Abstract
A multitarget molecular beacon-based real-time nucleic acid sequence-based amplification (NASBA) assay for the specific detection of Vibrio cholerae has been developed. The genes encoding the cholera toxin (ctxA), the toxin-coregulated pilus (tcpA; colonization factor), the ctxA toxin regulator (toxR), hemolysin (hlyA), and the 60-kDa chaperonin product (groEL) were selected as target sequences for detection. The beacons for the five different genetic targets were evaluated by serial dilution of RNA from V. cholerae cells. RNase treatment of the nucleic acids eliminated all NASBA, whereas DNase treatment had no effect, showing that RNA and not DNA was amplified. The specificity of the assay was investigated by testing several isolates of V. cholerae, other Vibrio species, and Bacillus cereus, Salmonella enterica, and Escherichia coli strains. The toxR, groEL, and hlyA beacons identified all V. cholerae isolates, whereas the ctxA and tcpA beacons identified the O1 toxigenic clinical isolates. The NASBA assay detected V. cholerae at 50 CFU/ml by using the general marker groEL and tcpA that specifically indicates toxigenic strains. A correlation between cell viability and NASBA was demonstrated for the ctxA, toxR, and hlyA targets. RNA isolated from different environmental water samples spiked with V. cholerae was specifically detected by NASBA. These results indicate that NASBA can be used in the rapid detection of V. cholerae from various environmental water samples. This method has a strong potential for detecting toxigenic strains by using the tcpA and ctxA markers. The entire assay including RNA extraction and NASBA was completed within 3 h.
Vibrio cholerae is the etiological agent of epidemic cholera, which causes watery diarrhea that can result in the rapid dehydration and death of infected persons. Coastal waters are an important reservoir of V. cholerae, and cholera is generally transmitted to humans via contaminated water or seafood (12, 13). Of more than 200 known serogroups of V. cholerae, the two well-known serogroups O1 and O139 have been associated with epidemic cholera (11). The serogroup O1 can be divided into three serotypes, Inaba, Ogawa, and Hikojima, and each serotype can be divided into two biotypes, classical and El Tor (30). The other serogroups of V. cholerae, collectively referred to as non-O1 and non-O139 serogroups, have not been associated with epidemics but have been associated with occasional outbreaks of cholera-like disease. These other serogroups are usually isolated from patients with mild diarrhea or from the environment (27, 39). Vibrio species such as V. parahaemolyticus, V. cholerae, and V. vulnificus are found in blue mussels harvested along the coastline of Norway (5).
The pathogenesis of cholera is a complex process, and the major virulence factors of V. cholerae are the cholera toxin (CT) encoded by the ctxAB genes and the toxin-coregulated pilus (TCP) encoded by the tcpA gene (30). CT leads to increased intestinal secretion of electrolytes and water into the lumen. TCP is a type IV pilus that is essential for intestinal colonization in humans and in animal models of cholera (26, 50). The ctxAB genes are located in the genome of the CTXΦ filamentous prophage, and tcpA is located in the large Vibrio pathogenicity island, which functions as a receptor for the CTXΦ prophage (31, 33, 53). The toxR gene encodes a transcription factor that directly regulates the expression of CT, among others (37). Most toxigenic strains and non-O1and non-O139 strains of V. cholerae possess the toxR gene (48). Another genetic target is the virulence factor hemolysin, which is proposed to cause diarrhea (29). The hemolysin produced by V. cholerae non-O1and non-O139 strains is identical to the hemolysin product of the V. cholerae El Tor strains (54). However, some El Tor strains do not express the hly gene. Still, the hlyA gene is detected in several nonhemolytic strains by PCR (48).
PCR has now become a frequently used detection method, and several PCR protocols have been developed for V. cholerae (24, 25, 35, 36, 48, 49). PCR detection confirms the presence of specific genetic regions in a target organism. However, RNA-based methods such as the nucleic acid sequence-based amplification (NASBA) method and reverse transcription-PCR may reveal the expression of various genes. Viable but nonculturable (VBNC) forms of bacterial cells are now recognized as a common phenomenon in many bacterial species, and these organisms may escape detection if only culture methods are used (28). It has been demonstrated that the viability of VBNC cells may be monitored by reverse transcription-PCR analysis (17).
The NASBA assay, initially introduced by Compton (14), is a sensitive, transcription-based amplification system specifically designed for detecting RNA. The technology relies on the simultaneous activity of three different enzymes: RNaseH, avian myeloblastosis virus reverse transcriptase, and T7 RNA polymerase. The presence of a T7 promoter sequence at the 5′ end of the forward primer is essential and is used by the T7 RNA polymerase during the synthesis of new RNA amplicons. In contrast to PCR and reverse transcription-PCR, the NASBA method is isothermal (41°C). At 41°C, genomic DNA remains double-stranded, and it is therefore not a substrate in NASBA. Although NASBA is specifically designed for the detection of RNA targets, in some NASBA systems DNA may be amplified (19, 42). However, the DNA amplification is ineffective and occurs only in the absence of RNA targets or in the case of a 1,000-fold excess of target DNA. Successful reverse transcription-PCR requires residual DNA to be eliminated in order to provide accurate detection of specific bacterial RNA sequences. Such extensive extraction of the nucleic acids may be time-consuming and may reduce the recovery of target RNA.
In NASBA, single-stranded RNA amplicons are produced, and these can easily be detected by hybridization with a sequence-specific probe such as a molecular beacon. Molecular beacons are single-stranded oligonucleotide hybridization probes designed such that they can form a stem-loop structure (51). The loop sequence is complementary to the target sequence to be detected, and the stem sequences are complementary to each other. A fluorescent label is linked to the end of one arm and a quencher to the other. Therefore, the molecular beacon is not fluorescent when it is free in a solution. A molecular beacon is designed so that its probe sequence is just long enough for a perfect complementary probe-target hybrid to be more stable than the stem hybrid. Consequently, the molecular beacon spontaneously forms probe-target hybrids in which the effect of the quencher is eliminated (19). Molecular beacon probes are designed according to specific rules (19), and the design is critical for the specificity and sensitivity of real-time NASBA (34).
NASBA has been extensively applied in clinical microbiology in detecting RNA viruses (19). However, NASBA is also used successfully for detection of microbial pathogens in food and environmental samples (16). Descriptions of such methods for the detection of Campylobacter spp. (52), Listeria monocytogenes (7), Salmonella enterica in various foods (15), Cryptosporidium parvum (3), and Escherichia coli in water (38) have been published. It has been shown that as few as 10 viable spores of Bacillus anthracis can be detected by NASBA (4).
