Abstract
We evaluated a ready-to-use real-time quantitative Legionella pneumophila PCR assay system by testing 136 hot-water-system samples collected from 55 sites as well as 49 cooling tower samples collected from 20 different sites, in parallel with the standard culture method. The PCR assay was reproducible and suitable for routine quantification of L. pneumophila. An acceptable correlation between PCR and culture results was obtained for sanitary hot-water samples but not for cooling tower samples. We also monitored the same L. pneumophila-contaminated cooling tower for 13 months by analyzing 104 serial samples. The culture and PCR results were extremely variable over time, but the curves were similar. The differences between the PCR and culture results did not change over time and were not affected by regular biocide treatment. This ready-to-use PCR assay for L. pneumophila quantification could permit more timely disinfection of cooling towers.
Legionellosis can be acquired by inhalation of Legionella pneumophila bacteria dispersed by environmental sources, such as hot-water systems and cooling towers. Legionellosis outbreaks are often associated with high mortality rates (15 to 20%) (10). Legionella pneumophila serogroup 1 is responsible for up to 80% of cases (9, 11, 30). The density of Legionella cells in water is theoretically associated with the risk of legionellosis (5, 15): cell densities above 104 to 105 CFU per liter of water have been shown to represent a potential health risk to humans (20, 23). Environmental Legionella monitoring is recommended in several countries (7, 16), and regular treatment of cooling tower installations is obligatory in France (7).
In France, conventional culture is the only approved technique for the detection and quantification of Legionella in water samples (2). However, definitive culture results take 10 days to obtain and may have decreased sensitivities due to Legionella growth characteristics (3, 6). Several authors have developed real-time-PCR-based methods for rapid detection of Legionella in water samples (3, 12, 27, 29). However, Joly et al. recently reported that quantitative real-time PCR is influenced by the type of water sample and that the results may be laboratory dependent (12). Several commercial real-time PCR kits are currently available, such as the iQ-check real-time PCR kit (Bio-Rad, France), the Aqua Screen Lp-qDual kit (Minerva Biolabs, Germany), and the GeneDisc Legionella kit (GeneSystems, France). The main differences between these kits are based on the degrees of standardization of the three critical steps: DNA extraction, PCR preparation, and data analysis.
In this study, we compared a standardized real-time quantitative PCR assay system with the conventional culture method. The PCR system, developed by GeneSystems (Bruz, France), is the first ready-to-use PCR instrument dedicated to routine Legionella detection in water samples that includes a dedicated filtration unit and DNA extraction instrument. The two methods were both applied to 136 hot-water-system samples and to 49 cooling tower water samples. In addition, we used both methods to monitor L. pneumophila density in the same industrial cooling tower for 13 months.
MATERIALS AND METHODS
Sample collection.
From April 2004 to August 2005, 190 water samples were collected from 75 sites in France. Each site corresponded to a distinct water distribution system or cooling tower. A maximum of five samples was taken from each site. Each sample was collected in a sterile 2-liter plastic bottle and was divided into two equal parts for culture and PCR. Five hot-water-system samples, collected from five different sites and yielding non-L. pneumophila legionellae by culture, were excluded, as the version of the GeneSystems PCR method used detects only L. pneumophila. Thus, 185 water samples were included in the study: 136 hot-water-system samples from 55 sites and 49 cooling tower samples from 20 sites.
In addition, from January 2005 to February 2006, L. pneumophila density was monitored in a cooling tower of an industrial plant. One hundred four water samples were analyzed (500 ml for culture and 500 to 1,000 ml for PCR). In keeping with French legislation (7), this cooling tower was treated with HOBr-isothiozalone for the last 5 days of each month. No mechanical cleaning of the tower was performed during the period of investigation.
Culture.
