Abstract
Bioaugmentation of soil polluted with polycyclic aromatic hydrocarbons (PAHs) is often disappointing because of the low survival rate and low activity of the introduced degrader bacteria. We therefore investigated the possibility of priming PAH degradation in soil by adding 2% of bioremediated soil with a high capacity for PAH degradation. The culturable PAH-degrading community of the bioremediated primer soil was dominated by Mycobacterium spp. A microcosm containing pristine soil artificially polluted with PAHs and primed with bioremediated soil showed a fast, 100- to 1,000-fold increase in numbers of culturable phenanthrene-, pyrene-, and fluoranthene degraders and a 160-fold increase in copy numbers of the mycobacterial PAH dioxygenase gene pdo1. A nonpolluted microcosm primed with bioremediated soil showed a high rate of survival of the introduced degrader community during the 112 days of incubation. A nonprimed control microcosm containing pristine soil artificially polluted with PAHs showed only small increases in the numbers of culturable PAH degraders and no pdo1 genes. Initial PAH degradation rates were highest in the primed microcosm, but later, the degradation rates were comparable in primed and nonprimed soil. Thus, the proliferation and persistence of the introduced, soil-adapted degraders had only a marginal effect on PAH degradation. Given the small effect of priming with bioremediated soil and the likely presence of PAH degraders in almost all PAH-contaminated soils, it seems questionable to prime PAH-contaminated soil with bioremediated soil as a means of large-scale soil bioremediation.
Various studies have investigated the possibility of bioaugmentation of polycyclic aromatic hydrocarbon (PAH)-polluted soil with PAH-degrading strains or consortia. In vitro experiments often show improved degradation of soil PAHs when PAH-degrading lab strains or consortia are added. However, scaling up of this type of experiment is often highly disappointing. For instance, a recent microcosm study of bacterial community dynamics and PAH degradation during bioremediation and bioaugmentation of creosote-contaminated soil did not detect any effect of the introduced degrader consortium either on community profiles or on PAH degradation (26). Likewise, a pilot-scale study using a PAH-degrading Mycobacterium strain and an enrichment culture did not result in increased numbers of pyrene degraders or increased PAH degradation compared to creosote-polluted soil simply treated with N, P, and K (15).
The poor results may be caused by the focus on the metabolic capacity of the introduced strains. However, bioaugmentation should be more than only the addition of a metabolic function; the introduced degrader cells should also show high persistence and be able to compete with the indigenous bacteria for space and resources (13).
The natural PAH-degrading communities of bioremediated soil with high PAH degradation potential may have these properties. These degrader cells have proven to be efficient in surviving and obtaining PAHs under natural conditions and may therefore constitute an appropriate inoculum for bioaugmentation.
The aim of our study was to use such degrader-enriched, bioremediated soil to prime “freshly polluted” pristine soil simply by adding bioremediated clay soil suspended in water and then to follow PAH degradation and the degrader communities in microcosms over time.
MATERIALS AND METHODS
Chemicals and reagents.
All chemicals were of analytical grade. [9-14C]Phenanthrene (>98% purity) and [3-14C]fluoranthene (>95% purity) were obtained from Sigma-Aldrich (Copenhagen, Denmark). [4,5,9,10-14C]pyrene (>95% purity) was obtained from Amersham Biosciences (Hillerød, Denmark). The cell proliferation reagent WST-1 {4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate} was obtained from Roche Molecular Biochemicals (Mannheim, Germany).
Soils.
Sandy, pristine soil was sampled from an A-horizon at Dronningmølle (Sealand, Denmark) in March 2004. The soil was sieved to 4 mm and stored for 2 days at 15°C. Bioremediated clay soil from a previous project (6) was used as a PAH degrader inoculum. This soil originated from the Ringe Asphalt and Tar Production Plant (Ringe, Denmark) and was excavated from a depth of 2 to 3 m, followed by homogenization and fertilization with NPK in piles of 170 kg for 4 years (6). One sample of the bioremediated soil was used for characterization of the degrader community (14C-PAH mineralization, most probable numbers [MPN], and isolation), and another was used in the microcosms.
Microcosms.
Phenanthrene, fluoranthene, and pyrene were selected as model compounds because they are found in high concentrations in soil polluted with pyrogenic PAHs. The contents of the microcosms are shown in Table 1. Subsamples of PAH-contaminated soil were prepared by adding phenanthrene, fluoranthene, and pyrene in acetone solution to pristine soil (200 mg kg−1). The acetone was evaporated at 15°C for 24 h. These subsamples were then mixed with pristine soil to give final PAH concentrations of 50 mg kg−1. Control soil contained acetone only. Four different microcosms were prepared in 4.0-liter stainless-steel containers containing 1.5 kg of soil. The water content was adjusted to 21% (wt/wt). The bioremediated clay soil (inoculum) was mixed with this water, whereby the suspendable fraction was added to the microcosms. A control microcosm used to account for abiotic processes was poisoned by the addition of sodium azide to a final porewater concentration of 5 g liter−1. The microcosms were sealed with polypropylene film (cling film) because this film is permeable to oxygen. Subsamples of 30 g each for chemical and microbiological analyses were taken at increasing time intervals after homogenization of the microcosms.
TABLE 1.
