Abstract
Cryptococcus gattii has recently emerged as a primary pathogen of humans and wild and domesticated animals in British Columbia, particularly on Vancouver Island. C. gattii infections are typically infections of the pulmonary and/or the central nervous system, and the incidence of infection in British Columbia is currently the highest reported globally. Prior to this emergence, the environmental distribution of and the extent of colonization by C. gattii in British Columbia were unknown. We characterized the environmental sources and potential determinants of colonization in British Columbia. C. gattii was isolated from tree surfaces, soil, air, freshwater, and seawater, and no seasonal prevalence was observed. The C. gattii concentrations in air samples were significantly higher during the warm, dry summer months, although potentially infectious propagules (<3.3 μm in diameter) were present throughout the year. Positive samples were obtained from many different areas of British Columbia, and some locations were colonization “hot spots.” C. gattii was generally isolated from acidic soil, and geographic differences in soil pH may influence the extent of colonization. C. gattii soil colonization also was associated with low moisture and low organic carbon contents. Most of the C. gattii isolates recovered belonged to the VGIIa genetic subtype; however, sympatric colonization by the VGIIb strain was observed at most locations. At one sampling site, VGIIa, VGIIb, VGI, and the Cryptococcus neoformans serotype AD hybrid all were coisolated. Our findings indicate extensive colonization by C. gattii within British Columbia and highlight an expansion of the ecological niche of this pathogen.
The basidiomycete fungal pathogens Cryptococcus neoformans and Cryptococcus gattii cause life-threatening diseases of the pulmonary and central nervous systems in humans and other animals. In contrast to C. neoformans, which has a global distribution, C. gattii has until recently been regarded as a tropical and subtropical organism (37, 38). Immunocompetent individuals infected with C. gattii living on the east coast of southern Vancouver Island, British Columbia, were identified in 1999, but whether C. gattii significantly colonized this region prior to that time is not known (32). The incidence of C. gattii infection in British Columbia is currently the highest reported globally (36 cases/million people/year) (45), and the median incubation period for human C. gattii infection in this region has been estimated at 6 to 7 months, with a range of 2 to 11 months (44).
Most of the C. gattii cases in British Columbia occurred in individuals who resided within or reported contact with geographic areas that are part of the coastal Douglas fir (CDF) biogeoclimatic zone. The CDF is one of 14 defined terrestrial ecosystems in British Columbia (47) and includes elevations below 150 m in a limited area of southeast Vancouver Island. Located in the rain shadow of the mountains on Vancouver Island and the Olympic peninsula of northwest Washington, the CDF also includes the southern Gulf Islands and a small part of the lower mainland of British Columbia. The CDF typically has warm, dry summers and mild, wet winters. Soils in the area are primarily brunisolic, mineral rich, and well drained. The CDF is bordered by the coastal western hemlock xeric maritime (CWHxm) zone, previously classified as a wet subzone of the CDF. The CWHxm is also characterized by low elevation, low precipitation, mild temperatures, and dry, fine-textured soil (47). Despite nomenclatural specificity to British Columbia, biogeoclimatic areas similar to the CDF and CWHxm also occur in the Pacific Northwest of the United States, including the San Juan Islands, the Puget Trough of Washington, and the Willamette Valley of Oregon (19).
Studies of the ecological niches of C. gattii were pioneered by Australian researchers. Ellis and Pfeiffer first isolated C. gattii from Eucalyptus camaldulensis (river red gum) in 1989, the distribution of which broadly corresponded to that of clinical cases of infection (15). This report was soon followed by the isolation of the fungus from Eucalyptus tereticornis (forest red gum) (52). The tree host range in Australia has since been expanded to include Syncarpia glomulifera (turpentine gum), Eucalyptus microcorys (tallowwood), Eucalyptus grandis (flooded gum), Eucalyptus rudis (West Australian flooded gum), Eucalyptus gomphocephala (tuart), and Eucalyptus blakelyi (Blakely's red gum) from all areas of the country (35, 52a, 61).
C. gattii has also been isolated from eucalypts in California, Colombia, Argentina, Brazil, India, and Egypt (7, 10, 16, 25, 28, 46, 49, 51), as well as Ficus soatensis, Croton bogotanus, Croton funckianus, Coussapoa sp., Cupressus lusitanica (Mexican cypress), Pinus radiata (Monterey pine), Acacia decurrens (black wattle), and Terminalia catappa (almond tree) in Colombia (5, 25) and Moquilea tomentosa (pottery tree), Cassia grandis (pink shower tree), and an Amazonian “quina-quina” tree (Guettarda acreana) in Brazil (18, 41). The presence of C. gattii outside of Australia has been speculatively attributed to the export of eucalypts from Australia in the form of seeds or cuttings (7, 14, 20, 21, 64).
Environmental sampling for C. gattii in British Columbia from October 2001 to June 2002 was described previously (32). In that study, more than 700 samples were collected at 16 sites across Vancouver Island, but only 62 samples from two sites were positive. The environmental isolates from these samples have been extensively characterized (20, 22, 31, 32) but may not be representative of the overall British Columbian C. gattii population because they were obtained within a very limited geographic area and time frame. In fact, most of these isolates were collected on a single day, and many were associated with a single tree. Our present study describes a large-scale evaluation of C. gattii isolates collected from environmental sources at many locations on Vancouver Island and in the Pacific Northwest over more than 4 years.
Genetic characterization of C. gattii isolates from many areas of the world has revealed a heterogeneous taxon comprising four major molecular types, designated VGI, VGII, VGIII, and VGIV (29, 48, 55). British Columbia C. gattii isolates from the earlier study were mostly (95%) of the VGII molecular type and were grouped into subtypes VGIIa and VGIIb at a ratio of approximately 9:1 (32). The VGII molecular type also has been identified in other areas of the world, including parts of South America and Australia and sporadically in the United States (2, 4, 17, 20, 29, 31-33, 48, 60). C. gattii of the VGI molecular type has caused at least six human and animal infections in British Columbia and was recently obtained from two environmental samples. However, these environmental isolates were genetically distinct from clinical VGI isolates in British Columbia (31; S. E. Kidd and K. H. Bartlett, unpublished data). The low representation of VGI in clinical and environmental sources in British Columbia is interesting given that VGI is the dominant C. gattii molecular type in other areas of the world (1, 8, 9, 29, 48, 55).
Based on multilocus sequence typing (MLST) profiles, British Columbian VGIIa strains and other C. gattii isolates from North America are identical, as are the VGIIb strain and isolates from Australia and Southeast Asia, thus providing clues to the potential origins of C. gattii in British Columbia (20, 31). The existence of a single VGIIa clinical isolate (NIH444) from Seattle ca. 1971 raised the possibility that VGIIa has been present in the Pacific Northwest region of North America for several decades (20, 31, 32). However, the residence and travel history of the patient from which this strain was isolated is not known, and no environmental samples were obtained in association with this case.
Our objective in this study was to isolate and characterize C. gattii samples obtained over an extended period of time from many areas of Vancouver Island, the mainland of British Columbia, and parts of the northwestern United States. The incidence of C. gattii in the Pacific Northwest is unprecedented given the temperate climate. Therefore, a thorough understanding of the C. gattii distribution and areas of endemicity in this region may provide insight into the ecological adaptability of C. gattii.
MATERIALS AND METHODS
Environmental sampling techniques.
We sampled areas with and without an associated suspicion of colonization, although most samples were taken in the towns or cities with human or animal C. gattii cases, including sites on properties where the cases occurred and in nearby parks and wooded areas. In addition, some areas not suspected to be colonized by C. gattii (i.e., no reported cases) were sampled, in an attempt both to identify areas with the potential for infection and to assemble a comprehensive sampling library for the CDF and CWHxm regions. Many areas have been sampled multiple times since 2001 to determine if there is a seasonal pattern to the presence of C. gattii in the environment. Except in the case of such longitudinal analyses, all data presented in this paper are from results for only the first swab, soil, and air samples collected at each sampling point to minimize statistical artifacts due to oversampling.
Sterile transport swabs containing Amies medium without charcoal (Starplex Scientific, Etobicoke, Ontario, Canada) were used to swab trees, wheel wells of vehicles, and shoes. The swabs were stored in a cool (4 to 10°C), dark place until they were transferred to Staib medium (57). The limit of detection for C. gattii on swab samples was experimentally determined to be ∼100 CFU/swab. The sensitivity of the swabbing method for C. gattii detection was assessed in a pilot study (32), and swabbing is regarded as an efficient sampling method (25, 54).