In the present study, a real-time multitarget NASBA assay for the specific detection of V. cholerae cells has been developed. Primers and molecular beacons were targeted to five different genomic regions. These were the constitutively expressed housekeeping gene groEL, the hemolysin A virulence gene hlyA, the CT gene ctxA, the TCP gene tcpA, and the regulator for the expression of CT, the toxR gene. Serial dilutions of V. cholerae RNA were used to determine the detection limits of the single assays (50 CFU/ml), and RNA extraction and NASBA were performed within 3 h. The specificity of the assay was tested against several isolates of V. cholerae, Vibrio spp., and strains of Bacillus cereus, S. enterica, and E. coli. Furthermore, V. cholerae was detected in spiked environmental water samples (5 × 103 CFU/ml). This is to our knowledge the first report describing the use of NASBA for the specific detection of V. cholerae. Our results indicate that the ctxA and tcpA markers may be used to monitor the environmental population of V. cholerae strains expressing the virulence factors.
MATERIALS AND METHODS
Bacterial strains, media, and cultures.
The Vibrio spp. strains used in this study are shown in Table 1. The isolates, collected from clinical and environmental samples, were kindly provided by the Norwegian School of Veterinary Science, the Norwegian Institute of Public Health, and the Istituto Superiore di Sanità, Italy. The strains of B. cereus and the V. cholerae strain Cip 106855 O1 Inaba El Tor were obtained from the American Type Culture Collection, and the V. cholerae strain NCTC 8457 was from the National Collection of Type Cultures, United Kingdom. The strains of S. enterica and the E. coli strain CCUG 29188 were obtained from the Culture Collection, University of Göteborg, Sweden. The E. coli strain DSM 4230 was from the German Collection of Microorganisms and Cell Cultures. The Vibrio spp. strains were grown in tryptic soy broth (TSB; Merck, Germany). The E. coli and S. enterica strains were grown in Luria-Bertani broth (Merck, Germany), and the B. cereus strains were grown in brain hearth infusion broth (Acumedia Manufacturers Inc., Baltimore, MD). All cultures were grown aerobically at 37°C and harvested by centrifugation at 2,000 × g for 10 min in the logarithmic phase of growth. The cells were washed once in sterile phosphate-buffered saline (PBS). The numbers of structurally intact cells were determined with a phase-contrast microscope (Zeiss, Germany). Cultures of V. cholerae were serially diluted, and CFU were enumerated by plating 100 μl of each dilution onto TSB agar and incubating at 37°C for 24 h. RNAlater (QIAGEN) was added to aliquots of the cell pellets to be used in RNA isolation prior to storage at −80°C.
TABLE 1.
Bacterial strains used for NASBA
| Bacterium | Strain and serogroup | Source,e date of collection, and type of isolate | Detection of genetic targetsf:
|
||||
|---|---|---|---|---|---|---|---|
| tcpA | ctxA | groEL | hlyA | toxR | |||
| V. cholerae O1 | O1 Inaba El Tor Cip106855 | ATTC 393 | + | + | + | + | + |
| O1 Inaba El Tor ATTC 14033 | NCTC 8457 | + | + | + | + | + | |
| VC242, O1 Ogawa El Tor | Fecesa | + | + | + | + | + | |
| VC243, O1 Ogawa El Tor | Fecesa | + | + | + | + | + | |
| VC 5/77, O1 | Peru, 1991b | + | + | + | + | + | |
| VC 2/60, O1 | Albania, 1994b | + | + | + | + | + | |
| VC 2/70, O1 | Albania, 1994b | + | + | + | + | + | |
| VC 2/67, O1 | Albania, 1994b | + | + | + | + | + | |
| VC 2/57, O1 | Albania, 1994b | + | + | + | + | + | |
| VC 2/21, O1 | Italy, 1994b | + | + | + | + | + | |
| VC 2/23, O1 | Italy, 1994b | + | + | + | + | + | |
| VC 2/26, O1 | Italy, 1994b | + | + | + | + | + | |
| VC 2/32, O1 | Italy, 1994b | + | + | + | + | + | |
| VC 6/31, O1 | Italy, 2005b,c | + | + | + | + | + | |
| VC 4/53, O1 | Italy, 1973b | + | + | + | + | + | |
| VC 4/57, O1 | Italyb; environmental, ctx positive | +/− | +/− | + | + | + | |
| VC 6/23, O1 | Italyb; environmental, ctx negative | − | − | + | + | + | |
| V. cholerae non-O1 | VC 5/47, O2 | Italyb; septicemic | +/− | +/− | + | + | + |
| VC 5/42, O158 | Italyb; septicemic | − | − | + | + | + | |
| VC 4/76, O111 | Italyb; septicemic | − | − | + | + | + | |
| VC084, non-O1 | Scampid | − | − | + | + | + | |
| VC216, non-O1 | Indiad | − | − | + | + | + | |
| VC229, non-O1 | Wounda | − | − | + | + | + | |
| VC230, non-O1 | Fecesa | − | − | + | + | + | |
| VC246, non-O1 | Norwaya; mussel | − | − | + | + | + | |
| VC344, non-O1 | Norwaya; mussel | − | − | + | + | + | |
| VC503, non-O1 | Norwayd; water | − | − | + | + | + | |
| Other Vibrio speciesd | |||||||
| V. fluvialis | VF 062 | − | − | − | − | − | |
| V. mimicus | VM 034 | − | +/− | − | − | − | |
| VM 052 | − | − | − | − | − | ||
| VM 345 | − | − | − | − | − | ||
| V. metschnikovii | VM 116 | − | − | − | − | − | |
| V. parahaemolyticus | VP 160 | − | − | − | − | − | |
| VP 363 | − | − | − | − | − | ||
| VP 438 | − | − | − | − | − | ||
| V. alginolyticus | VA 054 | − | − | − | − | − | |
| VA 647 | − | − | − | − | − | ||
| Other bacterial species | |||||||
| B. cereus | ATCC 14579 | ATCC | − | − | − | − | − |
| ATCC 10987 | ATCC | − | − | − | − | − | |
| E. coli | DSM 4230 | DSMZ | − | − | − | − | − |
| ATCC 43888, O157:H7 | CCUG 29188 | − | − | − | − | − | |
| S. enterica | ATCC 13076 Cip 82.97 | CCUG 34136 T | − | − | +/− | − | − |
| ATCC 43971 Cip 60.62 | CCUG 43971 | − | − | − | − | − | |
Strain received from J. Lassen, The Norwegian Institute of Public Health, Norway.
Strain received from A. Carotolli, Instituto Superiore di Sanita, Italy.
Imported case of V. cholerae.
Strain(s) received from L. M. Rørvik, The Norwegian School of Veterinary Science, Norway.
ATCC, American Type Culture Collection; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen (German Collection of Microorganisms and Cell Cultures); NCTC, National Collection of Type Cultures (United Kingdom); CCUG, Culture Collection of the University of Göteborg (Sweden).
+, specific amplified product detected; −, no amplified product detected; +/−, product was amplified at a late stage of the NASBA assay with a high t value and a weak fluorescence signal.
Primers and molecular beacon probes.
