases secreted by B. subtilis. As shown in Fig. 4, a clear difference between the stabilities of the two proteins is visible. Whereas more than 50% of the β-toxin is still present after 1 h of incubation, almost all β-toxoid (>90%) has been degraded. As a control, we tested supernatant from B. subtilis WB800. In this strain, the genes for eight proteases have been deleted (35). Incubation with supernatant from a WB800 culture gave significantly less degradation, and about 60% β-toxoid was still detectable after 1 h of incubation (Fig. 4). The results show that β-toxoid is more prone to degradation than β-toxin, indicating that the amino acid substitutions do make the protein conformation less stable.
FIG. 4.
Degradation of β-toxin and β-toxoid by spent B. subtilis supernatants. Lactococcus lactis-produced β-toxin and β-toxoid cultures were incubated with spent supernatants of stationary-phase cultures of B. subtilis strains 168 and WB800. As a control, TY medium was used. (A) Typical Coomassie brilliant blue-stained polyacrylamide gel showing results of the degradation assay. The incubation time (T) in minutes is indicated. Left lanes show β-toxoid and β-toxin exposed to strain 168 culture supernatant; right lanes show β-toxoid and β-toxin exposed to strain WB800 culture supernatant. M, protein marker, 35-kDa band. (B) The amount of β-toxin/β-toxoid measured after 10 min was set to 100%. The remaining amounts of β-toxoid and β-toxin after 1 h were determined and plotted. Experiments were performed in duplicate; error bars depict standard errors.
Intermediate mutants between β-toxin and β-toxoid.
To examine the importance of the individual amino acid mutations for protein secretion, we constructed all six possible intermediate mutants. Expression and secretion of the different β-toxin variants was tested by harvesting cells and supernatant fractions 1.5 h after induction and analyzing them by SDS-PAGE and Western blotting (Fig. 5A). From these experiments, two classes emerged: those with β-toxin production levels (D54DK [β-toxin], D54DA, A54DA, and A54DK) and those with hardly any protein secreted (A54AA [β-toxoid], A54AK, D54AK, and D54AA). This screening indicated that mutations at residues 54 and 56 do not have a significant effect on secretion efficiency, yet residue D55 is pivotal when it comes to efficient secretion of toxin.
FIG. 5.
Secretion and secretion stress of β-toxin, β-toxoid, and intermediate mutants. (A) Coomassie brilliant blue-stained 12% SDS-polyacrylamide gel containing 10× concentrated supernatant of B. subtilis strain NZ8900 1.5 h after induction of the inducible βtox plasmids. (B) The average PhtrA-GFP expression per cell in arbitrary units, measured 1.5 h after induction, is shown. Experiments were performed in duplicate; error bars depict standard errors. DDK, WT β-toxin (D54DK); AAA, β-toxoid (A54AA); all other intermediate mutants (mut) are likewise indicated.
The mean lethal doses of the β-toxin (D54DK) and the β-toxoids (D54DA, A54DA, and A54DK) were assessed by intravenous injection, in sterile phosphate-buffered saline, into mice weighing approximately 25 g. All three β-toxoids had a similar level of toxicity which is approximately one-fifth of that of the toxin. The original β-toxoid (A54AA) has a toxicity that is approximately 30-fold lower than that of the wild-type toxin (24). The poorly secreted β-toxoids (A54AK, D54AK, and D54AA) were not tested, since production levels were too low.
We also measured the effect of these six intermediate mutants on htrA expression. In accordance with the previous results, all mutants with an aspartic acid at position 55 showed relatively low, β-toxin-like htrA expression levels, whereas the mutants with an alanine at this position showed a strong upregulation of htrA, comparable to that for β-toxoid production (Fig. 5B). Clearly there is a strong relationship between poor β-toxoid secretion and the induction of the secretion stress regulon.