We tested 3,071 different trees for C. gattii by swabbing between October 2001 and January 2006. These trees were typically native to British Columbia, including coastal Douglas fir (Pseudotsuga menziesii var. menziesii; n = 1,085), western red cedar (Thuja plicata; n = 425), red alder (Alnus rubra; n = 316), maple (Acer sp.; n = 293), arbutus (Arbutus menziesii var. menziesii; n = 141), Garry oak (Quercus garryana; n = 123), pine (Pinus spp.; n = 100), western hemlock (Tsuga heterophylla; n = 62), fir (Abies sp.; n = 10), western birch (Betula occidentalis var. inopina; n = 46), spruce (Picea spp.; n = 46), poplar (Populus spp.; n = 39), black cottonwood (Populus balsamifera subsp. trichocarpa; n = 29), willow (Salix spp.; n = 16), dogwood (Cornus spp.; n = 9), bitter cherry (Prunus emarginata; n = 7), western yew (Taxus brevifolia; n = 2), and juniper (Juniperus sp.; n = 1) trees. Trees that were not native to British Columbia included various fruit trees (n = 53), Eucalyptus spp. (n = 25), various ornamental trees (n = 18), and oak (Quercus spp.; n = 9), horse chestnut (Aesculus hippocastanum; n = 8), and Oregon ash (Fraxinus latifolia; n = 7) trees. Also swabbed were stumps (n = 102), cut logs (n = 31), a variety of shrubs (n = 20), trees of undetermined species (n = 47), and nonliving structures, e.g., fence posts and animal bedding (n = 57). A recreational global positioning system receiver (eTrex; Garmin International Inc., Olathe, KS) was used to record the latitude and longitude positions of trees and other sampling points to within 15 m. Tree data, including local flowering or pollination times, were obtained from British Columbia Flora (University of British Columbia Botanical Gardens and Centre for Plant Research, Vancouver, British Columbia, Canada; http://www.bcflora.org/).
Soil samples were obtained from within the root zones of 1,584 trees, typically within 1 m of the base of the tree and within 15 cm of the surface, by collecting the soil in clean ziplock bags (1,481 of these trees were swabbed as described above). A further 222 soil samples were obtained from locations that were not associated with a tree. Thirty-centimeter core samples of soil (n = 24) were collected with a soil sampler (Oakfield Apparatus Company, Oakfield, WI) at eight different sampling sites in February and March of 2004. Each core sample was divided into two, corresponding to the top 15 cm and the lower 15 cm of soil. In addition, 41 samples of small tree debris, mulch, and wood chips were collected in ziplock bags. For all types of soil, mulch, or debris samples described here, approximately 2 g of each sample was suspended in 10 ml of sterile distilled water by vortexing. The sediment was allowed to settle for approximately 10 min, and then 100-μl aliquots of the supernatant were spread onto duplicate Staib agar plates. The limit of detection for soil, debris, and wood chip sampling was 25 CFU per gram.
Air samples were taken with either an RCS-Plus air sampler (Biotest Hycon, Denville, NJ) (56) or a six-stage sampler (Andersen Instruments, Smyrna, GA) using an Air-Con2 pump (Gilian Instruments, West Caldwell, NJ). Staib medium was used for cryptococcal isolation in both of the air-sampling systems. Initial surveys for airborne cryptococci were made with the RCS-Plus system, due to its high sampling efficiency for 3- to 4-μm particles (42). When colonized areas were identified, the Andersen six-stage system was used to characterize the range of sizes of airborne cryptococcal propagules. The RCS-Plus system was programmed to take in 200 liters (0.2 m3) of air at 50 liters/min, and the limit of detection for this method is 5 CFU per m3. The Andersen six-stage system was programmed to take in 283 liters (0.283 m3) at 28.3 liters/min, and the limit of detection for this system is 6 CFU per m3.
Water samples of >500 ml were collected in clean ziplock bags. Aliquots were filtered through 0.45-μm-pore-size nitrocellulose membranes (Fisher Scientific, Ottawa, Ontario, Canada), which were then incubated on Staib agar at 30°C for up to 10 days. At least five replicate plates were assessed for each water sample. The limit of detection for water sampling was 0.2 CFU per 100 ml, assuming that five replicates were performed.
Identification and genotyping of C. gattii isolates.
Cryptococci were initially identified by their characteristic dark brown appearance on Staib agar (57). These isolates were subcultured on malt extract agar until a pure culture was obtained. Isolates were confirmed as C. gattii by culturing on canavanine-glycine-bromothymol blue medium (39) and/or serotyping (CryptoChek; Iatron Laboratories, Tokyo, Japan). Samples were scored as positive or negative for the presence of C. gatti, and the concentration of C. gattii detected in each positive sample was noted. The molecular types of selected isolates were identified by using restriction fragment length polymorphisms and MLST methods as previously described (31, 32, 48).
Characterization of soil properties.
Soil was transported to the laboratory in sealed plastic bags. The moisture content was determined by measuring the weight of ∼5 ml of soil in a crucible, followed by the dry weight after incubation at 55°C for 48 h and storage in a desiccation chamber. The percentage of moisture content was calculated from the difference in weight between the wet and dried soil. Soil pH was measured by mixing ∼1 g of dry soil with 5 ml of distilled water and stirring for 20 min. After the sediment was allowed to settle, the pH of the supernatant was recorded. To determine the organic carbon content, dry soil (1.23 ml) was weighed in a crucible and ashed at 500°C for 2 h, left to cool overnight, and then heated at 55°C for 1 h. The ashed soil was weighed, and the loss of soil weight on ignition (LOI) was calculated as a percentage of the dry weight of the soil. The organic carbon content of the soil was determined by using the following expression: (0.7 × loss of weight on ignition) − 0.23 (59). For analyses based on soil properties, the C. gattii concentration was normalized by moisture content so that the concentration was expressed in terms of the dry weight of the soil.
Water survival assays.
C. gattii VGII isolate KB152A-6 was used to determine the potential for survival of C. gattii in solutions of various osmolalities. This isolate was chosen as a representative, potentially infectious C. gattii propagule since it was sufficiently small to penetrate the human respiratory system, being obtained from the sixth stage of an Andersen air sample from Vancouver Island (32). The isolate was inoculated onto malt extract agar (MEA) from a frozen glycerol stock and grown at 30°C for 48 h. Suspensions of the culture in distilled water, unfiltered ocean water (collected at the surface of deep water in the Strait of Georgia), filter-sterilized (0.45-μm-pore-size filter) ocean water, 10% NaCl, 15% NaCl, and 20% NaCl were prepared. The initial (time zero) concentration of each suspension was ∼1 × 106 to 2 × 106 CFU per ml. The suspensions were stored at room temperature (21 to 23°C) or at 4°C. Survival was assessed approximately monthly for 1 year by inoculating MEA plates and calculating numbers of CFU per ml.
Data analyses.
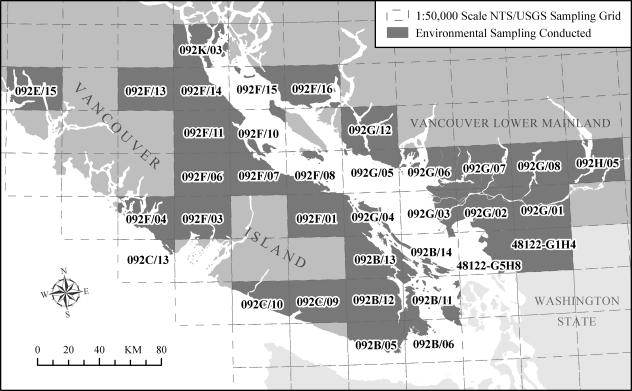
Environmental sampling data were compiled using Access 2002 (Microsoft Corporation, Redmond, WA), and statistical analyses were performed using SPSS 14 for Windows (SPSS Inc., Chicago, IL), except for the calculation of η2 values (43). The detected C. gattii concentrations among air, soil, and water samples were expressed as geometric means and geometric standard deviations. ArcGIS 9.0 and ArcView GIS 3.2 (Environmental Systems Research Institute Inc., Redlands, CA) were used to map the geographic locations of sites of environmental sampling by using global positioning system coordinates or a street address. The residential addresses of infected humans and animals were mapped against a reference street network data file (CanMap Streetfile 6.0, British Columbia; DMTI Spatial Inc., Markham, Ontario, Canada) or six-digit postal code (postal code conversion file, June 2001; Statistics Canada, Ottawa, Ontario, Canada). Ecosystem (Biogeoclimatic Zones of British Columbia 5.0, British Columbia Ministry of Forests, Victoria, British Columbia) and topographic (GTOPO30 elevation data, United States Geological Survey [USGS], Reston, VA) data were overlaid. A 1:50,000 scale sampling grid system was created to report environmental sampling results by merging National Topographic System of Canada (NTS) and USGS quadrangle map grids for British Columbia, Washington, and Oregon (Fig. 1). Climate data were obtained from the Environment Canada National Climate Archive (http://www.climate.weatheroffice.ec.gc.ca) as recorded at the Nanaimo A weather station (latitude, 49°03′N; longitude, 123°52′W; elevation, 28.4 m) between 1971 and 2000.