We used the AFNOR-T90-431 culture method, which includes species and serogroup identification (2). Briefly, 200 μl of each 1-liter sample was plated directly on selective glycine-vancomycin-polymyxin B-cycloheximide medium (Oxoid, Wesel, Germany). The samples were then filtered through 0.45-μm polycarbonate filters (Millipore, St. Quentin-en-Yvelines, France). The filters were placed in 5 ml of sterile water and sonicated for 2 min (Fischer Bioblock scientific sonicator; Illkirch, France) at 35 kHz. Then, 100 μl of the concentrate was plated on glycine-vancomycin-polymyxin B-cycloheximide medium. All samples were subjected to standard heat and acid treatments. The plates were incubated at 36°C ± 2°C, and colonies were counted after 3, 5, and 10 days. According to the AFNOR method, the result from the highest reliable plate type was chosen. Up to five colonies per sample, chosen for their heterogeneities and their times of emergence, were identified by means of direct immunofluorescence with polyclonal rabbit sera (National Reference Center for Legionella, Lyon, France) and Legionella-specific latex reagents (Oxoid, Hampshire, England) (Veolia Environnement and CARSO-LSEHL laboratories). The detection limit (DL) of the culture method was 50 CFU/liter, and the quantification limit (QL) was 250 CFU/liter (2).
Sample preparation for PCR.
Following the XP-T90-471 method (1), 500 to 1,000 ml of each sample was prepared with a GeneExtract instrument (GeneSystems, France), a dedicated DNA extraction system simultaneously handling five water samples and one negative control (1 liter of a distilled sample). The GeneExtract instrument uses a commercial DNA extraction pack, including all reagents and consumables required for water sample filtration, cell lysis, and DNA purification (GeneSytems, France). The GeneExtract instrument includes a filtration unit with a 0.45-μm-pore-size polycarbonate membrane. The membrane was transferred to a tube containing lysis buffer and was subjected to mechanical treatments (sonication for 20 min, using one sonotrode per sample, and then heating at 100°C for 10 min) in order to enhance DNA recovery from aggregates. Finally, DNA was purified by adsorption on a silica column and was eluted in a final volume of 200 μl of elution buffer.
Quantitative PCR.
Quantitative PCR was performed with a GeneDisc-Cycler apparatus (GeneSystems, France) and a dedicated consumable, GeneDisc Legionella pneumophila, a ready-to-use molecular biology device. GeneDisc Legionella pneumophila incorporates six analytical sectors for the analysis of five DNA extracts from water samples and one negative control. Each sector consists of six PCR wells preloaded with specific primers and a dual-fluorescence, dye-labeled probe (FAM [6-carbofluorescein]-TAMRA [6-carboxytetramethylrhodamine]). (i) Three wells are used for L. pneumophila detection, (ii) two wells are used for both positive and internal inhibitor controls with a calibrated synthetic DNA sequence which contained at each end the specific primer oligonucleotides for L. pneumophila detection, and (iii) one well is used for the negative PCR control. Each analytical well is filled with a mixture of DNA extract (6 μl) and master mix solution (6 μl) from GeneSystems. Quantitative PCR takes 45 min. GeneDisc software uses the cycle threshold and the positive control fluorescence value to detect the presence of inhibitor molecules. For each sample, the GeneDisc-Cycler indicates the final result as the number of genome units (GU) per liter. For each batch of GeneDiscs, linearity was confirmed by constructing an external five-point standard curve corresponding to between 25 and 2.5 × 105 GU of L. pneumophila serogroup 1 (ATCC 33152) per well.
The DL of this PCR assay, corresponding to the smallest number of GU yielding positive results in 90% of runs, was 5 GU per PCR mix, corresponding to 167 GU per liter. The QL of the PCR assay, corresponding to the smallest number of GU yielding a coefficient of variation below 25%, was 25 GU per reaction mixture (equivalent to 833 GU per liter). When PCR inhibition occurred (nonamplification of the positive control), the sample DNA was retested after 2- to 10-fold dilution in ultrapure H2O.
Interlaboratory reproducibility.
L. pneumophila serogroup 1 strain ATCC 33152 bacteria were grown overnight with vigorous agitation at 37°C in buffered yeast extract broth supplemented with 0.4 mg of l-cysteine and 0.25 mg of ferric pyrophosphate per milliliter (Oxoid, France) and then counted by measuring DAPI (4′,6′-diamidino-2-phenylindole) epifluorescence (24). The culture was serially diluted 10-fold, and 1-milliliter aliquots (containing 2 × 105 CFU) were concentrated by centrifugation at 5,500 × g for 20 min. The supernatants were discarded, and the cell pellets were shipped to four different laboratories.
In each laboratory, the cell pellet was resuspended in 10 ml of sterile distilled water, and 2 ml of this suspension was added to 2 liters of commercially available mineral water (Evian, France). After thorough shaking, five 250-ml portions (each theoretically containing 5 × 103 CFU) were analyzed with the GeneSystems method. A negative control (250 ml of noninoculated mineral water) was always run simultaneously.