Composition of microcosms
| Microcosm | Amt of pristine soil (g) | Amt of PAH-polluted soil (g) | Amt of solvent control soil (g) | Amt of inoculum (bioremediated soil) (g) | Amt of NaN3 (g liter−1) |
|---|---|---|---|---|---|
| PAH polluted, primed | 1,170 | 300 | 0 | 30 | 0 |
| PAH polluted, not primed | 1,200 | 300 | 0 | 0 | 0 |
| Nonpolluted, primed | 1,170 | 0 | 300 | 30 | 0 |
| PAH polluted, poisoned | 1,200 | 300 | 0 | 0 | 5 |
Soil PAH content.
Soil samples (5 g) were thoroughly homogenized and were spiked with eight deuterated PAHs as internal standards. The frozen samples (−18°C) were freeze-dried. All soil samples were then microwave extracted for 25 min at 110°C in 35 ml acetone:dichloromethane:water (3:3:1) using a commercial microwave system designed for extraction of organic compounds. The extracts were dried with Na2SO4 and concentrated by evaporation of the solvent under a stream of nitrogen. The extracts were loaded onto SiO2 columns and eluted with dichloromethane. All extracts were spiked with 10-[D]-chrysene as a PAH recovery standard. Compounds were identified and quantified with a Finnigan TRACE DSQ single-quadrupole gas chromatographer/mass spectrometer in the selected ion mode.
PAH degrader MPN.
The MPN of phenanthrene, pyrene, or fluoranthene degraders in the soil was determined for the bioremediated soil used as an inoculum and for subsamples from the microcosms using a microplate method with a fourfold dilution series, four parallel rows, and 4 weeks of incubation as described previously (7). In this assay, growth is determined by respiratory reduction of the tetrazolium compound WST-1 in active cells. The initial microcosm MPNs were calculated from the MPNs of the soils used. The optimum incubation time for the MPN plates was determined for the bioremediated soil (inoculum) by initially incubating plates for 2, 3, 4, and 5 weeks before determination of growth-positive wells.
Quantification of PAH dioxygenase genes by real-time q-PCR.
Whole-community DNA was extracted from 0.5-g samples of soil by application of a bead-beating procedure using the FastDNA spin kit for soil (BIO 101, Vista, CA) as described by de Lipthay et al. (5). Conventional PCR detection of nah-like naphthalene dioxygenase genes (e.g., nahAc and pahA) was performed using the NAH primers described by Baldwin et al. (2). Primers described by Johnsen et al. (9) were used for the detection of pdo1 PAH dioxygenase genes (17). Quantitative real-time PCR (q-PCR) was performed using a Bio-Rad iCycler (Bio-Rad, Hercules, CA) as described previously (8). The numbers of bacterial cells containing pdo1 and nah-like genes in the soil samples were determined from standard curves of DNA derived from Pseudomonas putida OUS82 (pahAc) (2, 24) and Mycobacterium sp. strain 6PY1 (pdo1) (17). All standard curves were made from DNA extracts corresponding to 3 × 102 to 3 × 107 CFU per PCR. Following all q-PCRs, melting-curve analysis and conventional agarose gel electrophoresis were done to confirm the validity of the PCR products. To estimate the extraction efficiency of template DNA in soil, 10 μl of dilutions of Mycobacterium sp. strain 6PY1 cells were mixed with 0.5-g samples of the pristine soil or with 0.5 ml water, followed by DNA extraction and q-PCR detection of pdo1.
Potential 14C-PAH mineralization of inoculum soil.
Ottawa sand was sterilized for 1 h at 200°C. Sterile Ottawa sand (1.5 g) was placed in 10-ml sterile glass scintillation vials. One hundred microliters of 14C-labeled PAH in acetone solution (60 μg ml−1; ≈2,500 Bq ml−1) was added, and the acetone was evaporated under a stream of nitrogen. Three grams (dry weight) of fresh soil was added to the vials and mixed with the 14C-PAH-contaminated sand to give a final PAH addition of 2 μg g−1 soil. Two subsamples were spiked for each PAH. Filter paper was placed at the bottom of sterile 250-ml Blue Cap flasks, and 2 ml of NaH2PO4 buffer (0.1 M, pH ≈4.8) was added to keep the air water saturated. The vials and test tubes containing 2 ml NaOH (1 M) to trap 14CO2 were placed in the flasks, and the flasks were closed with Teflon-lined caps and incubated at 15°C in the dark. The NaOH was replaced at increasing time intervals. The trapped amounts of 14CO2 were quantified by mixing the NaOH with 10 ml HiSafe 3 scintillation cocktail (Perkin-Elmer, Boston, MA) and counted on a Wallac 1409 liquid scintillation counter.
Isolation of PAH degraders in the inoculum soil.