FIG. 1.
Overview of geographic zones designated according to the NTS and the USGS in which environmental sampling was conducted. Sampling grid regions 44123-A1B4, 45122-C1D4, and 45122-C5D8, located in the state of Oregon, are beyond the extent of this map.
RESULTS
Overview of environmental sampling.
Of 5,704 first-attempt swab, soil, air, and water samples collected between 2001 and early 2006, 519 were positive for C. gattii (Fig. 2). Air sampling appeared to be associated with seasonal patterns of positivity (Table 1). However, no seasonal pattern of C. gattii detection could be discerned from the positivity patterns of swab, soil, or water samples. Of the trees, shrubs, stumps, and cut logs tested by swabbing, 8% were positive for C. gattii colonization on the first sampling attempt while 10% of soil samples were positive for C. gattii on the first sampling attempt (Table 2).
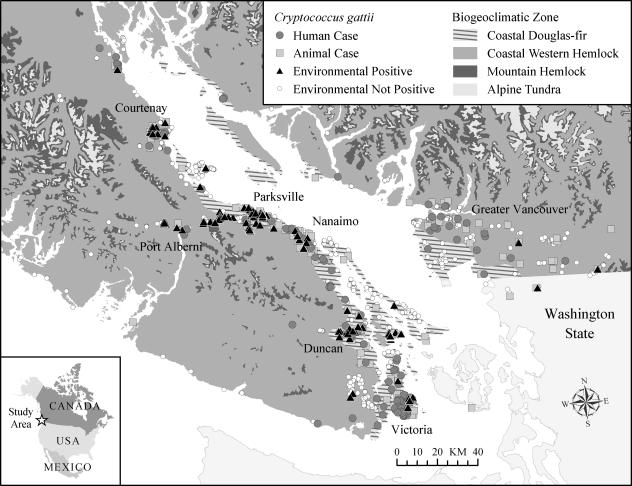
FIG. 2.
Map of Vancouver Island, the Gulf Islands, the lower mainland of British Columbia, and northern Washington state, showing the distribution of reported human and animal cases up to December 2005 as well as that of sampling sites from which positive and nonpositive environmental samples were obtained between October 2002 and January 2006.
TABLE 1.
Comparison of swab (all surfaces), soil, water, and air sample rates of positivity for C. gattii during different months of the year
| Month | Amt of sunshine (h)a | Mean maximum daily temp (°C)a | Mean rainfall (mm)a | Relative humidity (%)a,b | No. or % of samples of the indicated type
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Swab
|
Soil
|
Water
|
Airc
|
|||||||||
| Negative | Positive (%) | Negative | Positive (%) | Negative | Positive (%) | Negative | Positive (%) | |||||
| Jan. | 57 | 6.2 | 140 | 81 | 50 | 4.0 | 51 | 24 | 46 | 6.5 | 6 | 33 |
| Feb. | 80 | 8.2 | 120 | 73 | 224 | 6.3 | 153 | 16 | 6 | 33 | 7 | 0 |
| Mar. | 130 | 11 | 110 | 65 | 173 | 4.0 | 106 | 8.5 | 15 | 33 | 16 | 0 |
| Apr. | 180 | 14 | 63 | 59 | 148 | 4.7 | 85 | 2.4 | 10 | 0 | 6 | 33 |
| May | 230 | 18 | 50 | 58 | 293 | 9.6 | 117 | 14 | 15 | 27 | 15 | 27 |
| June | 220 | 21 | 45 | 58 | 352 | 6.5 | 261 | 12 | 4 | 50 | 54 | 44 |
| July | 290 | 24 | 26 | 53 | 281 | 11 | 190 | 8.9 | 8 | 25 | 72 | 28 |
| Aug. | 270 | 24 | 32 | 53 | 467 | 17 | 259 | 5.4 | 14 | 36 | 43 | 30 |
| Sept. | 200 | 21 | 39 | 57 | 351 | 3.7 | 184 | 6.5 | 12 | 25 | 17 | 41 |
| Oct. | 130 | 15 | 97 | 68 | 557 | 6.1 | 321 | 5.9 | 8 | 13 | 44 | 20 |
| Nov. | 66 | 9.1 | 190 | 79 | 101 | 0 | 73 | 22 | 23 | 30 | 15 | 6.7 |
| Dec. | 50 | 6.1 | 170 | 84 | 138 | 2.2 | 39 | 13 | 6 | 33 | 15 | 6.7 |
| Total | 1,903 | 14.7d | 1,082 | 66d | 3,135 | 7.7 | 1,839 | 9.6 | 167 | 22 | 310 | 27 |
Data are based on climate norms as recorded at the Nanaimo A weather station between 1971 and 2000 (http://climate.weatheroffice.ec.gc.ca/).
Relative humidity as recorded each day at 1500 h local standard time.
Air samples collected systematically between 2002 and 2004.
The mean value is presented.
TABLE 2.
Results for the first attempted swab and soil samples from trees of different species in the Pacific Northwest
| Host tree or sample source | Total no. of swabs | Positive swabs (%) | Total no. of soil samples | Positive soil samples (%) |
|---|---|---|---|---|
| Alder | 316 | 9.2 | 155 | 11 |
| Arbutus | 141 | 11 | 65 | 20 |
| Bitter cherry | 7 | 29 | 4 | 50 |
| Douglas fir | 1,085 | 9.3 | 503 | 11 |
| Eucalyptus | 25 | 0 | 2 | 0 |
| Garry oak | 123 | 14 | 61 | 23 |
| Hemlock | 62 | 0 | 14 | 0 |
| Maple | 293 | 4.8 | 191 | 9.4 |
| Pine | 100 | 4.0 | 50 | 2.0 |
| Red cedar | 425 | 10 | 250 | 11 |
| Spruce | 46 | 4.3 | 25 | 12 |
| Other trees | 248 | 1.2 | 171 | 2.3 |
| Shrubs (various) | 20 | 0 | 13 | 7.7 |
| Cut logs | 31 | 6.5 | 19 | 0 |
| Stumps | 102 | 2.9 | 44 | 4.5 |
| Unknown | 47 | 4.3 | 17 | 0 |
| Total | 3,071 | 7.7 | 1,584 | 10 |
Tree sampling.
C. gattii colonization of trees was assessed through swabbing as well as using soil associated with the root zone. Among those trees for which only the swab or only the soil sample was positive for C. gattii (n = 106), positive soil samples were approximately twice as common (65%) as positive swab samples (35%). Tree types that were associated with high rates of positive swab and soil samples on the first sampling attempt included Douglas fir, alder, arbutus, red cedar, and Garry oak (Table 2), all of which are native to the CDF. Less frequently, positive swab and soil samples were obtained from bitter cherry, maple, pine, and spruce trees and logs and stumps. There were no positive swab or soil samples obtained from hemlock or eucalyptus trees upon either the primary or subsequent samplings of these trees.
Based on the first swab and soil sample taken from each tree, geographic zones with the highest rates of positive tree samples were located in NTS grid regions 092B/13, 092B/14, 092F/01, 092F/07, and 092F/08 (Fig. 1; Table 3). Soil samples from NTS grid regions 092B/13 and 092B/14 contained the highest mean detected C. gattii concentrations among positive soil samples. The area represented by grid 092B/14 covers Saltspring Island and other Gulf Islands but not Vancouver Island. Geographic zones associated with a consistent but comparatively lower rate of positive tree samples included NTS grid regions 092B/06, 092F/10, and 092F/11. Occasional positive swab or soil samples were associated with NTS grid regions 092B/11, 092B/12, and 092G/04 and USGS grid region 48122-G1H4 in Washington state.
TABLE 3.