Statistical analysis.
PCR results were analyzed with the integrated GeneDisc software (GeneSystems, Bruz, France). SPSS 12.0 software was used to compare PCR and culture data with those for parametric tests (determination coefficient [r2] and Student's t) and nonparametric tests (Wilcoxon and Mann-Whitney), as appropriate. For the PCR method, we calculated the positive and negative predictive values (PPV and NPV, respectively), which corresponded to the ratio between the number of positive (or negative) samples by both methods and the number of positive (or negative) samples by PCR.
RESULTS
Interlaboratory comparison.
Four different laboratories used the GeneSystems PCR method to analyze L. pneumophila suspensions containing 5 × 103 CFU in 250 ml, corresponding to 5 × 103 GU (log10 5,000 = 3.7) (Table 1) . The difference between the experimental result and the target value (3.7 log10) was always below 0.5 log10, except for assay 4 in laboratory 3.
TABLE 1.
Results of interlaboratory comparisons of use of the GeneSystems PCR system (five repeated assays) on spiked water samples containing 5.0 × 103 CFU L. pneumophila in 250 ml
| Assay | Result for indicated laboratory and logarithma
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
|||||
| Log(Q) | ΔLog | Log(Q) | ΔLog | Log(Q) | ΔLog | Log(Q) | ΔLog | |
| 1 | 3.85 | −0.15 | 3.33 | 0.37 | 3.68 | 0.02 | 3.33 | 0.37 |
| 2 | 3.85 | −0.15 | 3.70 | 0.00 | 3.36 | 0.34 | 3.88 | −0.18 |
| 3 | 3.75 | −0.05 | 3.55 | 0.14 | 3.43 | 0.27 | 3.59 | 0.11 |
| 4 | 3.43 | 0.27 | 3.68 | 0.02 | 3.07 | 0.63 | 3.75 | −0.05 |
| 5 | 3.62 | 0.08 | 3.68 | 0.02 | 3.52 | 0.18 | 3.52 | 0.18 |
ΔLog is the difference between the experimental result and the target value (3.7 = log10 5,000 GU). Log(Q) is the log value of the experimental result.
Hot-water samples.
Among the 136 hot-water samples, 4 (2.9%) samples, collected from four different sites, contained PCR inhibitors (with or without 10-fold dilution in ultrapure H2O) and were excluded from the PCR/culture comparison. Two of these four samples were quantifiable by culture (5.6 × 103 and 3.7 × 105 CFU/liter). PCR results below the detection limit (5 GU/assay) were considered negative and those above this limit positive. Among the remaining 132 samples, 87 (65.9%) were positive by PCR and 59 (44.7%) were positive by culture (Table 2) . Among the PCR-positive samples, 50 (37.9%) were quantifiable (>25 GU/well). Forty (30.3%) of the culture-positive samples were quantifiable (>250 CFU L. pneumophila per liter), and 19 (14.4%) were not quantifiable (between 50 and 250 CFU/liter). The PPV and NPV of PCR for hot-water-system samples were 57.5% and 80.0%, respectively.
TABLE 2.
PCR and culture results for 132 hot-water-system samples and 46 cooling tower water samples
| Result groupa | No. (%) of samples for indicated sample group and method
|
|||
|---|---|---|---|---|
| Hot water systems
|
Cooling towers
|
|||
| PCR | Culture | PCR | Culture | |
| Negative | ||||
| <DL | 45 (34.1) | 73 (55.3) | 18 (39.1) | 31 (67.4) |
| Positive | ||||
| DL ≤ x ≤ QL | 37 (28.0) | 19 (14.4) | 8 (17.4) | 6 (13.0) |
| >QL | 50 (37.9) | 40 (30.3) | 20 (43.5) | 9 (19.6) |
| Total | 132 (100) | 132 (100) | 46 (100) | 46 (100) |
DL, 5 GU/well for PCR and 50 CFU/liter for culture; QL, 25 GU/well for PCR and 250 CFU/liter for culture. Results below the detection limit were considered negative.