The primers and probes used in this study were targeted to the constitutively expressed housekeeping gene groEL, the virulence gene hlyA, the CT gene ctxA, the TCP gene tcpA, and the toxR gene. V. cholerae-specific sequences of the mentioned genes were identified by performing BLASTP and BLASTN database searches. The results were shown as multiple alignments, and regions unique to V. cholerae could be identified and used in primer and probe design using the Primer3 program (43). Primers and molecular beacon probes were synthesized by Invitrogen and Eurogentec (Belgium) (Table 2). The structures of the probes were tested using mfold, a DNA folding program (Zucker program; http://www.bioinfo.rpi.edu/applications/mfold/rna/form1.cgi). The sequences of the target genes were selected from gi9657252:c3783-3667 (groEL), gi40548577:c268-403 (non-O1, non-O139 El Tor hemolysin gene, hlyA), gi497317:c83-185 (tcpA), gi18448890:c180-331 (ctxA), and gi155248:c397-536 (toxR). The general guidelines described previously (19) and advice from personnel at NorChip AS (personal communication) were followed in primer and probe construction.
TABLE 2.
Primers and molecular beacon probes for the V. cholerae real-time NASBA assay
| Primer or MBa | Nucleotide sequence, 5′ → 3′b | Predicted size of amplicon (bp) |
|---|---|---|
| Pvc55-1 hlyA | aattctaatacgactcactatagggcAATCTCTTCCGTCCGATCAA | 135 |
| Pvc56-2 hlyA | TGATGCTGAAGGTCAAGCAG | |
| MBvc10-hlyA | ccgatcdTCAGAAAGGCTTATGGGGTGgatcgg | |
| Pvc62-1 tcpA | aattctaatacgactcactatagggCGCTGAGACCACACCCATA | 102 |
| Pvc60-2 tcpA | GAAGAAGTTTGTAAAAGAAGAACACG | |
| MBvc11-tcpA | ccgatcAGAAAACCGGTCAAGAGGGTgatcgg | |
| Pvc64-1 ctxA | aattctaatacgactcactatagggAGAAGGTGGGTGCAGTGGCTATAACA | 151 |
| Pvc61-2 ctxA | TGATCATGCAAGAGGAACTCA | |
| MBvc-12 ctxA | ccgatcTTGTTAGGCACGATGATGGAgatcgg | |
| Pvc65-1 groEL | aattctaatacgactcactatagggATGATGTTGCCCACGCTAGA | 116 |
| Pvc66-2 groEL | GGTTATCGCTGCGGTAGAAG | |
| MBvc13-groEL | ccgatcCTGTCTGTACCTTGTGCCGAgatcgg | |
| Pvc69-1 toxR | aattctaatacgactcactatagggCGGAACCGTTTTGACGTATT | 139 |
| Pvc72-2 toxR | CTCGCAATGATTTGCATGAC | |
| MBvc14-toxR | ccgatcTTAACCCAAGCCATTTCGACgatcgg |
MB, molecular beacon. 5′ ends of molecular beacons were labeled with 6-carboxyfluorescein, and 3′ ends were labeled with dabsyl.
Nucleotide sequences of primers and molecular beacons are in capital letters; lowercase letters represent surrounding sequences.
T7 RNA polymerase promoter sequence.
Stem sequence.
RNA isolation.
Unless otherwise stated, the nucleic acids (DNA and RNA) used in all experiments were extracted from the toxigenic V. cholerae strain Cip 106855 O1 Inaba El Tor. RNA was isolated from cell pellets (as described above) according to the method for the NucliSens basic kit (bioMèrieux Ltd., Boxtel, The Netherlands) (8). Briefly, 900 μl of lysis buffer was added to a cell pellet from either 100 or 500 μl of bacterial cells grown as described above. Fifty microliters of silica suspension was added to the lysis buffer to bind the nucleic acids. The silica was washed with washing buffer two times and air dried. Nucleic acids were eluted with 50 or 100 μl of elution buffer, and the eluates were aliquoted and stored at −80°C. In some experiments, pure RNA was isolated using the RNeasy kit according to the procedure of the manufacturer (QIAGEN).
NASBA.
The NASBA reaction was carried out by using the NucliSens basic NASBA kit according to the guidelines of the manufacturer (BioMèrieux Ltd., Boxtel, The Netherlands) and the personnel at NorChip AS (personal communication). The NASBA assays were carried out with a total of 25 μl containing the reagent mix, primers (0.2 μM [each]), the molecular beacon (0.25 μM), KCl (70 mM), and the template (5 μl). Samples were incubated at 65°C for 4 min before the enzymes were added. The samples were then incubated at 41°C for 90 min. Each NASBA assay included a deionized H2O blank as a negative control. The RNA amplicons were identified in real-time and online by using specific fluorescent molecular beacon probes (Table 2). Real-time measurements with these probes were made with a SmartCycler thermocycler (Cepheid). The amplification of a specific NASBA product is indicated by the detection time in minutes (the t value). The t value represents the time point at which the fluorescence value crosses a fixed threshold that is 10 times the standard deviation of the baseline intensity. In some experiments, the RNA amplicons were analyzed by electrophoresis on a Bioanalyzer (Agilent Technology, Germany). The predicted sizes of the amplicons were between 102 and 151 nucleotides (Table 2). The expression of the selected target genes was tested at different time points during the logarithmic growth phase and in the late stationary growth phase. These results provided a basis for further experiments in which cells were grown to mid-logarithmic phase (about 4 × 107 CFU/ml) before harvesting. A variety of primers were tested, but only the optimal primer pair is shown in this report. Several isolates of Vibrio spp. and non-Vibrio spp. were used to test the specificities of the primers and the molecular beacons in NASBA (Tables 1 and 2).
Isolated nucleic acids were subjected to DNase or RNase treatment. Aliquots (100 μl) of nucleic acids were incubated with 10 U of RNase (EC 3.1.27.5; Sigma chemical Co.) in Tris (10 mM)-EDTA (1 mM) buffer, pH 7.5, at 37°C for 2 h or 120 U of DNase (EC 232-667-0; Sigma Chemical Co.) in sodium acetate (100 mM) buffer, pH 5.25, containing MgCl2 (5 mM) for 2 h at 25°C. These treatments were followed by phenol-chloroform extraction and NASBA.
Sensitivity.
The detection limit and the reproducibility were determined by amplifying samples from 10-fold serial dilutions of V. cholerae RNA. Each dilution, which corresponded to cell concentrations ranging from 5 to 5 × 105 CFU/ml, was tested in triplicate, and two separate experiments were performed. RNA was also isolated from a 10-fold-dilution series of V. cholerae cells ranging from 5 to 5 × 105 CFU/ml, and isolated nucleic acids were amplified by NASBA.
Analysis of spiked environmental water samples.