DISCUSSION
β-Toxoid, the genetically altered variant of Clostridium perfringens β-toxin, is not efficiently produced by Bacillus subtilis. To improve the secretion yield of the β-toxoid protein, we swapped signal sequences, used several expression systems, and tested protease-deficient hosts (21). None of the tested methods resulted in an appreciable increase in yield. However, wild-type β-toxin could be secreted much better than β-toxoid, in yields exceeding 50% of the total secreted protein fraction. To identify the bottleneck that was causing this difference in secretion yields, we applied a genome-wide expression analyses of the two production strains, hoping to find genes or processes responsible for this large production difference.
The DNA microarray analysis revealed that the differences can be largely attributed to the CssRS regulon, an indication of unfolded protein stress. The most upregulated gene in our array study, ykoJ, appeared to be part of the CssRS regulon as well. A deletion of the CssS sensor, effectively preventing induction of htrA, htrB (7), and ykoJ, did not improve secretion. We tried to overproduce YkoJ, but this proved to be lethal. Next to the CssRS regulon, two genes present in the liaRS regulon (15) were expressed significantly higher in the β-toxoid mutant. This effect was also found in another secretion stress study (2). Recently, it has been shown that LiaRS is activated by cell envelope stress (15). Only liaI and liaH, the genes that are generally much more highly expressed than the other genes in the regulon (15), were significantly upregulated in our study. The other genes that are part of this regulon were not found, indicating that the LiaRS induction differences are minor in our transcriptome comparison. We therefore did not characterize the effect of liaRS on β-toxoid production. Several of the purine biosyntheses genes were found to be downregulated, indicating a slight decrease in growth rate, which was missed in the growth rate determination but is picked up by the more sensitive microarray analysis.
Since altering the production host to increase secretion of β-toxoid was so far not successful, we looked more closely at β-toxoid itself, as the differences in yield between β-toxin and β-toxoid were striking. The stretch of three amino acid substitutions that morphs β-toxin into β-toxoid is not located in the secretion signal peptide where point mutations can have large effects on secretion efficiency (37). Furthermore, levels of intracellular retention of both the β-toxin and the β-toxoid are similar, indicating that no stalling problems occur when the protein gets secreted over the cytoplasm membrane via the Sec translocon.
Proteins secreted via the Sec secretion pathway are generally thought to be secreted in an unfolded state and are folded only after secretion over the plasma membrane (31). The current model of the β-toxin protein suggests that the point mutations introduced in β-toxoid might interfere with the correct folding or the rate of folding of β-toxoid after secretion. Upon induction of β-toxoid, a secretion stress response is observed, most likely induced by unfolded, secreted protein (5, 13). These results suggest that β-toxoid is reaching the outside of the membrane. The exact signal sensed by the CssS secretion stress sensor is as yet unknown, as it could also be the breakdown products of the malfolded and degraded protein that trigger the system.
The changed residues in β-toxoid are most likely affecting optimal folding kinetics, and therefore the β-toxoid protein is much more prone to degradation. Our experiments validated this assumption and showed that β-toxoid was much more prone to proteolysis than β-toxin, indicating that β-toxoid is in a folded conformation that is less stable than the WT β-toxin. The tested β-toxoid was produced and secreted by L. lactis, which could have influenced the folding of this protein. However, this is likely to be equally true for the β-toxin, which also was produced by L. lactis and which justifies this comparison. Incubation with the supernatant of B. subtilis strain WB800, which lacks the genes for seven extracellular proteases and the cell wall protease WprA, resulted in considerably less breakdown of β-toxoid. However, expression of β-toxoid by B. subtilis WB800 resulted in only a minimal improvement of β-toxoid secretion (21). This demonstrates that in the case of β-toxoid, most of the secreted protein is degraded before it is targeted by WprA or the other extracellular proteases deleted in WB800.
The constructed intermediate mutants of β-toxin demonstrate that only the aspartic acid at position 55 is necessary for the high secretion of the β-toxin. Although residues 54 and 56 also are charged and locate at the outside of the protein, they seem to be unimportant for secretion efficiency. They do play a role in the toxicity of the β-toxin, since the alterations of these residues does lower toxicity fivefold. The reason for this we do not know; possibly future structural studies might clarify this.