The numerically dominant phenanthrene-, pyrene-, and fluoranthene-degrading strains were isolated from the inoculum soil (Ringe). Phenanthrene, pyrene, or fluoranthene was dissolved in dimethyl sulfoxide (25 mg ml−1). Phosphate minimal medium without carbon was prepared as described previously (10) except that 20 g liter−1 Noble Agar (Difco, Detroit, MI) was added as a solidifying agent. PAH agar was prepared by placing the still-warm medium (1 liter, 50°C) on a magnetic stirrer (900 rpm). Four milliliters of dimethyl sulfoxide-PAH was slowly added using a pipette with the tip placed below the surface of the medium to produce turbid suspensions of PAH microcrystals. The PAH medium was poured into petri dishes. The phenanthrene, fluoranthene, and pyrene degraders were MPN enumerated. Scrapings from the bottom of MPN microplate wells at the highest growth-positive dilutions were streaked on the relevant PAH agar using 1-μl inoculation loops. The inoculated PAH-agar plates were incubated for 3 to 4 weeks at 20°C in a fume hood. Colonies, often very small, of the two most dominant morphologies were picked and pure streaked two to four times on PAH agar. Liquid cultures were prepared by adding 0.4 mg PAH in acetone solution (4 mg ml−1) to 20 ml sterile glass scintillation vials, followed by evaporation of the acetone in a sterile cabinet. Two milliliters of phosphate minimal medium was added, and the vials were inoculated with colonies from the PAH-agar plates, closed with sterile lids with aluminum foil inserts, and incubated on a rotary shaker for up to 5 weeks. When turbid, subsamples of the liquid cultures were stored in glycerol (30% [vol/vol], final concentration) at −80°C. The purity of the isolates was confirmed by streaking on 20%-strength tryptic soy broth agar.
PAH degradation capacity of isolates.
The isolates were tested for the ability to mineralize the PAH they were isolated on. Fifty microliters of acetone containing 0.2 mg 14C-labeled PAH (≈125 Bq) was added to 25-ml sterile glass scintillation vials. The acetone was evaporated, and 2 ml phosphate minimal medium was added to give final PAH spikes of 100 mg liter−1. 14CO2 was collected in 3-ml sterile glass tubes containing 1 ml NaOH (1 M) placed inside the scintillation vials. Growth on phenanthrene, pyrene, and fluoranthene as sole sources of carbon and energy was tested by a previously published microplate method, based on respiratory reduction of the tetrazolium compound WST-1 in growth-positive wells after 12 to 14 days of incubation (7).
Identification of isolates by partial 16S rRNA gene sequencing.
Approximately 530 bp of the 16S rRNA gene was amplified by PCR as described previously (28). The PCR products were sequenced by MWG-Biotech (Ebersberg, Germany). Only results for one isolate were reported when two isolates from the same microplate well gave similar 16S sequences. Eight randomly chosen isolates were tested for the presence of mycobacterium-like PAH dioxygenase by q-PCR using the pdo1 primers as described above.
RESULTS AND DISCUSSION
Optimization of PAH degrader MPN enumeration.
The optimum incubation time for PAH MPN counts was determined by preparing identical MPN plates, containing dilution series of the bioremediated Ringe soil, which were then incubated for 2, 3, 4, or 5 weeks before addition of the respiration indicator WST-1 for detecting growth-positive wells. The MPN estimates increased during the first 4 weeks, and 4 weeks was therefore used as the standard incubation time in the rest of the study.
Characterization of PAH degraders of the inoculum soil.
Bioremediated clay soil from a previous project (6) was used as a PAH degrader inoculum. This soil had been subjected to homogenization and fertilization with NPK for 4 years. During the first year, the content of 18 selected polycyclic aromatic compounds had dropped from 1,250 mg kg−1 to 380 mg kg−1 (6), followed by much slower degradation during the following years.
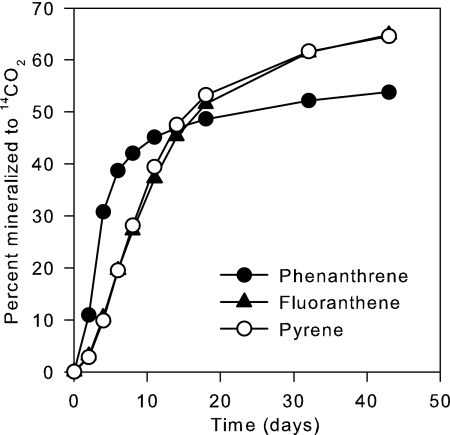
The PAH mineralization potentials and the numbers of PAH degraders in the bioremediated Ringe soil were determined before the microcosms were set up. “Fresh” [14C]phenanthrene, [14C]pyrene, and [14C]fluoranthene were readily mineralized (Fig. 1). Duplicate MPNs of PAH degraders showed 1.5 × 106 and 5.8 × 106 cells g−1 for phenanthrene, 1.5 × 106 and 2.6 × 106 cells g−1 for pyrene, and 1.8 × 106 and 2.9 × 106 cells g−1 for fluoranthene. Similar MPNs for the three PAHs suggested that many of the degrader cells could grow on more than one of the PAHs.
FIG. 1.
Mineralization of 14C-labeled PAHs (2 mg kg−1) by bioremediated subsoil used for priming of microcosms. PAH mineralization was estimated in duplicate as the recovery of 14CO2.
The numerically dominant PAH degraders in the Ringe soil were isolated from the highest growth-positive dilutions of the MPN plates. The isolates were identified by partial 16S rRNA gene sequencing, and their ability to grow on phenanthrene, pyrene, and fluoranthene was determined (Table 2). Surprisingly, all but one isolate were mycobacteria affiliated with the species Mycobacterium frederiksbergense, Mycobacterium austroafricanum, Mycobacterium gilvum, Mycobacterium aurum, Mycobacterium pyrenivorans, and Mycobacterium vaccae. A previous study showed, on the basis of 13C incorporation of phenanthrene-derived carbon into polar lipid fatty acids, that the phenanthrene degraders of the Ringe soil at that time were dominated by sphingomonads and an unclassified beta-proteobacterium (14). However, that study was carried out right after the initial, fast PAH degradation. Together, this supports the hypothesis of Leys et al. (18) that initial degradation of PAHs in soil is done by sphingomonads and other fast-growing bacteria (r-strategists), whereas later, the sphingomonads are outcompeted by relatively slow-growing mycobacteria (K-strategists). The dominance of mycobacteria in aged, PAH-polluted soil is also in accordance with the results reported by Uyttebroek et al. (25).