Environmental samples were collected from the Oslo fjord (marine) and from two different freshwater lakes. These samples were spiked with V. cholerae cells (Cip 106855 O1 Inaba El Tor), and RNA was isolated using the NucliSens basic kit. Briefly, 100 μl of V. cholerae cells was added to 0.9 ml of the environmental samples to final concentrations of 5 × 103 or 5 × 105 CFU/ml. The cell suspensions were centrifuged at 2,000 × g for 10 min, the pellets were suspended in 900 μl of lysis buffer, and RNA was isolated as described above. RNA isolated from one of the freshwater lake samples spiked with 5 × 105 CFU/ml was amplified using all five primer sets and molecular beacons. RNA from the other freshwater lake sample and from the seawater sample (spiked with 5 × 105 and 5 × 103 CFU/ml) was amplified using primers and molecular beacons targeting the groEL and tcpA genes. Spiked samples of PBS were used as positive controls for RNA isolation and amplification. Environmental water samples not spiked with V. cholerae cells were used as negative controls, and neither of the targets was amplified.
Viability testing.
Viable V. cholerae cells were investigated in comparison with nonviable cells. Samples of V. cholerae cells (5 × 105 CFU/ml) were subjected to the following treatments: autoclaving (121°C) for 20 min and further incubation at room temperature for 4 or 24 h and heating at 98°C for 20 min and further incubation at room temperature for 4 or 24 h. As controls, cells were kept on ice but otherwise treated similarly. The treatments were followed by the isolation of nucleic acids and subsequent analysis with all NASBA primers and beacons immediately after the treatment and after 4 h and 24 h. The viability was tested by plating samples onto TSB agar and incubating at 37°C for 24 h. Nucleic acids isolated from cells heated at 98°C were subjected to DNase and RNase treatment (as described above) to confirm that RNA and not DNA was amplified.
Real-time PCR.
Real-time PCR was performed using a LightCycler (Roche, Germany) real-time PCR instrument. The NASBA primers and molecular beacons were used for PCR. The real-time PCR assays were carried out as described previously (22) using a Lithos qPCR kit (Eurogentec, Belgium). Briefly, the reaction mixture contained deoxynucleoside triphosphates, Taq DNA polymerase and reaction buffer, MgCl2 (4.5 mM), primers (1 μM [each]), and the probe (0.75 μM) in a final volume of 20 μl. The PCR program was as follows: initial denaturation at 95°C for 3 min and then 40 cycles at 95°C for 5 s, 58°C for 10 s, and 72°C for 15 s.
RESULTS
NASBA.
Five different genetic markers (groEL, toxR, ctxA, tcpA, and hlyA) were used for the specific detection of V. cholerae. The selection of target genes was based on previous work (41, 48). The DNA sequences of the groEL, ctxA, tcpA, toxR, and hlyA markers showed no significant similarity, with one exception, to sequences of other genetic regions as determined using BLASTN. The toxR genetic target was 98% similar to the Serratia marcescens toxR gene.
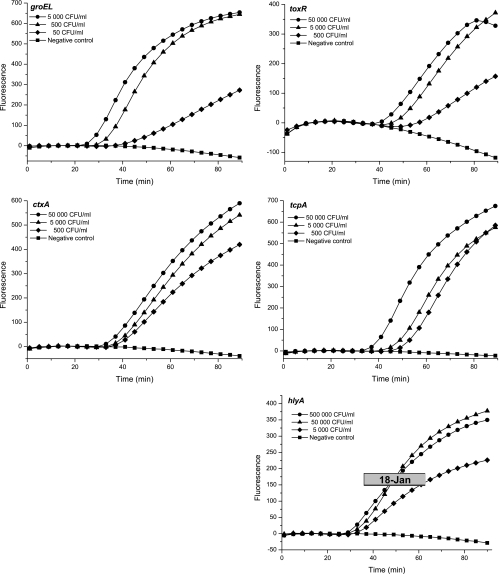
The five different molecular beacons were evaluated using serial dilutions of nucleic acids isolated from V. cholerae cells. All target sequences were amplified by the corresponding primers, and the amplicons were detected by molecular beacons as a significant increase in fluorescence (Fig. 1). The results were confirmed by the amplification of an RNA sample, containing no DNA, that had been isolated by using the RNeasy kit (results not shown). The sensitivity of the single-target NASBA assays was evaluated using 10-fold dilutions of nucleic acids extracted from V. cholerae cells. Two separate assays were performed, and each assay was run in triplicate. Consistent detection (Table 3) was obtained at 50 CFU/ml for groEL and tcpA, 500 CFU/ml for toxR and ctxA, and 5 × 105 CFU/ml for hlyA. As shown in Fig. 1 and Table 3, lower concentrations were occasionally detected, but these levels of detection could not be reproduced in all NASBA experiments (Table 3).
FIG. 1.
Real-time NASBA of V. cholerae RNA. The five different molecular beacons were evaluated by using a 10-fold dilution series of RNA isolated from V. cholerae cells (Cip 106855 O1 Inaba El Tor). Data shown are representative of results from multiple experiments. Fluorescence is measured in relative units.
TABLE 3.
Sensitivity and reproducibility of the single-target NASBA assaysa
| Target | No. of specific NASBA products detected/no. of total reactions for concn (CFU/ml) of:
|
||||||
|---|---|---|---|---|---|---|---|
| 5 | 50 | 250 | 500 | 5 × 103 | 5 × 104 | 5 × 105 | |
| groEL | 4/6 | 6/6 | NT | 6/6 | 3/3 | 3/3 | NT |
| tcpA | 2/6 | 6/6 | NT | 6/6 | 6/6 | NT | NT |
| ctxA | No signal | 3/6 | 3/3 | 6/6 | 6/6 | 6/6 | NT |
| toxR | No signal | 5/6 | 3/3 | 6/6 | 6/6 | 6/6 | NT |
| hlyA | No signal | No signal | No signal | No signal | 1/6 | 3/6 | 6/6 |
Two separate NASBA experiments, each run in triplicate, were performed. NT, not tested.
The sensitivity of NASBA detection is also dependent on the yield of RNA extracted from cells. The groEL and tcpA targets were easily detected even when RNA was isolated from V. cholerae cultures containing only 50 CFU/ml. This result indicates that the NucliSens basic kit was able to effectively isolate RNA even from a low cell concentration. Furthermore, a prerequisite for NASBA detection is the expression of the selected target gene. The groEL, toxR, ctxA, tcpA, and hlyA targets were detected in all samples collected from early logarithmic to late stationary phase, indicating that these targets are all expressed during these growth phases.
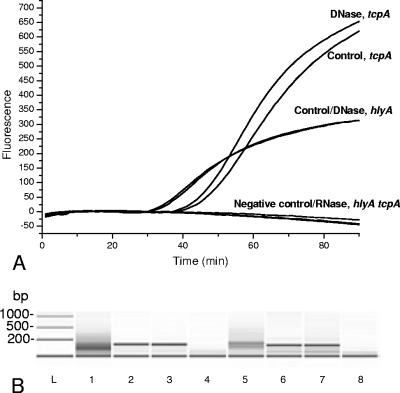
NASBA is specifically designed to amplify RNA. However, it has been shown that DNA can also be amplified (19, 42). To confirm that the origin of the NASBA signal was RNA, the nucleic acids were subjected to RNase and DNase treatment as described in Materials and Methods. In the presence of RNase, no targets were amplified, whereas DNase treatment did not affect NASBA. This result was further confirmed with the electrophoresis of all amplicons (Bioanalyzer). These results are illustrated for the hlyA and tcpA targets in Fig. 2. Similar results were obtained for all other targets. However, for toxR, a slight decrease in the level of RNA amplicons was observed after DNase treatment. The decrease in RNA did not affect the t value, but a small decrease in the fluorescence intensity was observed for toxR after DNase treatment. These results show that V. cholerae RNA was detected by NASBA in a DNA-contaminated background.