The responses of the htrA and ykoJ promoters to the overproduction of β-toxoid is indicative of extracellular folding stress. As proposed by Westers et al. (34), the expression of PhtrA or PhtrB can be utilized to monitor protein secretion. This study has added the ykoJ promoter to the possible indicators of secretion stress. A screening method using this promoter and site-directed/random mutagenesis of the secreted substrate should provide a rapid method to improve heterologous protein secretion.
With this study, we present a case where the bacterial host can be adapted in many ways without a significant yield improvement of secreted heterologous protein. The bottleneck turned out to be the secreted protein itself, where one point mutation made a crucial difference. In many cases, the intrinsic properties of the heterologous protein can be a main cause of the limited production yields, and increased attention to optimizing the protein itself rather than only the expression host is required.
Acknowledgments
We thank Paul Vermeij for supplying the pBtox1 plasmid and the anti-β-toxin monoclonal antibody and for helpful discussions. Keith Redhead is acknowledged for the determination of the mean lethal doses of the newly constructed β-toxoids. Hein Trip and Patricia van der Veek are gratefully acknowledged for construction of strain HT100A. We thank Ólafur S. Andrésson for the gift of strain XB10. Wiep Klaas Smits is acknowledged for critical reading of the manuscript. Tsjerk Wassenaar is gratefully acknowledged for assistance with understanding the β-toxin protein model.
This work was supported by Intervet International B.V. (Boxmeer, The Netherlands).
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis, A., H. Tjalsma, H. E. Smith, A. de Jong, R. Meima, G. Venema, S. Bron, and J. M. van Dijl. 1999. Evaluation of bottlenecks in the late stages of protein secretion in Bacillus subtilis. Appl. Environ. Microbiol. 65:2934-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongers, R. S., J.-W. Veening, W. Van Wieringen, O. P. Kuipers, and M. Kleerebezem. 2005. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71:8818-8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, P., G. Gerritse, J. M. van Dijl, and W. J. Quax. 1999. Improving protein secretion by engineering components of the bacterial translocation machinery. Curr. Opin. Biotechnol. 10:376-381. [DOI] [PubMed] [Google Scholar]
- 6.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. 1. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Hengst, C. D., P. Curley, R. Larsen, G. Buist, A. Nauta, D. van Sinderen, O. P. Kuipers, and J. Kok. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 12.Harwood, C. R. 1992. Bacillus subtilis and its relatives: molecular biological and industrial workhorses. Trends Biotechnol. 10:247-256. [DOI] [PubMed] [Google Scholar]
- 13.Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, C. L., K. Stephenson, S. T. Jorgensen, and C. Harwood. 2000. Cell-associated degradation affects the yield of secreted engineered and heterologous proteins in the Bacillus subtilis expression system. Microbiology 146:2583-2594. [DOI] [PubMed] [Google Scholar]
- 15.Jordan, S., A. Junker, J. D. Helmann, and T. Mascher. 2006. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuipers, O. P., P. G. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Leenhouts, K. J., J. Kok, and G. Venema. 1989. Campbell-like integration of heterologous plasmid DNA into the chromosome of Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:394-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leloup, L., E.-A. Haddaoui, R. Chambert, and M. F. Petit-Glatron. 1997. Characterization of the rate-limiting step of the secretion of Bacillus subtilis alpha-amylase overproduced during the exponential phase of growth. Microbiology 143:3295-3303. [DOI] [PubMed] [Google Scholar]
- 20.Lewis, P. J., and A. L. Marston. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101-110. [DOI] [PubMed] [Google Scholar]
- 20a.Lulko, A. T., G. Buist, J. Kok, and O. P. Kuipers. 2007. Transcriptome analysis of temporal regulation of carbon metabolism by CcpA in Bacillus subtilis reveals additional target genes. J. Mol. Microbiol. Biotechnol. 12: 82-95. [DOI] [PubMed] [Google Scholar]
- 21.Nijland, R. Lindner, C. Van Hartskamp, M., Hamoen, L. W., and O. P. Kuipers. 2007. Heterologous production and secretion of Clostridium perfringens β-toxoid in closely related Gram-positive hosts. J. Biotechnol. 127:361-372. [DOI] [PubMed] [Google Scholar]
- 22.Pieper, U., N. Eswar, H. Braberg, M. S. Madhusudhan, F. P. Davis, A. C. Stuart, N. Mirkovic, A. Rossi, M. A. Marti-Renom, A. Fiser, B. Webb, D. Greenblatt, C. C. Huang, T. E. Ferrin, and A. Sali. 2004. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 32:D217-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 24.Sergers, R. P. A. M., N. R. Waterfield, P. L. Frandsen, and J. M. Wells. Jan. 20, 1999. Clostridium perfringens vaccine. EUR patent EP0892054.