PCR results were on average 4.5-fold higher than culture results. Among the 45 PCR-negative samples, 9 (6.8%) were positive by culture; of these, 1 contained 1.0 × 104 CFU/liter, 5 contained between 450 and 1,500 CFU/liter, and 3 contained less than 250 CFU/liter (the quantification limit). For these nine PCR-negative samples, any partial or total inhibition was detected by the positive and internal inhibitor controls. Among the 73 culture-negative samples, 37 (28.0%) were positive by PCR, of which 19 (14.4%) were below the quantification limit (QLPCR), while the remaining 18 samples (13.6%) contained 8.7 × 102 to 2.8 × 105 GU/liter. An acceptable correlation was found between the PCR and culture results for hot-water-system samples (r2 = 0.732).
Cooling tower samples.
Three (6.1%) PCR-inhibitory samples, collected from three different sites, were found among the 49 cooling tower water samples (with and without 10-fold dilution in ultrapure H2O). One of these three samples was quantifiable by culture (900 CFU/liter). Among the remaining 46 samples, 15 (32.6%) were positive by culture, of which 9 (19.6%) contained >250 CFU L. pneumophila per liter (Table 2). Among these 46 samples, 28 (60.9%) were positive by PCR and 20 (43.5%) were quantifiable. The PCR PPV was 53.6% for cooling tower samples, and the NPV was 100% (no PCR-negative samples were culture positive).
PCR values were on average 3.1-fold higher than culture values. Among the 31 culture-negative samples, 13 (28.3%) were positive by PCR, of which 5 (10.9%) were below the QLPCR and 8 (17.4%) contained between 1.1 × 103 and 3.2 × 104 GU/liter. A weak correlation was obtained between the PCR and culture results for cooling tower samples (r2 = 0.187).
Longitudinal study. (i) Comparison of cooling tower results.
To determine whether the selected cooling tower was representative of the 49 cooling towers at the 20 sites analyzed from April 2004 to August 2005, we compared the differences between the PCR and culture results for the different sites by using Student's t test. No significant difference was found, showing that the selected tower was representative.
(ii) Qualitative and quantitative L. pneumophila assay for serial cooling tower samples.
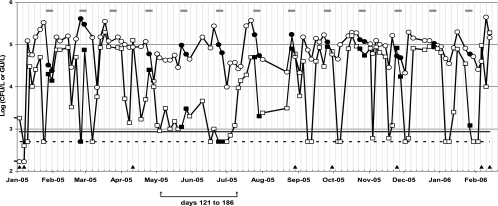
As shown in Fig. 1, PCR and culture (in log GU/liter and CFU/liter values, respectively) gave similar curves for the 104 serial samples collected from the selected water tower. PCR results were significantly higher than culture results (mean, 3.0-fold; up to 44.4-fold; P < 0.05 in the Wilcoxon test). The largest difference between the two methods occurred between day 121 and day 186 (May to July).
FIG. 1.
Follow-up of Legionella pneumophila cell density by quantitative PCR (GU/liter) and conventional culture (CFU/liter), from January 2005 to February 2006, in a single industrial cooling tower. When the culture or PCR value was below the detection or quantification limit, the sample was assigned a value equal to the corresponding limit. Values for log CFU/liter (open squares), log GU/liter (open circles), QLPCR (solid line), and QLculture (dotted line) and dates when PCR results were below culture results (filled triangles) are shown. Gray bars over the curves indicate the 5-day treatment periods, and values obtained during these periods are indicated by filled squares and circles.
Eight samples (7.7%) gave lower PCR results than culture results (up to 2.4-fold lower). Two samples (1.9%) were below the QLPCR, and 21 samples (20.2%) were below the QLculture. Culture results (mean, 3.7 × 104 ± 4.5 × 104 CFU/liter; standard deviation [SD], 122%) were more variable than PCR results (mean, 1.1 × 105 ± 8.8 × 104 GU/liter; SD, 80%). Statistical analysis showed no correlation between the results for the two methods (r2 = 0.120).
(iii) Treatment effects.
Nineteen of the 104 samples were taken during HOBr-isothiozalone decontamination. The differences between the PCR and culture results for these 19 samples were not significantly different from those for the other 85 samples (Mann-Whitney test, P = 0.850). The PCR results were significantly higher than the culture results during all periods (with and without treatment) (Wilcoxon test, P < 0.05).