Results for the first attempted swab and soil samples from trees in different geographic (NTS or USGS) grid regions in the Pacific Northwest
| Geographical grid designationa | Total no. of swabs | Positive swabs (%) | Total no. of soil samples | Positive soil samples (%) | Mean C. gattii concn (CFU/m3)b | SDc |
|---|---|---|---|---|---|---|
| 092B/05 | 101 | 0 | 25 | 0 | ||
| 092B/06 | 255 | 5.9 | 95 | 11 | 180 | 4.4 |
| 092B/11 | 44 | 0 | 21 | 9.5 | 28 | 1.5 |
| 092B/12 | 149 | 0.7 | 80 | 1.3 | 93 | |
| 092B/13 | 233 | 7.7 | 132 | 15 | 870 | 8.6 |
| 092B/14 | 86 | 22 | 92 | 32 | 750 | 14 |
| 092C/09 | 8 | 0 | 0 | |||
| 092C/10 | 5 | 0 | 0 | |||
| 092F/01 | 137 | 14 | 59 | 24 | 180 | 3.4 |
| 092F/03 | 3 | 0 | 3 | 0 | ||
| 092F/06 | 12 | 0 | 11 | 0 | ||
| 092F/07 | 265 | 9.8 | 140 | 16 | 120 | 6.7 |
| 092F/08 | 578 | 22 | 197 | 23 | 190 | 6.2 |
| 092F/10 | 230 | 5.2 | 154 | 5.8 | 550 | 7.4 |
| 092F/11 | 55 | 3.6 | 38 | 7.9 | 24 | 1.1 |
| 092F/14 | 32 | 0 | 5 | 0 | ||
| 092F/15 | 3 | 0 | 3 | 0 | ||
| 092F/16 | 3 | 0 | 3 | 0 | ||
| 092G/01 | 135 | 0 | 101 | 0 | ||
| 092G/02 | 166 | 0 | 111 | 0 | ||
| 092G/03 | 47 | 0 | 21 | 0 | ||
| 092G/04 | 214 | 0.5 | 81 | 3.7 | 110 | 5.7 |
| 092G/05 | 11 | 0 | 10 | 0 | ||
| 092G/06 | 79 | 0 | 52 | 0 | ||
| 092G/07 | 51 | 0 | 38 | 0 | ||
| 092G/08 | 3 | 0 | 2 | 0 | ||
| 092G/12 | 2 | 0 | 0 | |||
| 092H/05 | 2 | 0 | 0 | |||
| 092K/03 | 12 | 0 | 0 | |||
| 103J/08 | 5 | 0 | 0 | |||
| 092F/04 | 14 | 0 | 0 | |||
| 092C/13 | 1 | 0 | 0 | |||
| 092E/15 | 3 | 0 | 0 | |||
| 092F/13 | 5 | 0 | 0 | |||
| 48122-G5H8 | 9 | 0 | 2 | 0 | ||
| 48122-G1H4 | 21 | 0 | 20 | 5.0 | 71 | |
| 44123-A1B4 | 32 | 0 | 31 | 0 | ||
| 45122-C1D4 | 5 | 0 | 5 | 0 | ||
| 45122-C5D8 | 55 | 0 | 52 | 0 | ||
| Total | 3,071 | 7.7 | 1,584 | 10 | 270 | 8.3 |
See Materials and Methods and the legend to Fig. 1 for an explanation of designated geographic zones.
Among positive air samples only.
SD, standard deviation.
Since March 2002, 1,213 environmental samples have been collected longitudinally from 150 trees, as well as 32 samples not associated with trees in a provincial park in NTS grid region 092F/08 on Vancouver Island. These included 791 tree swabs and 265 soil, 180 air, and 9 water samples (Fig. 3). Seventy-nine of the 150 trees tested positive at least once by swab or soil sample. These trees usually were located in or near areas of the park with comparatively low tree densities, such as parking lots and a beach. Those trees that never (or transiently) tested positive usually were located in areas of the park with greater tree and rhizosphere densities.
FIG. 3.
Distribution of environmental sampling sites within a provincial park in geographic zone 092F/08. Positive samples were typically obtained from areas with relatively low tree densities, e.g., along the entrance road, around parking lots, and in the beach area.
Soil colonization.
Of 24 core samples of soil, 15 were positive for C. gattii in either the upper-15-cm or lower-15-cm section of the sample. Of the 15 positive core samples, all were positive in the upper section of soil but only 6 were also positive in the lower section, which may in part be attributed to the transfer of some soil and colonizing organisms downwards as an artifact of the core sampling process. In addition, the C. gattii concentration was significantly greater (P < 0.01) in the top section of the core sample (geometric mean, 500 ± 1.2 CFU/g) than in the lower part (7.2 ± 27 CFU/g).
The pH ranges for both positive (n = 586) and nonpositive (n = 929) soil samples were wide, although the range was slightly narrower for positive samples (pH 4.3 to 7.5) than for nonpositive ones (pH 3.5 to 8.2), with mean pH values (5.95 ± 0.62 and 6.05 ± 0.68, respectively) that were not significantly different (P ≥ 0.05). However, when results were stratified according to the location of sampling, mean differences in pH were observed between samples from locations that were heavily colonized and those from locations in which C. gattii was detected only transiently (data not shown). For example, the mean pH of soil samples from a heavily colonized park within NTS grid region 092F/08 was 0.46 lower (P < 0.01) than that of samples from parks in NTS grid region 092B/06 (∼150 km apart), where C. gattii was less commonly isolated.
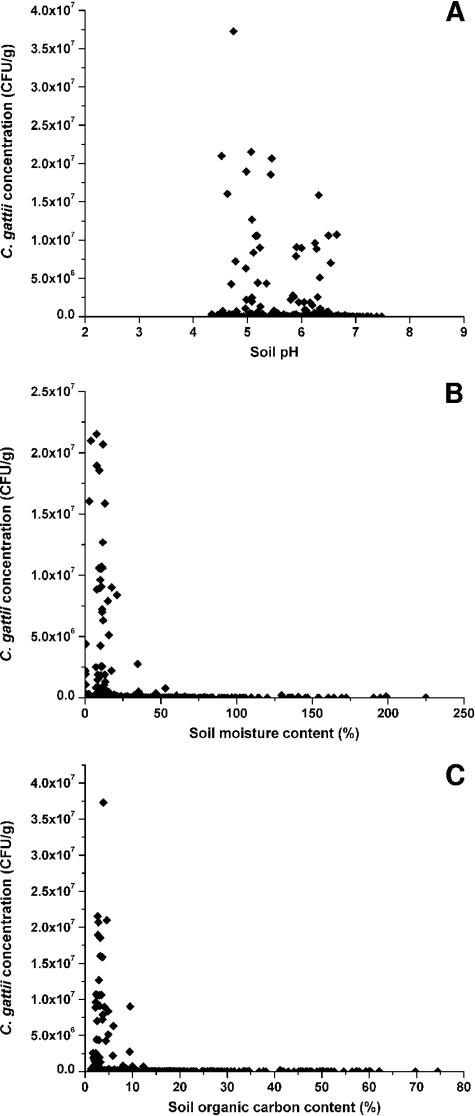
There were significant differences in the mean moisture and organic carbon contents of positive (n = 510) and nonpositive samples (n = 700), although the ranges were similar; positive soil samples had moisture contents of 0.3 to 280% and organic carbon contents of 1.1 to 75%, while nonpositive soil samples had moisture contents of 1.0 to 300% and organic carbon contents of 0.8 to 68%. The detection of C. gattii was associated with lower moisture contents (mean difference, 14%; P < 0.05) and lower organic carbon contents (mean difference, 8.0%; P < 0.05). Furthermore, high concentrations of C. gattii detected in soil samples were significantly associated with both low moisture (η2 = 0.96) and a low organic carbon content (η2 = 0.83) (Fig. 4), but these factors may not be independent of each other (R2 = 0.31).
FIG. 4.
Properties of soil samples colonized by C. gattii. Concentrations of C. gattii are expressed with respect to the dry weight of the soil.
Water sampling.
Samples were taken from both freshwater (n = 166) and seawater (n = 19) between May 2003 and January 2006. Twenty-seven of 130 samples from lakes were positive (C. gattii concentration, 5.1 ± 11 CFU/100 ml). Six of 36 samples from rivers or creeks were positive (C. gattii concentration, 1.1 ± 12 CFU/100 ml). Positive freshwater samples were associated with positive soil samples in the area, although this result requires reconfirmation through a random study design. Four of the 19 seawater samples were positive for C. gattii. The positive seawater samples were obtained from the shores of Vancouver Island in areas of endemicity within NTS grid regions 092F/08 (n = 2) and 092F/10 (n = 1), as well as from a Gulf Island in NTS grid region 092B/14 (n = 1), and yielded an overall C. gattii concentration of 2.2 ± 2.0 CFU/100 ml. Nonpositive seawater samples were obtained from these and other areas where C. gattii is endemic or not endemic.