TABLE 2.
Characterization of numerically dominant, culturable PAH degraders in the bioremediated soil used as an inoculum
| PAH used for isolation | Isolate | GenBank accession no. | Closest relative based on partial 16S rRNA gene sequence (% homology) | Growth on PAHs, determined as reduction of WST-1 (A450-630)a
|
|||
|---|---|---|---|---|---|---|---|
| Hex | Phe | Fla | Pyr | ||||
| Phe | Ri456a | EF012722 | Arthrobacter oxydans (99.8) | 0.01 | 0.86 | 0.01 | 0.01 |
| Phe | Ri463b | EF012740 | M. frederiksbergense (99.8) | 0.27 | 1.69 | 0.22 | 1.62 |
| Phe | Ri452b | EF012741 | M. austroafricanum (97.6) | 0.07 | 1.66 | 0.06 | 0.52 |
| Phe | Ri465a | EF012742 | M. austroafricanum (100) | 0.05 | 0.79 | 0.10 | 0.35 |
| Phe | Ri455 | EF012743 | M. gilvum (99.7) | 0.07 | 2.04 | 0.01 | 0.48 |
| Phe | Ri457 | EF012744 | M. gilvum (100) | 0.03 | 0.73 | 0.01 | 0.31 |
| Phe | Ri469 | EF185792 | M. aurum (99.8) | 0.24 | 1.22 | 0.24 | 0.36 |
| Phe | Ri464 | EF012746 | M. aurum (100) | 0.17 | 1.36 | 0.43 | 0.63 |
| Phe | Ri458a | EF012747 | M. vaccae (100) | 0.02 | 0.57 | 0.11 | 0.18 |
| Phe | Ri460 | EF012723 | M. vaccae (100) | 0.04 | 0.61 | 0.50 | 0.39 |
| Phe | Ri466 | EF012725 | M. vaccae (99.5) | 0.07 | 1.36 | 0.84 | 1.18 |
| Pyr | Ri483a | EF012726 | M. aurum (100) | 0.09 | 0.56 | 0.09 | 0.26 |
| Pyr | Ri477 | EF012727 | M. frederiksbergense (99.5) | 0.31 | 2.07 | 0.20 | 1.67 |
| Pyr | Ri470a | EF012728 | M. gilvum (99.8) | 0.03 | 1.14 | 0.02 | 0.26 |
| Pyr | Ri484b | EF012729 | M. gilvum (99.8) | 0.12 | 0.73 | 0.10 | 0.54 |
| Pyr | Ri481b | EF012730 | M. gilvum (100) | 0.03 | 1.09 | 0.02 | 0.26 |
| Pyr | Ri489 | EF012731 | M. gilvum (100) | 0.10 | 0.77 | 0.12 | 0.46 |
| Pyr | Ri471c | EF012724 | M. vaccae (99.8) | 0.02 | 0.44 | 0.18 | 0.26 |
| Pyr | Ri486ba | EF012732 | M. vaccae (99.8) | 0.03 | 0.59 | 0.19 | 0.29 |
| Pyr | Ri487 | EF012733 | M. vaccae (99.4) | 0.01 | 0.24 | 0.07 | 0.20 |
| Fla | Ri505b | EF012734 | M. pyrenivorans (99.8) | 0.01 | 0.01 | 0.05 | 0.01 |
| Fla | Ri506 | EF012735 | M. pyrenivorans (99.8) | 0.05 | 0.05 | 0.34 | 0.05 |
| Fla | Ri488a | EF012736 | M. aurum (100) | 0.14 | 0.07 | 0.37 | 0.14 |
| Fla | Ri494 | EF012737 | M. aurum (100) | 0.02 | 0.00 | 0.49 | 0.03 |
| Fla | Ri495 | EF012738 | M. aurum (100) | 0.07 | 0.02 | 0.18 | 0.03 |
| Fla | Ri496 | EF012739 | M. vaccae (100) | 0.08 | 0.74 | 0.26 | 0.23 |
Numbers in bold are significantly higher than those for the hexane control (one-sided t test, P < 0.05; n = 3). Phe, phenanthrene; Pyr, pyrene; Fla, fluoranthene; Hex, hexane control.
The Arthrobacter isolate grew only on phenanthrene, whereas the 20 mycobacteria that grew on phenanthrene could also grow on pyrene. Also, all pyrene-degrading isolates could grow on phenanthrene. If these mycobacteria degrade phenanthrene through the o-phthalate pathway described for mycobacteria (17, 20), then pyrene may be shuttled into the phenanthrene pathway through formation of 3,4-dihydroxyphenanthrene from pyrene in six metabolic steps (17). It therefore makes sense that these strains could grow on both phenanthrene and pyrene.
The degradation of fluoranthene to phthalate by mycobacteria, on the other hand, proceeds through other pathways, since fluoranthene contains a C5 ring. Mycobacterium sp. strain AP1 and strain PYR-1 both metabolize fluoranthene to phthalate through formation of either 9-fluorenone-1-carboxylic acid or acenaphthenone (16, 19). This may explain why 5 fluoranthene degraders grew neither on phenanthrene nor on pyrene and why 12 out of 20 mycobacterial phenanthrene/pyrene isolates did not grow on fluoranthene. The nine isolates that grew on all three PAHs presumably have both pyrene and fluoranthene pathways, like Mycobacterium sp. strain AP1 (19).