DISCUSSION
We compared conventional culture with a ready-to-use real-time quantitative PCR system dedicated to routine quantification of Legionella pneumophila in water samples.
Joly et al. observed variations dependent on the version of the PCR instrument running the same quantitative PCR technique for Legionella DNA quantification (in terms of extraction and the amplification target) in two different laboratories (12). Additional variability could be due to differences in the PCR tools and in the choice of the Legionella target gene(s) (14, 18, 28). The GeneSystems assay method standardizes both DNA extraction (on the GeneExtract instrument) and DNA quantification (on the GeneDisc-Cycler). In addition, the final result for a given sample is the mean for triplicate determinations, and an internal inhibitor control is used to detect the presence of PCR inhibitors. We first studied interlaboratory reproducibility with the GeneSystems real-time PCR system and found no laboratory-dependent differences in five repeated tests.
Quantitative PCR is much faster and more sensitive than conventional culture for L. pneumophila detection (3, 4, 8, 17, 27), as confirmed here. Previous studies have shown that (i) PCR results are usually higher than culture values (12, 27, 29) and that (ii) samples that are not quantifiable by PCR usually contain ≤250 CFU/liter (12, 22). Our results for 185 hot-water-system and cooling tower samples collected from 75 different sites confirm these reports: (i) our PCR results (GU/liter) were on average 4.5-fold and 3.1-fold higher than the culture results (CFU/liter) for hot-water-system and cooling tower samples, respectively, and (ii) we found that PCR had NPVs of 80% for the presence of L. pneumophila in hot-water-system samples and 100% for cooling tower waters. However, quantitative PCR has several limitations. First, the detection of both living and dead bacteria makes it difficult to evaluate the real health risk (14, 22, 28), and second, PCR inhibitors present in complex water systems can lead to false-negative results (14, 19, 26, 28). Among the 136 hot-water-system samples and 49 cooling tower water samples analyzed here, PCR inhibitors were present in 2.9% and 6.1% of cases, respectively. It has been reported that cooling tower waters frequently contain PCR inhibitors (up to 30.6% of samples) (21).
We found an acceptable correlation between the PCR and culture results for hot-water samples (r2 = 0.732), in keeping with previous reports (12, 27). Only a weak correlation was obtained for cooling tower samples (r2 = 0.187), again as previously described (12). This could be explained by a higher complexity of the matrix (water and its components) in cooling towers.
Unlike Joly et al. (12), we did not attempt to establish a PCR positivity cutoff and focused rather on the utility of the PCR method for monitoring Legionella-contaminated systems. This is the first long-term (13-month) follow-up study of L. pneumophila cell density in the same cooling tower, using both PCR and culture. Among the 104 water samples analyzed, 98.1% were quantifiable by PCR and 79.9% by culture. These 104 water samples were representative of the 46 cooling tower water samples from 20 sites used for the cross-sectional study in terms of differences between PCR and culture values. The PCR values remained significantly higher than the culture values during the follow-up, but the results showed similar patterns of change, suggesting that this PCR method is appropriate for monitoring L. pneumophila contamination. The culture results were extremely variable over time (mean, 3.7 × 104 ± 4.5 × 104 CFU/liter; SD, 122%), and so were the PCR values (mean, 1.1 × 105 ± 8.8 × 104 GU/liter; SD, 80%). The differences between the culture and PCR results were largest between days 121 and 186 (May to July) (Fig. 1). We found no explanation for these differences.
The PCR and culture values both tended to fall during periods of biocide treatment, but the differences during other periods were not significant. Interestingly, although HOBr-isothiozalone treatment kills bacteria and might generate more nonviable (and therefore nonculturable) cells, the differences between PCR and culture results were not more marked during periods of decontamination (13, 25). The fluctuations of a Legionella population could be monitored more rapidly by quantitative PCR than by culture, potentially leading to reduced biocide costs and ecological benefits.
In conclusion, this ready-to-go quantitative PCR system from GeneSystems appears suitable for monitoring Legionella pneumophila contamination, especially in cooling towers. The present system detects L. pneumophila only, but a new GeneDisc version (GDLSP-471) will detect the DNA of all Legionella species.
Acknowledgments
We are most grateful to Gerard Lina for his help with the statistical analyses; Philippe Joly for his help with sample collection; Thomas Garel for technical help; the Legionella National Reference Center, CARSO-LSEHL, and Veolia Environnement laboratories for their help with the culture method; and David Young for editing the manuscript.