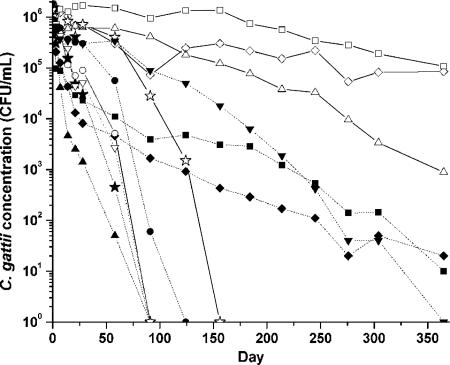
We used laboratory assays to evaluate the survival of C. gattii in different types of water and at different temperatures over the course of a year (Fig. 5). C. gattii survived best in filtered ocean water at room temperature, unfiltered ocean water at room temperature, and distilled water at room temperature (Fig. 5). C. gattii survived most poorly in distilled water at 4°C, 10% NaCl at 4°C, 20% NaCl at room temperature, and 15% NaCl at room temperature, all of which had no viable C. gattii cells by day 91.
FIG. 5.
Survival of a British Columbian C. gattii VGII isolate over 1 year in distilled water at room temperature (▵) and 4°C (▴), ocean water at room temperature (⋄) and 4°C (♦), filter-sterilized ocean water at room temperature (□) and 4°C (▪), 10% NaCl at room temperature (⋆) and 4°C (★), 15% NaCl at room temperature (○) and 4°C (•), and 20% NaCl at room temperature (▿) and 4°C (▾).
Air sampling.
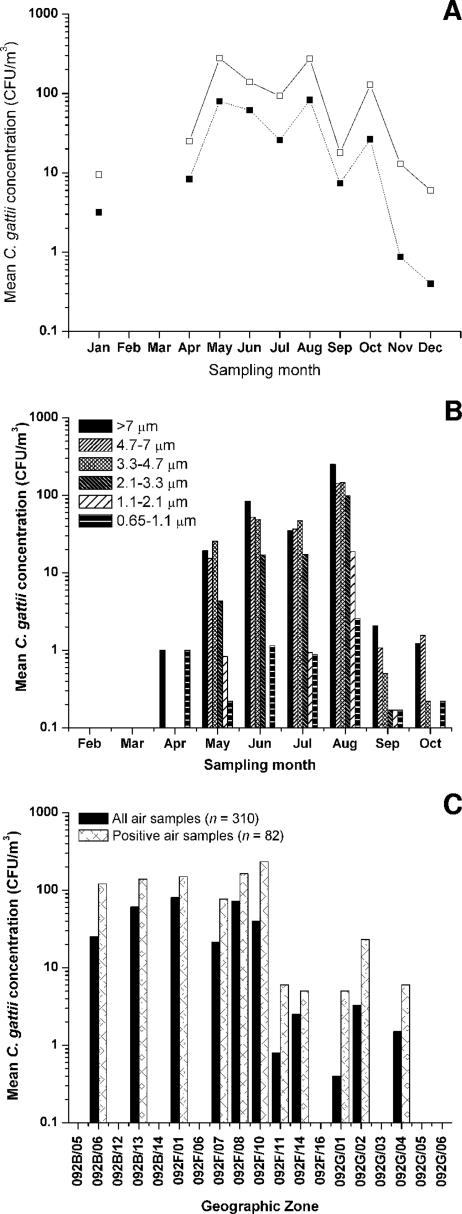
The concentration of airborne cryptococci (Fig. 6A) during the northern hemisphere spring and summer months peaked in August (mean, 275 ± 563 CFU/m3 for 13 positive samples), when the weather in southwest British Columbia is the warmest (average temperature, 11 to 24°C) and driest (mean monthly rainfall, 31.6 mm). In cooler, wetter months, e.g., December (average temperature, −1 to 6°C; mean monthly rainfall, 166.4 mm), the concentrations of airborne cryptococci declined, presumably because all of the propagules were washed out of the air. None of the air samples taken during periods of rain or shortly thereafter were positive. Relative humidity was negatively correlated with the concentration of airborne C. gattii, accounting for 4.3% of the observed variation (P < 0.05; data not shown). We observed no association between the flowering or pollination times of particular tree species and the detected concentrations of airborne cryptococci sampled from beneath these trees (Table 4).
FIG. 6.
Characterization of air samples collected systematically between 2002 and 2004. (A) Mean C. gattii concentrations among all air samples (closed squares; n = 310) and positive air samples (open squares; n = 82) collected at different months of the year. (B) Mean concentrations of C. gattii stratified by propagule size during different months of the year. Andersen air samples were collected from February through October, for which the C. gattii concentrations shown represent those among all attempted samples (n = 118). (C) Mean C. gattii concentrations detected in air samples collected from different geographic grid regions of British Columbia.
TABLE 4.
Months of peak concentrations of airborne C. gattii propagules associated with native British Columbian trees for which air samples were collected systematically and months of flowering or pollination
| Tree type | No. of air samples | Months of flowering/pollinationa | Month(s) of peak detected C. gattii concnb |
|---|---|---|---|
| Alder | 55 | March and April | August |
| Arbutus | 18 | April and May | June and July |
| Douglas-fir | 131 | April and May | August |
| Garry oak | 24 | April and June | October |
| Maple | 58 | March-June | August |
| Pine | 13 | May and June | No positive samples |
| Red cedar | 71 | April and May | July |
| Spruce | 7 | May and June | September |
Data were collected from http://www.bcflora.org.
Among positive air samples only.
We took 118 samples with an Andersen six-stage air sampler (Fig. 6B) throughout the year from February through October. Positive samples (n = 62) were obtained from April to October. Airborne cryptococci were more common in the upper sample stages (stages 1, 2, and 3, corresponding to particle sizes of >7 μm, 4.7 to 7 μm, and 3.3 to 4.7 μm, respectively) than in the lower stages (stages 4, 5, and 6, corresponding to particle sizes of 2.1 to 3.3 μm, 1.1 to 2.1 μm, and 0.65 to 1.1 μm, respectively). Cryptococcal propagules of all sizes were detected during all months in which positive air samples were obtained, although the concentrations varied with the same seasonal pattern observed for other air samples.
Geographically (Fig. 6C), the highest rate of positive first-attempt air samples corresponded to the contiguous geographic regions 092F/01 (7/13 samples) and 092F/08 (30/68 samples); soil samples from these regions contained C. gattii in high concentrations (Table 3). However, there was no significant variation in the airborne C. gattii concentrations detected among different geographical areas where positive air samples were obtained.
Genetic characterization of isolates.
All of the C. gattii isolates we recovered belonged to serotype B. We assessed the genotypes of 108 isolates from first-attempt samples (swab, soil, and air). Ninety-one (84%) belonged to the VGIIa genotype, 13 (12%) belonged to the VGIIb genotype, and 4 (4%) belonged to the VGI molecular type. We recovered VGI strains from air, soil, and swab samples associated with trees on Vancouver Island within NTS grid region 092B/13, which appeared to be genotypically distinct from those previously analyzed by MLST (31) from another Gulf Island (data not shown). Considering all sampling attempts in which isolates were genotyped, sympatric colonization of genotypes VGIIa and VGIIb was frequently observed in single samples or at single sampling points throughout Vancouver Island, despite apparent differences in the prevalence of these genotypes. At one sampling site within NTS grid region 092/B13, VGIIa, VGIIb, VGI, and serotype AD were consistently coisolated from a series of trees over a period of more than 3 years.
DISCUSSION
We isolated C. gattii from many different geographic zones in British Columbia and the Pacific Northwest of the United States. However, many geographic zones in this region yielded no C. gattii isolates, including the San Juan Islands (USGS grid regions E5F8 and C5D8 and NTS grid regions 092B/11 and 092B/06), which were sampled by another group (21). These results suggest that there are “hot spots” of C. gattii within the sampled areas. Some of these hot spots occur within the CWHxm as well as the CDF, which finding expands the biogeoclimatic range of colonization previously reported for C. gattii in British Columbia (32). The CDF and CWHxm share many characteristics, and the boundary between them, in practice, is rather diffuse.