PAH degradation in microcosms.
Recently, it was demonstrated that the clay fraction of a long-term PAH-polluted soil had a high potential for PAH degradation, as opposed to the sand and silt fractions (25). We therefore used a PAH-polluted, bioremediated clay soil as an inoculum in the primed microcosms.
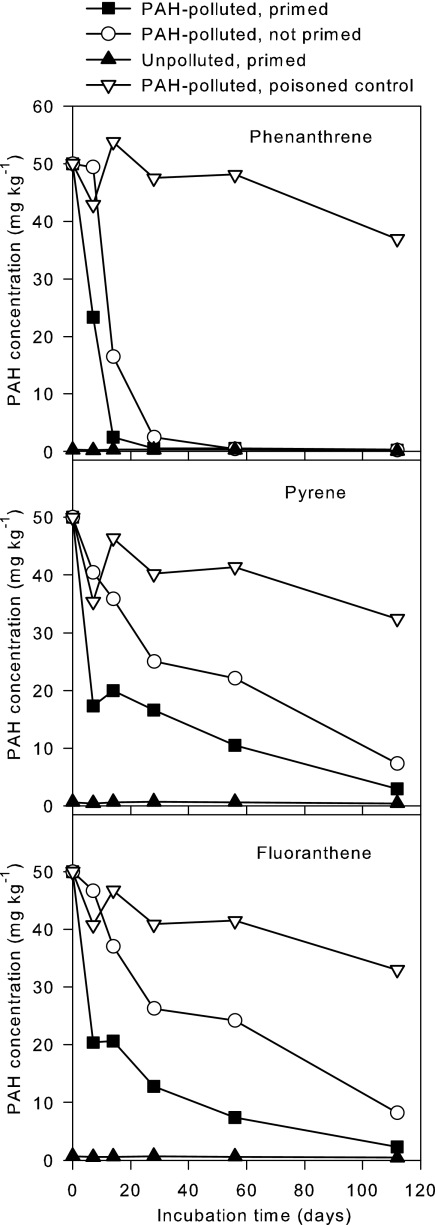
The PAH content of the pristine soil was below the limit of detection. The bioremediated soil contained, at the time of the experiment, 9.0 mg kg−1 of phenanthrene, 22.1 mg kg−1 of pyrene, and 22.4 mg kg−1 of fluoranthene and thus constituted less than 1 percent of the phenanthrene, fluoranthene, and pyrene in the polluted microcosms. The initial PAH concentrations were not measured but were calculated from the amounts added. Changes in PAH concentrations are depicted in Fig. 2. Phenanthrene disappeared rapidly within the first 2 to 4 weeks, whereas pyrene and fluoranthene were gradually degraded so that most had disappeared at week 16. The initial degradation rates were higher in the primed microcosm than in the nonprimed microcosm, but at later stages, the degradation rates were comparable.
FIG. 2.
Degradation of phenanthrene, pyrene, and fluoranthene in soil microcosms. PAH-polluted, polluted with 50 mg kg−1 of phenanthrene, pyrene and fluoranthene; primed, primed with 2% bioremediated soil.
The poisoned control microcosm failed to remain inactive (high numbers of CFU). This probably explains the reduction in PAH concentrations for the poisoned control at the end of the incubation; however, it cannot be excluded that some PAH disappearance might have been due to abiotic processes.
PAH-degrading populations.
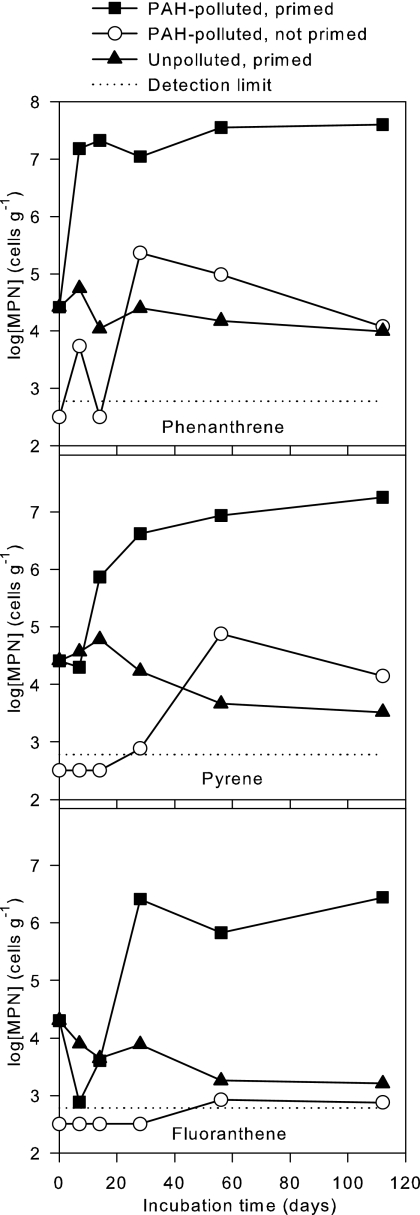
The PAH degraders of the bioremediated inoculum soil showed high survival rates in the control microcosm without PAHs, and 34% of phenanthrene degraders, 13% of pyrene degraders, and 8% of fluoranthene degraders were still culturable after 16 weeks of incubation (Fig. 3). The high survival rate demonstrates the greater fitness of soil-adapted degrader cells compared to the generally fast decline of laboratory strains introduced into soil microcosms. For comparison, a classic survival study showed that eight bacterial strains introduced into unsterile loam soil declined 1 to 4 orders of magnitude within 1 week due to starvation and protozoan predation (1).