FIG. 2.
Detection of RNA isolated from V. cholerae cells (Cip 106855 O1 Inaba El Tor) after RNase and DNase treatment of the nucleic acids isolated with the NucliSens basic kit. Fluorescence is measured in relative units. (A) Real-time NASBA of the hlyA and tcpA gene targets. (B) Electrophoresis (Bioanalyzer) of the hlyA and tcpA real-time NASBA products. Lanes: L, molecular weight standard; 1, hlyA negative control; 2, hlyA control; 3, hlyA DNase; 4, hlyA RNase; 5, tcpA negative control; 6, tcpA control; 7, tcpA DNase; 8, tcpA RNase.
Specificity.
The specificity of the NASBA assay was determined by amplifying specific RNA targets from several Vibrio spp. strains, those of V. cholerae (27 strains), V. mimicus (3 strains), V. parahaemolyticus (3 strains), V. alginolyticus (2 strains), V. fluvialis (1 strain), and V. metschnikovii (1 strain), and from strains of other bacterial species, B. cereus (2 strains), S. enterica (2 strains), and E. coli (2 strains). The results are summarized in Table 1. In general, the ctxA and tcpA RNA was detected in all O1 (clinical) isolates of V. cholerae but not in the environmental non-O1 isolates. Although the ctxA and tcpA targets were detected in the VC 4/57 O1 environmental isolate and in the VC 5/47 strain of the O2 serogroup (Table 1), these potential target sequences were amplified at a late stage of the NASBA assay with high t values and low fluorescence intensities. The toxR, hlyA, and groEL gene targets were detected in all V. cholerae strains examined, and no fluorescence signal was detected for the other Vibrio spp. However, to our surprise, in two of three separate experiments the groEL target was detected in RNA from S. enterica (ATCC 13076). The t value was 49 min for S. enterica compared to 20 min for the V. cholerae groEL target. The maximum fluorescence levels (F) were 100 and 600 relative units, respectively. The ctxA target in V. mimicus (VM034) was also amplified. In this case, the sequence was amplified at t of 59 min and F of 100 relative units compared to t of 34 min and F of 600 relative units for V. cholerae (Cip 106855). Electrophoresis of the nonspecific RNA amplicons from S. enterica and V. mimicus showed that these products were larger than the specific products from V. cholerae (not shown). Furthermore, real-time PCR using the same molecular beacon probes was also used to verify the specificity of the groEL and ctxA targets. These DNA targets were not amplified in the S. enterica (ATCC 13076) and V. mimicus (VM034) strains. In conclusion, specific NASBA detection of V. cholerae was obtained using a combination of groEL and toxR as general primers and beacons and the ctxA and tcpA primers and beacons for detecting toxigenic strains.
Spiked environmental water samples.
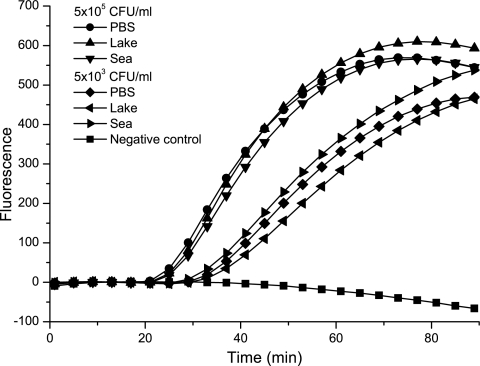
Highly sensitive detection of V. cholerae from environmental samples is dependent on efficient RNA extraction and the removal of potential NASBA inhibitors, as well as other factors. RNA was extracted and amplified from three different water samples, one from seawater and two from different freshwater lakes, spiked with V. cholerae cells. The genetic targets were specifically detected in V. cholerae, and no inhibition was observed. The results are illustrated by the amplification of the groEL target from seawater, freshwater, and a positive control sample (PBS) (Fig. 3). Similar results were obtained with all environmental samples and for all genetic targets (Table 4).
FIG. 3.
Real-time NASBA detection (groEL) of V. cholerae RNA isolated from environmental water samples spiked with V. cholerae cells (Cip 106855 O1 Inaba El Tor). Seawater, freshwater, and a positive control (PBS) were spiked with 5 × 103 and 5 × 105 CFU/ml, and RNA was isolated using the NucliSens basic kit. Fluorescence is measured in relative units.
TABLE 4.
Detection of V. cholerae cells in spiked water samplesa
| Sample | Amplification of genetic target from V. cholerae cells at:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 × 103 CFU/ml
|
5 × 105 CFU/ml
|
|||||||||
| groEL | tcpA | ctxA | hlyA | toxR | groEL | tcpA | ctxA | hlyA | toxR | |
| Seawater | + | + | NT | NT | NT | + | + | NT | NT | NT |
| Freshwater lake 1 | + | + | NT | NT | NT | + | + | NT | NT | NT |
| Freshwater lake 2 | NT | NT | NT | NT | NT | + | + | + | + | + |
| PBS | + | + | NT | NT | NT | + | + | + | + | + |
+, specific amplified product detected; NT, not tested. Environmental water samples without spiking had no effect on amplification.
Viability tests.
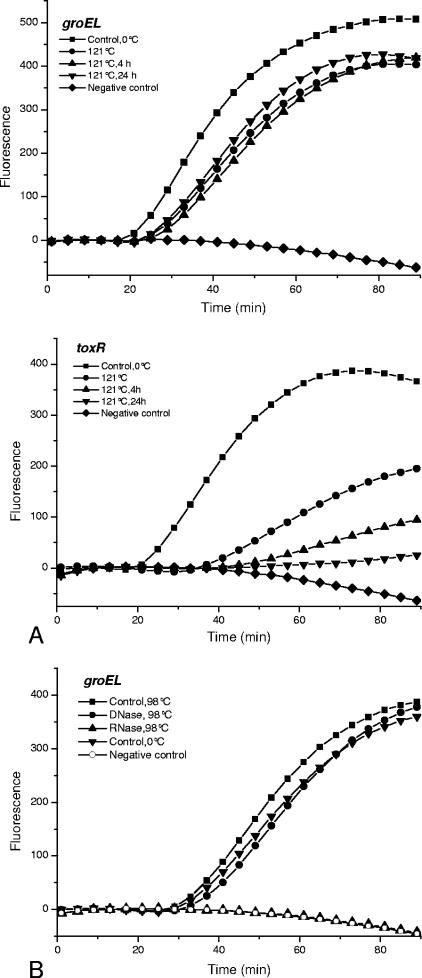
Previous studies have shown that NASBA may be used to detect viable cells by using mRNA as the amplification target. In order to verify this possibility, V. cholerae cells were subjected to heat treatments at 98°C and 121°C, followed by further incubation for 4 or 24 h at room temperature. Cell death after these treatments was confirmed by CFU enumeration. The ctxA, hlyA, and toxR targets were not detected after treatment at 121°C and further incubation at 24 h. After 4 h, no hlyA and ctxA amplicons were detected, but toxR was still detectable. In contrast, the groEL and tcpA targets were detected in the heat-killed (121°C) cells. These results are illustrated for the toxR and groEL targets in Fig. 4A. Heating of the cells at 98°C was less effective, resulting in the detection of all targets, and treatment with RNase and DNase confirmed that RNA and not DNA was amplified. These results are illustrated for groEL in Fig. 4B. These results indicate that care must be taken when genetic targets are selected if NASBA is to be used for the detection of viable cells.