The failure to isolate C. gattii from all of the sampled areas, as well as the failure to isolate this pathogen from the San Juan Islands (21), does not mean that the fungus is not present in these areas, but rather that if it is present it exists at levels below the detection limit. Further sampling of the mainland United States and the San Juan Islands over an extended period of time would be of great value, particularly given the prevalence of C. gattii on nearby Vancouver Island and the British Columbian Gulf Islands, evidence of dispersal (30), and recent reports of human and animal C. gattii infections in this region (45; A. Upton, J. A. Fraser, S. E. Kidd, S. Raverty, C. Bretz, J. Heitman, K. H. Bartlett, and K. A. Marr, unpublished results).
Tree and soil sampling.
Previous studies of ecological reservoirs of C. gattii have focused predominantly on trees, particularly eucalypts (5-7, 10, 15, 17, 18, 25, 28, 33, 40, 46, 49, 51-53, 61). Our observation that soil samples were positive more often than swab samples for corresponding trees suggests that soil may be the principal reservoir of colonization. This finding contradicts the results of our pilot study, in which positive swab samples outnumbered positive soil samples ninefold (32). We think that the current data more accurately represent the present situation since the sampling sites in the previous study were overrepresented by duplicate and replicate swab samples from a limited number of trees.
It has been suggested that eucalyptus tree imports could be the route of introduction of C. gattii into the Pacific Northwest (20, 21), particularly since one isolate (CBS7750) with a genotype apparently identical to that of the dominant strain in British Columbia (20, 31) was obtained from a Eucalyptus camaldulensis tree in San Francisco in 1993. Numerous reports from Italy, Egypt, India, southern California, and parts of South America have suggested the coimportation of C. gattii and eucalyptus seeds or cuttings into these regions (6, 7, 10, 16, 28, 46, 50, 51). However, few of these studies have tested the hypotheses with genotypic data, and given the apparent global dispersal of C. gattii (31), it is difficult to demonstrate that C. gattii was absent from these areas prior to its isolation from imported trees. We have not found an association between C. gattii and eucalypts in British Columbia, although the eucalypt of origin may no longer exist. This finding is significant because it indicates that, at least in British Columbia, eucalypts may not be the primary ecological niche of C. gattii, as previously suggested (14, 15, 51, 52, 52a). Eucalypt imports in British Columbia in recent years have been in the form of seeds, and to our knowledge C. gattii has not been isolated from eucalyptus seeds (11, 36; D. H. Ellis, University of Adelaide, personal communication).
In an overall heavily colonized park on Vancouver Island, C. gattii was more prevalent in areas of less dense tree cover, particularly near open areas such as parking lots. This pattern could result from increased competition for nutrients in areas of greater microbial density associated with high tree and rhizosphere densities. Differences in concentrations also may reflect variability in C. gattii population stability as a result of introduction and redistribution through the movement of humans, animals, and traffic within the park (30). In addition, the possibility of antagonists influencing the extent of C. gattii colonization remains to be evaluated.
Soil colonization.
Our findings suggest that C. gattii, like most fungal species, colonizes primarily the top 15 cm of the humic layer of soil due to temperature, humidity, and nutrient requirements (3, 13, 24) and is presumably protected from UV exposure by its melanin production (63). In addition, our data also suggest that C. gattii is tolerant of acidic conditions, which is consistent with reports that tremellalean Cryptococcus spp. preferentially colonize soil with a lower pH (23, 62). Many other acidophilic and acid-tolerant fungi and yeasts have also been identified (27). Although soil moisture contents varied throughout the year, no significant seasonality in the C. gattii concentrations in soil was observed. These factors, combined with microclimate, probably are important determinants of colonization following the dispersal of C. gattii propagules and are consistent with the hypothesis that colonization is limited to portions of the CDF and CWHxm biogeoclimatic zones.
In contrast to our findings, analyses of C. gattii colonization in Colombia (25) did not reveal an association of cryptococci with lower soil moisture contents, possibly because soils containing C. neoformans and C. gattii were not distinguished or perhaps because the dominant strain observed in British Columbia (VGIIa) is uniquely adapted to thrive under these conditions. Presumably, the number of competing microorganisms is smaller in soil with lower moisture and organic contents, and the success of C. gattii under these conditions reflects the ability to adapt to specific ecological niches. The proliferation of C. gattii in British Columbia, outside its traditional tropical niches, may be due to increased fitness of the VGIIa strain resulting from either selection or sexual recombination (20).
C. gattii in water.
C. gattii was isolated at high frequencies from freshwater (27 of 132 samples [20%]) and seawater (4 of 19 samples [21%]). While other fungal pathogens, most notably Blastomyces dermatitidis, have been isolated from bodies of water (34), to our knowledge this is the first report of C. gattii isolation from water. Different mechanisms of inoculation of C. gattii into the water, as well as the extent of colonization of nearby trees and soil, probably influence the rate at which positive samples occur. The observed association between positive freshwater sampling sites and permanently colonized soil suggests that C. gattii may, in part, be inoculated into the water by soil runoff. The higher observed frequency of C. gattii isolation from lakes than from rivers or creeks may be due to reduced dispersal in still water, which makes measurable levels of the fungus easier to detect. The role of human activity in lake recreational areas also has been implicated in the presence of C. gattii in water (30).
We found that filtered and unfiltered ocean water supported the highest rate of C. gattii survival over 1 year. The seawater samples at room temperature and 4°C were broadly representative of natural seawater conditions in British Columbia, and the relatively high survival rates under these conditions are consistent with the survival of C. gattii propagules in the seawater surrounding Vancouver Island. Taken together, the relatively high number of C. gattii-positive samples from both fresh- and seawater samples and the survival patterns of C. gattii under laboratory conditions indicate that C. gattii could be dispersed through bodies of water. These findings also provide a plausible transmission mechanism for the previously reported cryptococcal infections in wild marine mammals in the Strait of Georgia (12, 58).
Seasonality of C. gattii detection.
C. gattii infection usually results from inhalation, and exposure is thought to be a function of the airborne concentration of C. gattii propagules. Airborne C. gattii has been associated with the seasonal flowering of eucalypts in the spring in Australia (15), but not in other locations (25, 40). There are numerous tree hosts for C. gattii in British Columbia that flower at different times, but we observed no association between detected concentrations of airborne C. gattii and the times of the flowering or pollination of these trees. Rather, we suggest that factors more likely to influence the concentration of airborne C. gattii include the extent of local colonization and meteorological conditions. In our study, air samples were more likely to be positive for C. gattii and to contain higher concentrations of propagules (Fig. 6A and B) during the summer months, which are associated with relatively warm and dry conditions with low relative humidity (Table 1).
An increase like that of the isolation of serotype B but not serotypes A or C from trees and soil in Colombia during conditions of high rainfall and relative humidity (25, 26) was not observed in our study. Similarly, a decrease like that of the isolation of C. gattii from trees in India during the extreme heat of summer (53) was not discernible among our data, although the temperate British Columbian climate is not comparable to that of India. We attribute an apparent increase in the proportion of positive swabs during the relatively dry and warm summer months to an artifact of increased sampling during those months and the fact that the selection of sampling sites was not randomized. In contrast to our air sampling data, longitudinal swab and soil sampling data (not shown) obtained from sites of endemicity in British Columbia over a 2- to 3-year period do not reflect a seasonality of C. gattii prevalence, which may indicate the uniqueness of the emergence of C. gattii in the temperate climate of British Columbia compared to the climates in similar studies. Our observations of increased airborne concentrations of C. gattii propagules (all serotype B) in British Columbia during periods of low rainfall and relative humidity are probably not comparable to the findings of Colombian studies since they did not report on air sample data.
Airborne C. gattii.
The cryptococci obtained at stages 4, 5, and 6 of the Andersen air sampler were considered sufficiently small (<3.3 μm) to penetrate the lower respiratory system, potentially causing infection. Given that the positive Andersen air samples obtained between April and October all included propagules of <3.3 μm, the risk of exposure to infectious C. gattii propagules is effectively continuous in the areas where these samples were collected.
No significant geographic distribution pattern of airborne C. gattii concentrations was observed. Given the seasonal variation, the observed pattern may be a sampling artifact, since not all sites were sampled every month. The windborne dispersal of the propagules also may affect the concentration detected at any given location.
Genotypic diversity among C. gattii.
The proportions of VGIIa and VGIIb subtypes we observed were similar to those observed previously (32), and their sympatric colonization occurred frequently throughout the geographic range of this study. This codistribution could indicate concomitant introduction, concomitant dispersal, or synergistic colonization. These hypotheses warrant further evaluation since interactions between the two genotypes could have public health implications. The consistent coisolation of VGIIa, VGIIb, VGI, and C. neoformans serotype AD from several sampling points at one site suggests that in some cases the populations could be very complex.