- 25.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinthorsdottir, V., V. Fridriksdottir, E. Gunnarsson, and O. S. Andresson. 1998. Site-directed mutagenesis of Clostridium perfringens beta-toxin: expression of wild-type and mutant toxins in Bacillus subtilis. FEMS Microbiol. Lett. 158:17-23. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson, K., N. M. Carter, C. R. Harwood, M. F. Petit-Glatron, and R. Chambert. 1998. The influence of protein folding on late stages of the secretion of alpha-amylases from Bacillus subtilis. FEBS Lett. 430:385-389. [DOI] [PubMed] [Google Scholar]
- 28.Stephenson, K., C. L. Jensen, S. T. Jorgensen, J. H. Lakey, and C. R. Harwood. 2000. The influence of secretory-protein charge on late stages of secretion from the Gram-positive bacterium Bacillus subtilis. Biochem. J. 350:31-39. [PMC free article] [PubMed] [Google Scholar]
- 29.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thwaite, J. E., L. W. J. Baillie, N. M. Carter, K. Stephenson, M. Rees, C. R. Harwood, and P. T. Emmerson. 2002. Optimization of the cell wall microenvironment allows increased production of recombinant Bacillus anthracis protective antigen from B. subtilis. Appl. Environ. Microbiol. 68:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tjalsma, H., H. Antelmann, J. D. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J. Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veening, J.-W., W. K. Smits, L. W. Hamoen, J. D. H. Jongbloed, and O. P. Kuipers. 2004. Visualization of differential gene expression by improved cyan fluorescent protein and yellow fluorescent protein production in Bacillus subtilis. Appl. Environ. Microbiol. 70:6809-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitikainen, M., H. L. Hyyrylainen, A. Kivimaki, V. P. Kontinen, and M. Sarvas. 2005. Secretion of heterologous proteins in Bacillus subtilis can be improved by engineering cell components affecting posttranslocational protein folding and degradation. J. Appl. Microbiol. 99:363-375. [DOI] [PubMed] [Google Scholar]
- 34.Westers, H., E. Darmon, G. Zanen, J. W. Veening, O. P. Kuipers, S. Bron, W. J. Quax, and J. M. van Dijl. 2004. The Bacillus secretion stress response is an indicator for alpha-amylase production levels. Lett. Appl. Microbiol. 39:65-73. [DOI] [PubMed] [Google Scholar]
- 35.Wu, S.-C., J. C. Yeung, Y. Duan, R. Ye, S. J. Szarka, H. R. Habibi, and S.-L. Wong. 2002. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl. Environ. Microbiol. 68:3261-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeats, C., N. D. Rawlings, and A. Bateman. 2004. The PepSY domain: a regulator of peptidase activity in the microbial environment? Trends Biochem. Sci. 29:169-172. [DOI] [PubMed] [Google Scholar]
- 37.Zanen, G., E. N. Houben, R. Meima, H. Tjalsma, J. D. Jongbloed, H. Westers, B. Oudega, J. Luirink, J. M. van Dijl, and W. J. Quax. 2005. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J. 272:4617-4630. [DOI] [PubMed] [Google Scholar]