FIG. 4.
Real-time NASBA of RNA isolated from heat-treated nonviable V. cholerae cells (Cip 106855 O1 Inaba El Tor). (A) NASBA of the toxR and groEL genes in cells treated at 121°C and further incubated for 4 and 24 h at room temperature. (B) NASBA of the groEL gene target in cells treated at 98°C and further incubated for 24 h at room temperature. To confirm that RNA and not DNA was amplified, the nucleic acids were treated with RNase and DNase as described in Materials and Methods. No amplification was detected after RNase treatment. Fluorescence is measured in relative units.
DISCUSSION
There is a need for rapid, accurate, and sensitive detection of target organisms responsible for food and water poisoning. To our knowledge, this paper describes for the first time a sensitive multitarget real-time NASBA application for the specific detection of V. cholerae cells in water.
In general, NASBA is a sensitive method, and more than 109 copies of RNA can be amplified in the assay in 90 min (14). In the present study, as little as 50 CFU/ml was detected using the groEL or tcpA primers and molecular beacons. The groEL marker is a general marker for detecting all V. cholerae strains. The tcpA marker is specific for the detection of toxigenic strains. The sensitivity of the presented method was in agreement with previous NASBA results for other bacterial species. Min and Baeumner (38) were able to detect 40 E. coli cells/ml of water, and as few as 10 spores of B. anthracis could be detected (4). In the latter case, the spores were inoculated into a culture medium for 30 min in order to germinate the spores and express target RNA. In a recent study, the levels of toxigenic V. cholerae cells in water in Mathbaria, Bangladesh, were monitored. Seasonal variation was observed, and the concentration of V. cholerae cells varied from <10 to 3.4 × 107 cells/liter (1). In the NASBA assay in the present study, 5 × 104 CFU/liter was detected. Preliminary studies in our laboratory indicate that 5 × 104 CFU/ml corresponds to 1 × 106 cells/liter. The sensitivity might be further increased by the filtration of large volumes of water, which has not been done in this study. The infectious dose of V. cholerae from water is assumed to be 103 to 106 bacteria (44). Thus, such doses may be detected using the present NASBA method either directly or after filtration of the water.
A report of the use of molecular beacons for the multiplex real-time PCR detection of V. cholerae cells has recently been published, stating a detection limit of 5 × 103 CFU/ml (25). The sensitivity of our assay proved to be 100-fold higher (50 CFU/ml). Previous reports revealed that real-time PCR using TaqMan probes or SYBR green can detect 10 to 106 CFU of V. cholerae/ml (24, 36). It is well known that the nucleotide sequences and the binding efficiencies of primers and probes for the target strand are of importance in obtaining high sensitivity and specificity for real-time PCR and NASBA assays. The kinetics of the NASBA reaction are mainly determined by the efficiency of primer binding and the extent of nonspecific product synthesis due to mispriming events (20). In this study, the sensitivity of the amplification of hlyA was lower than that of the other targets. This might be due to primer or probe construction or a lower level of expression of the hlyA marker compared to the other targets.
Theoretically, the possibility that the groEL primers and molecular beacon can amplify and detect nontarget strains is low, confirmed by BLAST analysis. Surprisingly, the amplification of RNA isolated from the S. enterica ATCC 13076 strain occurred in two out of three separate experiments. Sequence alignments of the groEL target gene of V. cholerae (>gi15640032:2832629-2834254) and the groEL gene of S. enterica (gi7527386) showed approximately 45% similarity at the DNA level, but the sequence of the groEL molecular beacon probe appeared to be unique (BLASTN analysis). Therefore, the detection of groEL in S. enterica was not expected. Furthermore, the amplification of S. enterica was not reproducible. Also, in a few experiments a background signal in the negative control was achieved with the groEL molecular beacon probe. These two observations might be due to the binding of the molecular beacon to primers (19). However, S. enterica ATCC 13076 was not detected at all using real-time PCR and molecular beacon probe detection.
Pathogenicity is dependent on the expression of virulence genes in a microorganism. The ctxA and tcpA genes are known to play a crucial role in maintaining the virulence of V. cholerae. These genes are usually associated with clinical strains of the O1 and O139 serogroups. However, outbreaks of cholera caused by non-O1 and non-O139 have also been reported (18, 45), and some of these strains produce CT or CT-like toxins (10). Interestingly, the ctxA and tcpA genes were detected by NASBA in a strain belonging to the O2 serogroup (VC 5/47). However, the t value was high and the fluorescence low. This result was most likely due to mispriming or a low yield of RNA. PCR studies have also revealed the presence of the ctxA and tcpA genes in various non-O1 and non-O139 serogroups of environmental V. cholerae strains (10). An environmental O1 isolate (VC 4/57) was also detected in this study. As said, the t value was high and the increase in the fluorescence signal was low and most likely due to mispriming or the level of expression. Furthermore, isolates of V. mimicus that express the two main virulence factors (CT and TCP) of V. cholerae have been identified (9), and this finding might explain why the V. mimicus (VM034) strain was detected using the ctxA primers and a molecular beacon. Thus, CT or CT-like proteins are associated with non-O1 and non-O139 serogroups of V. cholerae and strains of V. mimicus as well. Amplification of the ctxA gene target in environmental samples by NASBA indicates the detection of a potential pathogen. The potential nonspecific detection in environmental samples of, e.g., ctxA in V. mimicus or toxR in S. marcescens, found in water, soil, insects, plants, and vegetables (23), is not a problem since the specificity of the presented NASBA assay is based on the use of several genetic targets. Using a combination of groEL and tcpA, the presented multitarget NASBA assay is specific for the detection of V. cholerae cells. For the monitoring of strains expressing virulence factors, the ctxA and tcpA targets can be used. A direct comparison of the specificity of NASBA and real-time PCR using molecular beacons is difficult since different targets and different strains of bacteria were used. A real-time PCR assay using molecular beacons used four different genetic targets in a multiplex assay (rtxA, epsM, ompW, and tcpA) in order to detect V. cholerae cells; in this assay, V. mimicus was detected nonspecifically through ompW (25). The tcpA marker used in PCR was designed to detect the toxigenic El Tor strains. However, in one of these strains no amplification was detected (25). To our knowledge, these two studies are the only studies describing the use of molecular beacon probes for the detection of V. cholerae.