The environmental colonization by VGI in British Columbia is now more evident than in previous reports (31, 32). However, all VGI strains isolated thus far from environmental sources in British Columbia are genetically distinct from clinical isolates from this region (31; Kidd and Bartlett, unpublished). Nevertheless, the existence of environmental sources of VGI in British Columbia means that at least some of the British Columbian VGI infections could have been acquired locally rather than through travel to other areas of endemicity.
In conclusion, we found that different C. gattii genotypes colonized many areas of Vancouver Island and the Pacific Northwest, with the distribution of the fungus dependent, at least in part, on the soil properties. These data confirm the expansion of the ecological niche of C. gattii to temperate regions and fill significant gaps in our understanding of exposure to this fungus, as there have recently been some human and animal cases in British Columbia and the Pacific Northwest of the United States in individuals who are not known to have traveled to Vancouver Island (45). Furthermore, evidence for the human-mediated dispersal of C. gattii (30), suggests that further environmental colonization by and increased clinical cases of infection with this pathogen are possible.
Acknowledgments
We thank the members of the British Columbia Cryptococcal Working Group (http://www.cryptococcusgattii.ca), Caroline Chen, Andrea Griffiths, Tracy Kirkham, Tim Ma, Fred Rockwell, Hua Shen, and Clement Tsui for assistance with environmental sampling and laboratory processing and British Columbia Parks employees and Vancouver Island Environmental Health officers for their cooperation.
Funding was provided by the Michael Smith Foundation for Health Research, Canadian Institutes of Health Research, the British Columbia Lung Association, and the Workers' Compensation Board of British Columbia.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Abegg, M. A., F. L. Cella, J. Faganello, P. Valente, A. Schrank, and M. H. Vainstein. 2006. Cryptococcus neoformans and Cryptococcus gattii isolated from the excreta of psittaciformes in a southern Brazilian zoological garden. Mycopathologia 161:83-91. [DOI] [PubMed] [Google Scholar]
- 2.Barreto de Oliveira, M. T., T. Boekhout, B. Theleen, F. Hagen, F. A. Baroni, M. S. Lazera, K. B. Lengeler, J. Heitman, I. N. G. Rivera, and C. R. Paula. 2004. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. J. Clin. Microbiol. 42:1356-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisby, C. R., N. James, and M. I. Timonin. 1935. Fungi isolated from soil profiles in Manitoba. Can. J. Res. 13:47-65. [Google Scholar]
- 4.Boekhout, T., B. Theleen, M. Diaz, J. W. Fell, W. C. Hop, E. C. Abeln, F. Dromer, and W. Meyer. 2001. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology 147:891-907. [DOI] [PubMed] [Google Scholar]
- 5.Callejas, A., N. Ordonez, M. C. Rodriguez, and E. Castaneda. 1998. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Med. Mycol. 36:341-344. [PubMed] [Google Scholar]
- 6.Campisi, E., F. Mancianti, G. Pini, E. Faggi, and G. Gargani. 2003. Investigation in Central Italy of the possible association between Cryptococcus neoformans var. gattii and Eucalyptus camaldulensis. Eur. J. Epidemiol. 18:357-362. [DOI] [PubMed] [Google Scholar]
- 7.Chakrabarti, A., M. Jatana, P. Kumar, L. Chatha, A. Kaushal, and A. A. Padhye. 1997. Isolation of Cryptococcus neoformans var. gattii from Eucalyptus camaldulensis in India. J. Clin. Microbiol. 35:3340-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, S., T. Sorrell, G. Nimmo, B. Speed, B. Currie, D. Ellis, D. Marriott, T. Pfeiffer, D. Parr, K. Byth, and the Australasian Cryptococcal Study Group. 2000. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin. Infect. Dis. 31:499-508. [DOI] [PubMed] [Google Scholar]
- 9.Colom, M. F., S. Frases, C. Ferrer, A. Jover, M. Andreu, S. Reus, M. Sanchez, and J. M. Torres-Rodriguez. 2005. First case of human cryptococcosis due to Cryptococcus neoformans var. gattii in Spain. J. Clin. Microbiol. 43:3548-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davel, G., R. Abrantes, M. Brudny, S. Cordoba, L. Rodero, C. E. Canteros, and D. Perrotta. 2003. First environmental isolation of Cryptococcus neoformans var. gattii in Argentina. Rev. Argent. Microbiol. 35:110-112. (In Spanish.) [PubMed] [Google Scholar]
- 11.Dempsey, S. L., and K. L. McLachlan. 2000. Quarantine risks associated with eucalyptus seed imports, supplementary volume. National Office of Animal and Plant Health, Canberra, Australia.
- 12.Duncan, C. 2005. M.Sc. thesis. University of Saskatchewan, Saskatoon, Canada.
- 13.Ekelund, F., R. Ronn, and S. Christensen. 2001. Distribution with depth of protozoa, bacteria and fungi in soil profiles from three Danish forest sites. Soil Biol. Biochem. 33:475-481. [Google Scholar]
- 14.Ellis, D. H., and T. J. Pfeiffer. 1990. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet 336:923-925. [DOI] [PubMed] [Google Scholar]
- 15.Ellis, D. H., and T. J. Pfeiffer. 1990. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 28:1642-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escandon, P., E. Quintero, D. Granados, S. Huerfano, A. Ruiz, and E. Castañeda. 2005. Isolation of Cryptococcus gattii serotype B from detritus of eucalyptus trees in Colombia. Biomedica 25:390-397. [PubMed] [Google Scholar]
- 17.Escandón, P., A. Sánchez, M. Martínez, W. Meyer, and E. Castañeda. 2006. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 6:625-635. [DOI] [PubMed] [Google Scholar]
- 18.Fortes, S. T., M. S. Lazera, M. M. Nishikawa, R. C. Macedo, and B. Wanke. 2001. First isolation of Cryptococcus neoformans var. gattii from a native jungle tree in the Brazilian Amazon rainforest. Mycoses 44:137-140. [DOI] [PubMed] [Google Scholar]
- 19.Franklin, J. F., and C. T. Dyrness. 1973. Natural vegetation of Oregon and Washington. General technical report PNW-8. U.S. Department of Agriculture and Forest Services, Portland, OR.
- 20.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 21.Fraser, J. A., S. M. Lim, S. Diezmann, E. C. Wenink, C. G. Arndt, G. M. Cox, F. S. Dietrich, and J. Heitman. 2006. Yeast diversity sampling on the San Juan Islands reveals no evidence for the spread of the Vancouver Island Cryptococcus gattii outbreak to this locale. FEMS Yeast Res. 6:620-624. [DOI] [PubMed] [Google Scholar]
- 22.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gadanho, M., D. Libkind, and J. P. Sampaio. 2006. Yeast diversity in the extreme acidic environments of the Iberian Pyrite Belt. Microb. Ecol. 52:552-563. [DOI] [PubMed] [Google Scholar]
- 24.Garrett, S. D. 1955. Microbial ecology of the soil. Trans. Br. Mycol. Soc. 38:1-9. [Google Scholar]
- 25.Granados, D. P., and E. Castañeda. 2005. Isolation and characterization of Cryptococcus neoformans varieties recovered from natural sources in Bogota, Colombia, and study of ecological conditions in the area. Microb. Ecol. 49:282-290. [DOI] [PubMed] [Google Scholar]
- 26.Granados, D. P., and E. Castañeda. 2006. Influence of climatic conditions on the isolation of members of the Cryptococcus neoformans species complex from trees in Colombia from 1992-2004. FEMS Yeast Res. 6:636-644. [DOI] [PubMed] [Google Scholar]
- 27.Gross, S., and E. I. Robbins. 2000. Acidophilic and acid-tolerant fungi and yeasts. Hydrobiologia 433:91-109. [Google Scholar]
- 28.Gugnani, H. C., T. G. Mitchell, A. P. Litvintseva, K. B. Lengeler, J. Heitman, A. Kumar, S. Basu, and A. Paliwal-Joshi. 2005. Isolation of Cryptococcus gattii and Cryptococcus neoformans var. grubii from the flowers and bark of eucalyptus trees in India. Med. Mycol. 43:565-569. [DOI] [PubMed] [Google Scholar]
- 29.Kidd, S. E. 2003. Ph.D. thesis. University of Sydney, Sydney, Australia.