FIG. 3.
Development in populations of PAH degraders. MPN, most probable number of phenanthrene degraders.
The degrader populations of the polluted, primed microcosm responded rapidly by increases of 3 orders of magnitude for the phenanthrene and pyrene degrader MPNs and 2 orders of magnitude for the fluoranthene degrader MPNs (Fig. 3). For comparison, numbers of CFU of a PAH-degrading Sphingomonas lab strain introduced into highly PAH-polluted soil decreased 2 to 3 orders of magnitude within 20 days, depending on pretreatment of the inoculum (4).
The MPNs of phenanthrene degraders in the primed, polluted microcosm was close to the theoretical growth yield, assuming that the degraded phenanthrene (49.7 mg kg−1) was converted into biomass. The yield of Sphingomonas sp. strain LH128 on phenanthrene was 1.3 × 109 ± 0.1 × 109 cells mg−1 (9), corresponding to a cell density of 6.5 × 107 ± 0.5 × 107 cells g−1 in the microcosm. The maximum MPN of phenanthrene degraders was 4.0 × 107 cells g−1. This comparison does not take into account that the MPN technique detects only those cells that can grow in mineral medium with PAHs as the sole carbon source, that degraders in the microcosm may have been eaten by protozoa, or that they may have utilized more than one carbon source. Also, the yields of the microcosm strains may be different from those of Sphingomonas. However, the yield of mycobacteria could not be reliably estimated because the hydrophobic cells flocculated when grown on PAHs in mineral medium.
Interestingly, the phenanthrene MPN continued to increase after the complete disappearance of phenanthrene in the microcosm (Fig. 3, day 28 to day 112), suggesting that pyrene degraders and possibly some fluoranthene degraders could also grow on phenanthrene. Alternatively, the delayed growth may be explained by the utilization of temporarily excreted metabolites.
The general fast decline of bioaugmentation inocula, compared to the high survival rate for the priming community, may be caused by the genetic changes induced by the liquid techniques used for isolation and production of the inocula. Even a single overnight culture induces profound genetic changes (23). Repeated growth in liquid batch cultures is highly selective for the planktonic mode of living, and as a result, the bioaugmentation strains may lose the ability to excrete extracellular polymeric substances and to attach and form biofilm (3). However, attached-biofilm growth protects the cells against environmental stress and predation by protozoa (22). Biofilm formation directly on the PAH sources is also an important means for degrader bacteria to reduce the constraints of low PAH bioavailability (11, 27). In liquid cultures, the cells also become adapted to growth on organic and mineral nutrients in high concentrations, a situation rarely encountered in soil.
PAH degraders in the unpolluted soil were undetectable by MPN enumeration. MPN counts were initially below the limit of detection (<600 cells g−1) in the nonprimed, polluted microcosm (Fig. 3). The number of fluoranthene degraders remained rather low, whereas phenanthrene degraders peaked at 2.3 × 105 cells g−1 (day 28) and pyrene degraders peaked at 7.6 × 104 cells g−1 (day 56). These increases explain the PAH degradation that was achieved in this microcosm. The low initial densities, compared to those for primed soils, explain the slower onset of degradation, but once densities of 105 cells g−1 were reached, degradation occurred at rates similar to those in the primed soils. The maximum numbers of degraders were lower than in the primed microcosm, which may be explained in three ways. First, the PAH degraders may be less culturable in the nonprimed microcosm. Second, some PAH degradation in the nonprimed microcosm may be cometabolic and therefore not detected in the MPN counts. Third, protozoa may have continuously grazed the degraders. The last explanation seems the most likely, as suggested by the declining populations at the end of the experiment.
PAH dioxygenase genes.
The nah-like genes found in Pseudomonas spp. and related genera encode dioxygenases of the N.2.A subfamily that degrade low-molecular-weight PAHs (2). nah genes were initially present in the two primed microcosms at densities of 1.8 × 104 cells g−1 and 2.0 × 104 cells g−1 (day 0). However, they were detected by q-PCR neither in later samples (day 7 to day 56) nor in the nonprimed microcosm, suggesting that pseudomonads were of minor importance in all microcosms.
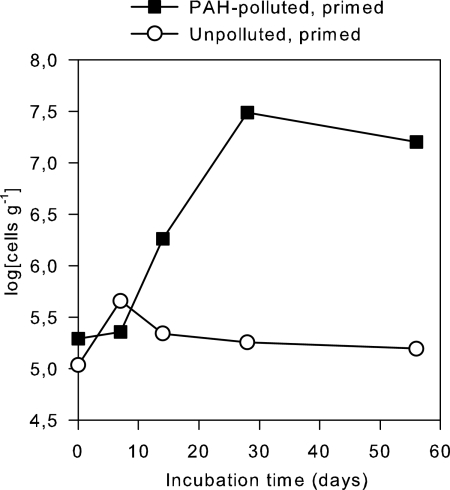
The eight Mycobacterium isolates tested (Ri452b, Ri465a, Ri463b, Ri477, Ri481b, Ri483a, Ri486ba, and Ri487) all gave pdo1 PCR products of the same size as that of the reference strain, Mycobacterium sp. 6PY1, on agarose gel. Also, the melting temperatures (q-PCR) of the Ri- isolates, as well as of 6PY1, were in the range of 89.5 to 90.0°C. Changes in copy numbers of pdo1 in the primed microcosms (Fig. 4) were similar to the development in pyrene degrader MPNs. The population containing pdo1 increased 160-fold in the PAH-polluted, primed microcosm, whereas the population remained almost constant in the nonpolluted, primed microcosm (Fig. 4). Melting points (89.5 to 90.0°C) and sizes of the PCR products were similar to those for the reference strain, Mycobacterium sp. 6PY1, and also were similar to those for the strains isolated from the soil used for priming. It was not possible to determine the efficiency of template DNA extraction, since different attempts gave different results. Numbers shown in Fig. 4 should therefore be regarded as relative rather than absolute.