Using NASBA for analyzing environmental samples requires efficient methods for sample preparation since the samples generally contain substances that inhibit NASBA (16). V. cholerae was detected by NASBA in spiked environmental seawater and freshwater samples. Potential inhibitors were effectively removed, and the whole analysis including RNA extraction and NASBA was completed within 3 h. If the samples contain large amounts of inhibitory substances, the extraction methods might require modifications. In contrast, conventional culture methods and subsequent bacterial identification are time-consuming and require several days to complete (21). Vibrio spp. in the environment are known to be in a VBNC state, a state that is described as a survival mechanism of bacteria facing environmental stress conditions (28). Preliminary results show that V. cholerae cells in the VBNC state could be detected by NASBA.
In principle, the presence of RNA in bacterial cells may serve as an indicator for viable cells (32), and mRNA species are supposed to have an average half-life of only minutes in metabolizing cells (2). However, in several studies it has been shown that mRNA species may persist in a detectable form for many hours after cell death (6, 46, 47). The persistence of mRNA species may vary depending on the method of cell death (46, 47) and the physiological state of the cell population before killing. In another study, only viable cells of E. coli were detected using NASBA (38). Other studies have also demonstrated that the selection of the genetic target is important for providing good correlations with cell viability (40, 55). This was also the case in this study. A correlation between cell viability and NASBA was demonstrated for the ctxA, hlyA, and toxR targets, whereas such correlation was not demonstrated for the groEL and tcpA targets. Our results therefore indicate that care must be taken in using NASBA for viability studies. This finding also highlights the need for increased knowledge of the persistence and decay characteristics of the potential mRNA targets. False-positive results due to the amplification of RNA from dead cells can be eliminated by preenrichment in a selective medium, and the analysis time might be shortened from days (culture methods) to hours (52). This strategy also solves the problems with NASBA inhibitors contained in environmental samples.
In conclusion, the described multitarget NASBA assay for the detection of V. cholerae is a novel, sensitive, specific, and rapid method in which nucleic acid extraction and NASBA can be completed in less than 3 h. The NASBA assay is run at a single temperature, and a thermocycler is not required. The advantages, including the use of a crude RNA extract, have a potential for being further improved for field applications. Environmental water samples spiked with V. cholerae cells were used to demonstrate that the combination of a rapid method for the extraction of nucleic acids (DNA and RNA) and NASBA could be a promising strategy for the rapid and sensitive detection of V. cholerae. The monitoring of particular strains expressing virulence factors known to cause human disease is of great value for hazard evaluation of water. Preliminary studies also show that the NASBA method has potential as a detection tool for VBNC cells isolated from water samples.
The groEL and toxR targets are general markers and detected all V. cholerae strains. The sensitivity of the groEL assay was 10-fold higher than that of the toxR assay. In order to detect toxigenic strains, we suggest that the tcpA and ctxA markers be used. However, the sensitivity of the tcpA assay was 10-fold higher than that of the ctxA assay. A combination of the groEL, toxR, tcpA, and ctxA markers provides maximum specificity for the detection of all V. cholerae strains.
Acknowledgments
Vibrio strains were kindly provided by L. M. Rørvik, Norwegian School of Veterinary Science, Department of Food Safety and Infection Biology; J. Lassen, Norwegian Institute of Public Health, Department of Food-borne Infections; and A. Carotolli, Department of Infectious Parasitic and Immuno-mediated Diseases, Istituto Superiore di Sanità, Italy. Environmental water samples were kindly provided by Anne Marie Holtet, NorAnalyse AS, Lillestrøm, Norway. We are also grateful to NorChip AS for introducing us to the NASBA technique.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Alam, M., M. Sultana, G. B. Nair, R. B. Sack, D. A. Sack, A. K. Siddique, A. Ali, A. Huq, and R. R. Colwell. 2006. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl. Environ. Microbiol. 72:2849-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arraiano, C. M., S. D. Yancey, and S. R. Kushner. 1988. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 170:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baeumner, A. J., M. C. Humiston, R. A. Montaga, and R. A. Durst. 2001. Detection of viable Cryptosporidium parvum following nucleic acid sequence-based amplification. Anal. Chem. 73:1176-1180. [DOI] [PubMed] [Google Scholar]
- 4.Baeumner, A. J., B. Leonard, J. McElwee, and R. A. Montagna. 2004. A rapid biosensor for viable B. anthracis spores. Anal. Bioanal. Chem. 380:15-23. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, A., Ø. Østensvik, M. Florvag, Ø. Ørmen, and L. M. Rørvik. 2006. Occurrence of Vibrio parahaemolyticus, V. cholerae, and V. vulnificus in Norwegian blue mussels (Mytilus edulis). Appl. Environ. Microbiol. 72:3058-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birch, L., C. E. Dawson, J. H. Cornett, and J. T. Keer. 2001. A comparison of nucleic acid amplification techniques for the assessment of bacterial viability. Lett. Appl. Microbiol. 33:296-301. [DOI] [PubMed] [Google Scholar]
- 7.Blais, B. W., G. Turner, R. Sookanan, and L. T. Malek. 1997. A nucleic acid sequence-based amplification system for detection of Listeria monocytogenes hlyA sequences. Appl. Environ. Microbiol. 63:310-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boom, R., C. J. A. Sol, E. M. M. Salimans, C. L. Hansen, P. M. E. Wertheim-van Dillen, and J. van der Norordaa. 1990. Rapid and simple method for the purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd, E. F., K. E. Moyer, L. Shi, and M. K. Waldor. 2000. Infectious CTXPhi and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68:1507-15134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cholera Working Group. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. [PubMed] [Google Scholar]
- 12.Colwell, R. R., J. Kaper, and S. W. Joseph. 1977. Vibrio cholerae and Vibrio parahaemolyticus and other vibrios: occurrence and distribution in Chesapeake Bay. Science 198:394-396. [PubMed] [Google Scholar]
- 13.Colwell, R. R., R. J. Seidler, J. Kaper, S. W. Joseph, S. Garges, H. Lockman, D. Maneval, H. Bradford, N. Roberts, E. Remmers, I. Huq, and A. Huq. 1981. Occurrence of Vibrio cholerae serotype O1 in Maryland and Louisiana estuaries. Appl. Environ. Microbiol. 41:555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Compton, J. 1991. Nucleic acid sequence-based amplification. Nature 350:91-92. [DOI] [PubMed] [Google Scholar]
- 15.Cook, N., J. Ellison, A. S. Kurdziel, S. Simpkins, and J. P. Hays. 2002. A nucleic acid sequence-based amplification method to detect Salmonella enterica serotype enteritidis strain PT4 in liquid whole egg. J. Food Prot. 65:1177-1178. [DOI] [PubMed] [Google Scholar]
- 16.Cook, N. 2003. The use of NASBA for the detection of microbial pathogens in food and environmental samples. J. Microbiol. Methods 53:165-174. [DOI] [PubMed] [Google Scholar]
- 17.Coutard, F., M. Pommepuy, S. Loaec, and D. Hervio-Heath. 2005. mRNA detection by reverse transcription-PCR for monitoring viability and potential virulence in a pathogenic strain of Vibrio parahaemolyticus in viable but nonculturable state. J. Appl. Microbiol. 98:951-961. [DOI] [PubMed] [Google Scholar]
- 18.Dalsgaard, A., M. J. Albert, D. N. Taylor, T. Shimada, R. Meza, O. Serichantalergs, and P. Echeverria. 1995. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 33:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deiman, B., P. vanAarle, and P. Sillekens. 2002. Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 20:163-179. [DOI] [PubMed] [Google Scholar]
- 20.Fahy, E., D. Y. Kwoh, and T. R. Gingeras. 1991. Self-sustained sequence replication (3SR): an isothermal transcription-based amplification system alternative to PCR. PCR Methods Appl. 1:25-33. [DOI] [PubMed] [Google Scholar]
- 21.Farmer, J. J., III, and F. W. Hickman-Brenner. 1992. The genera Vibrio and Photobacterium, p. 2952-3011. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The procaryotes. A handbook of the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed. Springer-Verlag, New York, NY.