- 30.Kidd, S. E., P. J. Bach, A. O. Hingston, S. Mak, Y. Chow, L. MacDougall, J. W. Kronstad, and K. H. Bartlett. 2007. Cryptococcus gattii dispersal mechanisms, British Columbia, Canada. Emerg. Infect. Dis. 13:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kidd, S. E., H. Guo, K. H. Bartlett, J. Xu, and J. W. Kronstad. 2005. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot. Cell 4:1629-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. Macdougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (BC, Canada). Proc. Natl. Acad. Sci. USA 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidd, S. E., T. C. Sorrell, and W. Meyer. 2003. Isolation of two molecular types of Cryptococcus neoformans var. gattii from insect frass. Med. Mycol. 41:171-176. [DOI] [PubMed] [Google Scholar]
- 34.Klein, B. S., J. M. Vergeront, A. F. DiSalvo, L. Kaufman, and J. P. Davis. 1987. Two outbreaks of blastomycosis along rivers in Wisconsin. Isolation of Blastomyces dermatitidis from riverbank soil and evidence of its transmission along waterways. Am. Rev. Respir. Dis. 136:1333-1338. [DOI] [PubMed] [Google Scholar]
- 35.Krockenberger, M. B., P. J. Canfield, and R. Malik. 2002. Cryptococcus neoformans in the koala (Phascolarctos cinereus): colonization by C. neoformans var. gattii and investigation of environmental sources. Med. Mycol. 40:263-272. [DOI] [PubMed] [Google Scholar]
- 36.Kularatne, H. A. G. C. 2004. Ph.D. thesis. RMIT University, Melbourne, Australia.
- 37.Kwon-Chung, K. J., and J. E. Bennett. 1984. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 120:123-130. [DOI] [PubMed] [Google Scholar]
- 38.Kwon-Chung, K. J., and J. E. Bennett. 1984. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentbl. Bakteriol. Mikrobiol. Hyg. A 257:213-218. [PubMed] [Google Scholar]
- 39.Kwon-Chung, K. J., I. Polacheck, and J. E. Bennett. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lazera, M. S., M. A. Cavalcanti, L. Trilles, M. M. Nishikawa, and B. Wanke. 1998. Cryptococcus neoformans var. gattii—evidence for a natural habitat related to decaying wood in a pottery tree hollow. Med. Mycol. 36:119-122. [PubMed] [Google Scholar]
- 41.Lazera, M. S., M. A. Salmito Cavalcanti, A. T. Londero, L. Trilles, M. M. Nishikawa, and B. Wanke. 2000. Possible primary ecological niche of Cryptococcus neoformans. Med. Mycol. 38:379-383. [DOI] [PubMed] [Google Scholar]
- 42.Lee, K. S., K. H. Bartlett, M. Brauer, G. M. Stephens, W. A. Black, and K. Teschke. 2004. A field comparison of four samplers for enumerating fungal aerosols. I. Sampling characteristics. Indoor Air 14:360-366. [DOI] [PubMed] [Google Scholar]
- 43.Levine, T. R., and C. R. Hullett. 2002. Eta squared, partial Eta squared, and misreporting of effect size in communication research. Hum. Commun. Res. 28:612-625. [Google Scholar]
- 44.MacDougall, L., and M. Fyfe. 2006. Emergence of Cryptococcus gattii in a novel environment provides clues to its incubation period. J. Clin. Microbiol. 44:1851-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacDougall, L., S. E. Kidd, E. Galanis, S. Mak, M. J. Leslie, P. R. Cieslak, J. W. Kronstad, M. G. Morshed, and K. H. Bartlett. 2007. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 13:42-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahmoud, Y. A. 1999. First environmental isolation of Cryptococcus neoformans var. neoformans and var. gattii from the Gharbia Governorate, Egypt. Mycopathologia 148:83-86. [DOI] [PubMed] [Google Scholar]
- 47.Meidinger, D., and D. Pojar. 1991. Ecosystems of BC. British Columbia Ministry of Forests, Victoria, British Columbia, Canada. http://www.for.gov.bc.ca/hfd/pubs/Docs/Srs/SRseries.htm.
- 48.Meyer, W., A. Castañeda, S. Jackson, M. Huynh, E. Castañeda, and the Ibero-American Cryptococcal Study Group. 2003. Molecular typing of Ibero-American Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montenegro, H., and C. R. Paula. 2000. Environmental isolation of Cryptococcus neoformans var. gattii and C. neoformans var. neoformans in the city of Sao Paulo, Brazil. Med. Mycol. 38:385-390. [DOI] [PubMed] [Google Scholar]
- 50.Nishikawa, M. M., M. S. Lazera, G. G. Barbosa, L. Trilles, B. R. Balassiano, R. C. Macedo, C. C. Bezerra, M. A. Perez, P. Cardarelli, and B. Wanke. 2003. Serotyping of 467 Cryptococcus neoformans isolates from clinical and environmental sources in Brazil: analysis of host and regional patterns. J. Clin. Microbiol. 41:73-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeiffer, T., and D. Ellis. 1991. Environmental isolation of Cryptococcus neoformans gattii from California. J. Infect. Dis. 163:929-930. [DOI] [PubMed] [Google Scholar]
- 52.Pfeiffer, T. J., and D. H. Ellis. 1992. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J. Med. Vet. Mycol. 30:407-408. [PubMed] [Google Scholar]
- 52a.Pfeiffer, T. J., and D. H. Ellis. 1996. Int. Meet. Exhibition Aust. N. Z. Soc. Microbiol., abstr. P5.6.
- 53.Randhawa, H. S., T. Kowshik, and Z. U. Khan. 2003. Decayed wood of Syzygium cumini and Ficus religiosa living trees in Delhi/New Delhi metropolitan area as natural habitat of Cryptococcus neoformans. Med. Mycol. 41:199-209. [DOI] [PubMed] [Google Scholar]
- 54.Randhawa, H. S., T. Kowshik, and Z. U. Khan. 2005. Efficacy of swabbing versus a conventional technique for isolation of Cryptococcus neoformans from decayed wood in tree trunk hollows. Med. Mycol. 43:67-71. [DOI] [PubMed] [Google Scholar]
- 55.Sorrell, T. C., S. C. Chen, P. Ruma, W. Meyer, T. J. Pfeiffer, D. H. Ellis, and A. G. Brownlee. 1996. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J. Clin. Microbiol. 34:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staib, F. 1985. Sampling and isolation of Cryptococcus neoformans from indoor air with the aid of the Reuter centrifugal sampler (RCS) and Guizotia abyssinica creatine agar. A contribution to the mycological-epidemiological control of Cr. neoformans in the fecal matter of caged birds. Zentbl. Bakteriol. Mikrobiol. Hyg. B 180:567-575. [PubMed] [Google Scholar]
- 57.Staib, F., M. Seibold, E. Antweiler, B. Frohlich, S. Weber, and A. Blisse. 1987. The brown colour effect (BCE) of Cryptococcus neoformans in the diagnosis, control and epidemiology of C. neoformans infections in AIDS patients. Zentbl. Bakteriol. Mikrobiol. Hyg. A 266:167-177. [DOI] [PubMed] [Google Scholar]
- 58.Stephen, C., S. Lester, W. Black, M. Fyfe, and S. Raverty. 2002. Multispecies outbreak of cryptococcosis on southern Vancouver Island, BC. Can. Vet. J. 43:792-794. [PMC free article] [PubMed] [Google Scholar]
- 59.Storer, D. A. 1984. A simple high sample volume ashing procedure for determination of soil organic matter. Commun. Soil Sci. Plant Anal. 15:759-772. [Google Scholar]
- 60.Trilles, L., M. Lazera, B. Wanke, B. Theleen, and T. Boekhout. 2003. Genetic characterization of environmental isolates of the Cryptococcus neoformans species complex from Brazil. Med. Mycol. 41:383-390. [DOI] [PubMed] [Google Scholar]
- 61.Vilcins, I., M. Krockenberger, H. Agus, and D. Carter. 2002. Environmental sampling for Cryptococcus neoformans var. gattii from the Blue Mountains National Park, Sydney, Australia. Med. Mycol. 40:53-60. [DOI] [PubMed] [Google Scholar]
- 62.Vishniac, H. S. 2006. A multivariate analysis of soil yeasts isolated from a latitudinal gradient. Microb. Ecol. 52:90-103. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Y., and A. Casadevall. 1994. Decreased susceptibility of melanized Cryptococcus neoformans to UV light. Appl. Environ. Microbiol. 60:3864-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zacharin, R. F. 1978. Emigrant eucalypts: gum trees as exotics. Melbourne University Press, Melbourne, Australia.