FIG. 4.
Development in populations of mycobacterial PAH degraders, estimated from quantification of the PAH dioxygenase gene pdo1.
Changes in copy numbers of pdo1 in the primed microcosms (Fig. 4) were similar to the development in pyrene degrader MPNs. This increase in pdo1 genes, together with the presence of pdo1 in the numerically dominant PAH degraders, indicates that mycobacteria became dominant degraders of heavy-molecular-weight PAHs in the primed microcosm. In the literature, we have not found any indication of lateral transfer of 4-ring PAH dioxygenases, which would have blurred the pdo1-Mycobacterium relationship. Also, there are no reports of pdo1 genes in bacteria that do not belong to the Mycobacterium-Nocardioides-Rhodococcus clade.
Interestingly, the number of pdo1 genes (Fig. 4) did not mirror the initial sharp rise in the phenanthrene degrader MPN in the primed, polluted microcosm (Fig. 3). This suggests that phenanthrene, which is the most bioavailable of the three PAHs, was initially metabolized by fast-growing, nonmycobacterium strains. It may also explain why the calculated growth yields based on results from Sphingomonas were surprisingly close to the observed number of phenanthrene degraders.
DNA from the nonprimed, polluted microcosm did not at any time yield PCR products with the pdo1 primers, suggesting that PAHs were not degraded by mycobacteria in that microcosm.
Perspectives.
The degrader community of the bioremediated soil used for priming had the expected qualities: fast multiplication in PAH-polluted soil, high survival and persistence, and the ability to utilize several PAHs as sources of carbon. However, though the culturable PAH-degrading community of the pristine soil was initially below the limit of detection, the bacterial community of that soil also responded rapidly to the addition of PAHs.
The PAH degradation potential of PAH-polluted soil is generally closely linked to the level of PAH pollution (12). In a bioremediation situation, the PAH-polluted soil would therefore in most cases already have a high capacity for PAH degradation, and the effect of inoculation would be less than that seen in the present study. These indigenous PAH degraders respond very fast when growth conditions are improved (6, 15). Also, the PAH bioavailability in aged PAH contaminations is often low, and PAH degradation would in many cases be controlled by intraparticle diffusion and not by the degrader community, because PAHs must diffuse to the surface of the soil particles to be bioavailable (21). This would further reduce the effect of inoculation.
On an industrial scale, it is important to consider the extra cost from the priming process compared to the cost of simply incubating the soil for a longer time at the treatment plants. Given the small effect of priming with a soil-adapted degrader community in the present study and the likely presence of PAH degraders in almost all “natural” PAH contaminations (e.g., gasworks soil and diffusely polluted city soil [12]), it seems questionable to inoculate PAH-polluted soil with PAH-degrading lab strains or bioremediated soil, since these bioaugmentation strategies may have only a marginal effect on PAH degradation.
Acknowledgments
This study was supported financially by the Danish Ministry of Science, Technology and Innovation through embedding of the Strategic Environmental Research Center BIOPRO.
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Acea, M. J., C. R. Moore, and M. Alexander. 1988. Survival and growth of bacteria introduced into soil. Soil Biol. Biochem. 20:509-515. [Google Scholar]
- 2.Baldwin, B. R., C. H. Nakatsu, and L. Nies. 2003. Detection and enumeration of aromatic oxygenase genes by multiplex and real-time PCR. Appl. Environ. Microbiol. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton, J. W., and H. M. Lappin-Scott. 1995. Introduction to microbial biofilms, p. 1-11. In J. W. Costerton and H. M. Lappin-Scott (ed.), Microbial biofilms. The Press Syndicate of the University of Cambridge, Cambridge, United Kingdom.