This work was supported by a grant from GeneSystems, Bruz, France.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Association Française de Normalisation. 2006. Qualité de l'eau. Détection et quantification des Legionella et/ou Legionella pneumophila par concentration et amplification génique par réaction de polymérisation en chaîne (PCR). XP T90-471. http://www.boutique.afnor.fr/boutique.asp.
- 2.Association Française de Normalisation. 2003. Water quality. Detection and enumeration of Legionella spp and Legionella pneumophila. Method by direct inoculation and after concentration by membrane filtration or centrifugation. AFNOR NF T90-431. http://www.boutique.afnor.fr/boutique.asp.
- 3.Ballard, A. L., N. K. Fry, L. Chan, S. B. Surman, J. V. Lee, T. G. Harrison, and K. J. Towner. 2000. Detection of Legionella pneumophila using a real-time PCR hybridization assay. J. Clin. Microbiol. 38:4215-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behets, J., P. Declerck, Y. Delaedt, B. Creemers, and F. Ollevier. 14 August 2006. Development and evaluation of a Taqman duplex real-time PCR quantification method for reliable enumeration of Legionella pneumophila in water samples. J Microbiol. Methods doi: 10.1016/j.mimet.2006.07.002. [DOI] [PubMed]
- 5.Best, M., V. L. Yu, J. Stout, A. Goetz, R. R. Muder, and F. Taylor. 1983. Legionellaceae in the hospital water-supply. Epidemiological link with disease and evaluation of a method for control of nosocomial legionnaires' disease and Pittsburgh pneumonia. Lancet ii:307-310. [DOI] [PubMed] [Google Scholar]
- 6.Boulanger, C. A., and P. H. Edelstein. 1995. Precision and accuracy of recovery of Legionella pneumophila from seeded tap water by filtration and centrifugation. Appl. Environ. Microbiol. 61:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conseil supérieur d'hygiène publique de France. 2005. Le risque lié aux légionelles. Guide d'investigation et d'aide à la gestion. Rapport du Conseil Supérieur d'Hygiène Publique de France Juillet 2005. Rapport du Conseil Supérieur d'Hygiène Publique de France. http://www.sante.gouv.fr/htm/pointsur/legionellose/guid2005.pdf.
- 8.Declerck, P., J. Behets, E. Lammertyn, I. Lebeau, J. Anne, and F. Ollevier. 2006. Detection and quantification of Legionella pneumophila in water samples using competitive PCR. Can. J. Microbiol. 52:584-590. [DOI] [PubMed] [Google Scholar]
- 9.Doleans, A., H. Aurell, M. Reyrolle, G. Lina, J. Freney, F. Vandenesch, J. Etienne, and S. Jarraud. 2004. Clinical and environmental distributions of Legionella strains in France are different. J. Clin. Microbiol. 42:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helbig, J. H., S. Bernander, M. Castellani Pastoris, J. Etienne, V. Gaia, S. Lauwers, D. Lindsay, P. C. Luck, T. Marques, S. Mentula, M. F. Peeters, C. Pelaz, M. Struelens, S. A. Uldum, G. Wewalka, and T. G. Harrison. 2002. Pan-European study on culture-proven Legionnaires' disease: distribution of Legionella pneumophila serogroups and monoclonal subgroups. Eur. J. Clin. Microbiol. Infect. Dis. 21:710-716. [DOI] [PubMed] [Google Scholar]
- 12.Joly, P., P. A. Falconnet, J. Andre, N. Weill, M. Reyrolle, F. Vandenesch, M. Maurin, J. Etienne, and S. Jarraud. 2006. Quantitative real-time Legionella PCR for environmental water samples: data interpretation. Appl. Environ. Microbiol. 72:2801-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, B. R., J. E. Anderson, S. A. Mueller, W. A. Gaines, and A. M. Kendall. 2002. Literature review: efficacy of various disinfectants against Legionella in water systems. Water Res. 36:4433-4444. [DOI] [PubMed] [Google Scholar]