- 22.Fykse, E. M., J. S. Olsen, and G. Skogan. 2003. Application of sonication to release DNA from Bacillus cereus for quantitative detection by real-time PCR. J. Microbiol. Methods 55:1-10. [DOI] [PubMed] [Google Scholar]
- 23.Grimont, F., and P. A. D. Grimont. 1992. The genus Serratia, p. 2822-2862. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The procaryotes. A handbook of the biology of bacteria: ecophysiology, isolation, identification, applications, 2nd ed. Springer-Verlag, New York, NY.
- 24.Gubala, A. J. 2006. Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 65:278-293. [DOI] [PubMed] [Google Scholar]
- 25.Gubala, A. J., and D. F. Proll. 2006. Molecular-beacon multiplex real-time PCR assay for detection of Vibrio cholerae. Appl. Environ. Microbiol. 72:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes, J. M., D. G. Hollis, E. J. Gangarosa, and R. E. Weaver. 1978. Non-cholera vibrio infections in the United States. Clinical, epidemiologic, and laboratory features. Ann. Intern. Med. 88:602-606. [DOI] [PubMed] [Google Scholar]
- 28.Huq, A., and R. R. Colwell. 1996. A microbiological paradox: viable but not culturable bacteria with special reference to Vibrio cholerae. J. Food Prot. 59:96-101. [DOI] [PubMed] [Google Scholar]
- 29.Ichinose, Y. K., N. Yamamoto, M. J. Nakasone, T. Tanabe, T. Takeda, T. Miwatani, and M. Iwanga. 1987. Enterotoxicity of El Tor-like hemolysin of non-O1 Vibrio cholerae. Infect. Immun. 55:1090-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 32.Keer, J. T., and L. Birch. 2003. Molecular methods for assessment of bacterial viability. J. Microbiol. Methods 53:175-183. [DOI] [PubMed] [Google Scholar]
- 33.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 34.Leone, G., H. van Schijndel, B. van Gemen, F. R. Kramer, and C. D. Schen. 1998. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 26:2150-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipp, E. K., I. N. G. Rivera, A. I. Gil, E. M. Espeland, N. Choopun, V. R. Louis, E. Russek-Cohen, A. Huq, and R. R. Colwell. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69:3676-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyon, W. J. 2001. TaqMan PCR for detection of Vibrio cholerae O1, O139, non-O1, and non-O139 in pure cultures, raw oysters, and synthetic seawater. Appl. Environ. Microbiol. 67:4685-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 30:271-279. [DOI] [PubMed] [Google Scholar]
- 38.Min, J., and J. Baeumner. 2002. Highly sensitive and specific detection of viable Escherichia coli in drinking water. Anal. Biochem. 303:186-193. [DOI] [PubMed] [Google Scholar]
- 39.Morris, J. G., Jr., R. Wilson, B. R. Davis, I. K. Wachsmuth, C. F. Riddle, H. G. Wathen, R. A. Pollard, and P. A. Blake. 1981. Non-O group 1 Vibrio cholerae gastroenteritis in the United States: clinical, epidemiologic, and laboratory characteristics of sporadic cases. Ann. Intern. Med. 94:656-658. [DOI] [PubMed] [Google Scholar]
- 40.Norton, D. M., and C. A. Batt. 1999. Detection of viable Listeria monocytogenes with a 5′ nuclease PCR assay. Appl. Environ. Microbiol. 65:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Làzaro, D., J. Lloyd, J. Ikonomopoulos, M. Pla, and N. Cook. 2004. Unexpected detection of DNA by nucleic acid sequence-based amplification technique. Mol. Cell. Probes 18:251-253. [DOI] [PubMed] [Google Scholar]
- 43.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 44.Seas, C., and E. Gotuzzo. 2005. Vibrio cholerae, p. 2536-2544. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, 6th ed., vol. 2. Elsevier Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 45.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheridan, G. E. C., E. A. Szabo, and B. M. Mackey. 1999. Effect of post-treatment holding conditions on detection of tufA mRNA in ethanol-treated Escherichia coli; implications for RT-PCR-based indirect viability tests. Lett. Appl. Microbiol. 29:375-379. [DOI] [PubMed] [Google Scholar]
- 48.Singh, D. V., M. H. Matte, G. R. Matte, S. Jiang, F. Sabeena, B. N. Shukla, S. C. Sanyal, A. Huq, and R. R. Colwell. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh, D. V., S. R. Isac, and R. R. Colwell. 2002. Development of a hexaplex PCR assay for rapid detection of virulence and regulatory genes in Vibrio cholerae and Vibrio mimicus. J. Clin. Microbiol. 40:4321-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyagi, S., D. Bratu, and F. R. Kramer. 1998. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 52.Uyttendaele, M., R. Schukkink, B. vanGemen, and J. Debevere. 1995. Detection of Campylobacter jejuni added to foods by using a combined selective enrichment and nucleic-acid sequence based amplification (NASBA). Appl. Environ. Microbiol. 61:1341-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto, K., M. Al-Omani, T. Honda, Y. Takeda, and T. Miwatani. 1984. Non-O1 Vibrio cholerae hemolysin: purification, partial characterization, and immunological relatedness to El Tor hemolysin. Infect. Immun. 45:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaron, S., and K. R. Matthews. 2002. A reverse transcriptase-polymerase chain reaction assay for detection of viable Esherichia coli O157:H7: investigation of specific target genes. J. Appl. Microbiol. 92:633-640. [DOI] [PubMed] [Google Scholar]