- 4.Cunliffe, M., A. Kawasaki, E. Fellows, and M. A. Kertesz. 2006. Effect of inoculum pretreatment on survival, activity and catabolic gene expression of Sphingobium yanoikuyae B1 in an aged polycyclic aromatic hydrocarbon-contaminated soil. FEMS Microbiol. Ecol. 58:364-372. [DOI] [PubMed] [Google Scholar]
- 5.de Lipthay, J. R., K. Johnsen, H.-J. Albrechtsen, P. Rosenberg, and J. Aamand. 2004. Bacterial diversity and community structure of a sub-surface aquifer exposed to realistic low herbicide concentrations. FEMS Microbiol. Ecol. 49:59-69. [DOI] [PubMed] [Google Scholar]
- 6.Hestbjerg, H., P. A. Willumsen, M. Christensen, O. Andersen, and C. S. Jacobsen. 2003. Bioaugmentation of tar-contaminated soils under field conditions using Pleurotus ostreatus refuse from commercial mushroom production. Environ. Toxicol. Chem. 22:692-698. [PubMed] [Google Scholar]
- 7.Johnsen, A. R., K. Bendixen, and U. Karlson. 2002. Detection of microbial growth on polycyclic aromatic hydrocarbons in microtiter plates using the respiration indicator WST-1. Appl. Environ. Microbiol. 68:2683-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnsen, A. R., J. R. de Lipthay, F. Reichenberg, S. J. Sørensen, O. Andersen, P. Christensen, M. L. Binderup, and C. S. Jacobsen. 2006. Biodegradation, bioaccessibility and genotoxicity of diffuse polycyclic aromatic hydrocarbon (PAH) pollution at a motorway site. Environ. Sci. Technol. 40:3293-3298. [DOI] [PubMed] [Google Scholar]
- 9.Johnsen, A. R., J. R. de Lipthay, S. J. Sørensen, F. Ekelund, P. Christensen, O. Andersen, U. Karlson, and C. S. Jacobsen. 2006. Microbial degradation of street dust PAHs in microcosms simulating diffuse pollution of urban soil. Environ. Microbiol. 8:535-545. [DOI] [PubMed] [Google Scholar]
- 10.Johnsen, A. R., M. Hausner, A. Schnell, and S. Wuertz. 2000. Evaluation of fluorescently labelled lectins for noninvasive localization of extracellular polymeric substances in Sphingomonas biofilms. Appl. Environ. Microbiol. 66:3487-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnsen, A. R., and U. Karlson. 2004. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons (PAHs). Appl. Microbiol. Biotechnol. 63:452-459. [DOI] [PubMed] [Google Scholar]
- 12.Johnsen, A. R., and U. Karlson. 2005. PAH degradation capacity of soil microbial communities—does it depend on exposure? Microb. Ecol. 50:488-494. [DOI] [PubMed] [Google Scholar]
- 13.Johnsen, A. R., L. Y. Wick, and H. Harms. 2005. Principles of microbial PAH-degradation in soil. Environ. Pollut. 133:71-84. [DOI] [PubMed] [Google Scholar]
- 14.Johnsen, A. R., A. Winding, U. Karlson, and P. Roslev. 2002. Linking of microorganisms to phenanthrene metabolism in soil by analysis of 13C-labeled cell lipids. Appl. Environ. Microbiol. 68:6106-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juhasz, A. L., N. Waller, C. Lease, R. Bentham, and R. Stewart. 2005. Pilot scale bioremediation of creosote contaminated soil—efficacy of enhanced natural attenuation and bioaugmentation strategies. Bioremed. J. 9:141-157. [Google Scholar]
- 16.Kelley, I., J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1993. Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 59:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leys, N. M., A. Ryngaert, L. Bastiaens, P. Wattiau, E. M. Top, W. Verstraete, and D. Springael. 2005. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51:375-388. [DOI] [PubMed] [Google Scholar]
- 19.López, Z., J. Vila, C. Minguillón, and M. Grifoll. 2006. Metabolism of fluoranthene by Mycobacterium sp. strain AP1. Appl. Microbiol. Biotechnol. 70:747-756. [DOI] [PubMed] [Google Scholar]
- 20.Moody, J. D., J. P. Freman, D. R. Doerge, and C. E. Cerniglia. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder, H., A. M. Breure, and W. H. Rulkens. 2001. Prediction of complete bioremediation periods for PAH soil pollutants in different physical states by mechanistic models. Chemosphere 43:1085-1094. [DOI] [PubMed] [Google Scholar]
- 22.Queck, S.-H., M. Weitere, A. M. Moreno, S. A. Rice, and S. Kjelleberg. 2006. The role of quorum sensing mediated developmental traits in the resistance of Serratia marcescens biofilms against protozoan grazing. Environ. Microbiol. 8:1017-1025. [DOI] [PubMed] [Google Scholar]
- 23.Rainey, P. 2004. Bacterial populations adapt genetically by natural selection even in the lab! Microbiol. Today 31:160-162. [Google Scholar]
- 24.Takizawa, N., N. Kaida, S. Torigoe, T. Moritani, T. Sawada, S. Satoh, and H. Kiyohara. 1994. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J. Bacteriol. 176:2444-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uyttebroek, M., P. Breugelmans, M. Janssen, P. Wattiau, B. Joffe, U. Karlson, J. J. Ortega-Calvo, L. Bastiaens, A. Ryngaert, M. Hausner, and D. Springael. 2006. Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons (PAHs) among different size fractions of a long-term PAH-contaminated soil. Environ. Microbiol. 8:836-847. [DOI] [PubMed] [Google Scholar]
- 26.Viñas, M., J. Sabaté, M. J. Espuny, and A. M. Solanas. 2005. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 71:7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wick, L. Y., A. Ruis-de-Munain, D. Springael, and H. Harms. 2002. Responses of Mycobacterium sp. 501T to the low bioavailability of solid anthracene. Appl. Microbiol. Biotechnol. 58:378-385. [DOI] [PubMed] [Google Scholar]
- 28.Willumsen, P. A., J. E. Johansen, U. Karlson, and B. M. Hansen. 2005. Isolation and taxonomic affiliation of N-heterocyclic aromatic hydrocarbon transforming bacteria. Appl. Microbiol. Biotechnol. 67:420-428. [DOI] [PubMed] [Google Scholar]