- 14.Koide, M., A. Saito, N. Kusano, and F. Higa. 1993. Detection of Legionella spp. in cooling tower water by the polymerase chain reaction method. Appl. Environ. Microbiol. 59:1943-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kool, J. L., D. Bergmire-Sweat, J. C. Butler, E. W. Brown, D. J. Peabody, D. S. Massi, J. C. Carpenter, J. M. Pruckler, R. F. Benson, and B. S. Fields. 1999. Hospital characteristics associated with colonization of water systems by Legionella and risk of nosocomial Legionnaires' disease: a cohort study of 15 hospitals. Infect. Control Hosp. Epidemiol. 20:798-805. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J. V., and C. Joseph. 2002. Guidelines for investigating single cases of Legionnaires' disease. Commun. Dis. Public Health 5:157-162. [PubMed] [Google Scholar]
- 17.Levi, K., J. Smedley, and K. J. Towner. 2003. Evaluation of a real-time PCR hybridization assay for rapid detection of Legionella pneumophila in hospital and environmental water samples. Clin. Microbiol. Infect. 9:754-758. [DOI] [PubMed] [Google Scholar]
- 18.Liu, H., Y. Li, X. Huang, Y. Kawamura, and T. Ezaki. 2003. Use of the dnaJ gene for the detection and identification of all Legionella pneumophila serogroups and description of the primers used to detect 16S rDNA gene sequences of major members of the genus Legionella. Microbiol. Immunol. 47:859-869. [DOI] [PubMed] [Google Scholar]
- 19.Maiwald, M., K. Kissel, S. Srimuang, M. von Knebel Doeberitz, and H. G. Sonntag. 1994. Comparison of polymerase chain reaction and conventional culture for the detection of legionellae in hospital water samples. J. Appl. Bacteriol. 76:216-225. [DOI] [PubMed] [Google Scholar]
- 20.Meenhorst, P. L., A. L. Reingold, D. G. Groothuis, G. W. Gorman, H. W. Wilkinson, R. M. McKinney, J. C. Feeley, D. J. Brenner, and R. van Furth. 1985. Water-related nosocomial pneumonia caused by Legionella pneumophila serogroups 1 and 10. J. Infect. Dis. 152:356-364. [DOI] [PubMed] [Google Scholar]
- 21.Miyamoto, H., H. Yamamoto, K. Arima, J. Fujii, K. Maruta, K. Izu, T. Shiomori, and S. Yoshida. 1997. Development of a new seminested PCR method for detection of Legionella species and its application to surveillance of legionellae in hospital cooling tower water. Appl. Environ. Microbiol. 63:2489-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng, D. L., B. B. Koh, L. Tay, and B. H. Heng. 1997. Comparison of polymerase chain reaction and conventional culture for the detection of legionellae in cooling tower waters in Singapore. Lett. Appl. Microbiol. 24:214-216. [DOI] [PubMed] [Google Scholar]
- 23.Patterson, W. J., D. V. Seal, E. Curran, T. M. Sinclair, and J. C. McLuckie. 1994. Fatal nosocomial Legionnaires' disease: relevance of contamination of hospital water supply by temperature-dependent buoyancy-driven flow from spur pipes. Epidemiol. Infect. 112:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 25.Skaliy, P., T. A. Thompson, G. W. Gorman, G. K. Morris, H. V. McEachern, and D. C. Mackel. 1980. Laboratory studies of disinfectants against Legionella pneumophila. Appl. Environ. Microbiol. 40:697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villari, P., E. Motti, C. Farullo, and I. Torre. 1998. Comparison of conventional culture and PCR methods for the detection of Legionella pneumophila in water. Lett. Appl. Microbiol. 27:106-110. [DOI] [PubMed] [Google Scholar]
- 27.Wellinghausen, N., C. Frost, and R. Marre. 2001. Detection of legionellae in hospital water samples by quantitative real-time LightCycler PCR. Appl. Environ. Microbiol. 67:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto, H., Y. Hashimoto, and T. Ezaki. 1993. Comparison of detection methods for Legionella species in environmental water by colony isolation, fluorescent antibody staining, and polymerase chain reaction. Microbiol. Immunol. 37:617-622. [DOI] [PubMed] [Google Scholar]
- 29.Yáñez, M. A., C. Carrasco-Serrano, V. M. Barberá, and V. Catalán. 2005. Quantitative detection of Legionella pneumophila in water samples by immunomagnetic purification and real-time PCR amplification of the dotA gene. Appl. Environ. Microbiol. 71:3433-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]