Abstract
Lactic acid bacteria, such as Lactococcus lactis, are attractive hosts for the production of plant-bioactive compounds because of their food grade status, efficient expression, and metabolic engineering tools. Two genes from strawberry (Fragaria x ananassa), encoding an alcohol acyltransferase (SAAT) and a linalool/nerolidol synthase (FaNES), were cloned in L. lactis and actively expressed using the nisin-induced expression system. The specific activity of SAAT could be improved threefold (up to 564 pmol octyl acetate h−1 mg protein−1) by increasing the concentration of tRNA1Arg, which is a rare tRNA molecule in L. lactis. Fermentation tests with GM17 medium and milk with recombinant L. lactis strains expressing SAAT or FaNES resulted in the production of octyl acetate (1.9 μM) and linalool (85 nM) to levels above their odor thresholds in water. The results illustrate the potential of the application of L. lactis as a food grade expression platform for the recombinant production of proteins and bioactive compounds from plants.
Plants produce a wide variety of secondary metabolites with a broad range of functionalities that are of industrial interest, including antimicrobial, antifungal, antioxidant, flavor-enhancing, or health-promoting properties (33). One obvious approach for harvesting these compounds is their isolation from plants. In many cases, however, productivities of target compounds are low and they may have to be isolated from complex raw materials requiring advanced downstream processing procedures. In recent years, there has been growing interest in using recombinant microbial systems as alternative production platforms for the efficient production of specific bioactive plant compounds. Microbial production systems offer the possibility for production of target compounds in a clean and simple metabolic background that minimizes the risk of formation of unwanted side products. Moreover, additional metabolic engineering strategies aimed at increasing the availability of precursors or the addition of functional groups that increase bioactivity, as for instance through the addition of glycosyl groups, may be applied. Various groups have described the construction of Escherichia coli strains producing carotenoids, terpenoids, flavonoids, and flavanones after the introduction of the respective metabolic pathways of Rhodotorula rubra, plant, or fungal origin (1, 27, 31, 41, 43). Similarly, yeast strains have been constructed producing taxol or sesquiterpenes (15, 28, 54).
In recent years, Lactococcus lactis has gained a strong position as an alternative cell factory for the production of proteins and bioactive compounds (reference 45 and references therein). This has been facilitated by the development of efficient expression systems such as the nisin-controlled expression (NICE) system. The NICE system consists of a bacterial host with the nisRK regulatory genes integrated into the chromosome and an expression vector carrying the gene of interest under the control of the nisA promoter. Using this system, expression can be efficiently controlled through the addition of nisin (34).
This system has several interesting properties, including the use of a food grade inducer molecule, a linear dose-response curve, and the absence of formation of inclusion bodies and endospores (46). Moreover, the relatively simple metabolism of L. lactis allows efficient rerouting of metabolic fluxes, enabling the rational increase of production levels of desired products. Finally, its food grade status favors its application as a host for the production of plant metabolites that are used as food ingredients. Recently, Martinez-Cuesta et al. (42) reported the first example of the functional expression of a plant protein, coumarate:coenzyme A (CoA) ligase from Arabidopsis thaliana, in L. lactis.
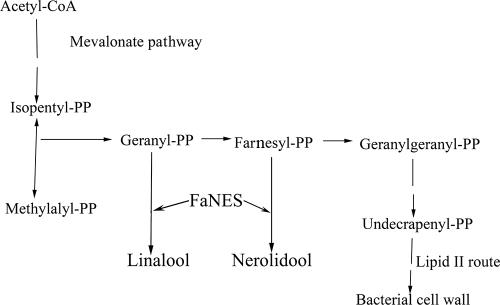
In the current paper, we report on the expression of genes from strawberry (Fragaria x ananassa) in L. lactis. We focus on enzymes involved in the production of two major classes of fruit flavor metabolites: terpenoids and esters. Terpenoids are an important class of biologically active molecules, including flavor and fragrance compounds, pheromones, medically active compounds, growth regulators, and vitamins (2, 44). Terpenoid classification is based on the number of C5 moieties (isopentyl diphosphate and dimethylallyl diphosphate) used to build the carbon skeleton of the molecule. Monoterpenes are the simplest terpenes, composed of two of these moieties, whereas compounds containing three moieties are referred to as sesquiterpenes. Linalool is a monoterpene directly derived from geranyl diphosphate (GPP) and a key flavor compound in strawberry and other fruits (1, 5, 38). This acyclic monoterpene has a sweet, floral, and citrus-like odor, and recently, various linalool synthases have been characterized (1, 17, 29, 40) and used for the enhancement of volatile production in transgenic plants (38). Another component of strawberry flavor is nerolidol, a sesquiterpene with a floral and woody odor that plays a role in the defense response of plants against arthropod pests (32). The production of linalool and nerolidol occurs via the same pathway, where the last biosynthetic step(s) is catalyzed by the Fragaria x ananassa linalool/nerolidol synthase (FaNES) (9) (Fig. 1).
FIG. 1.
Partial view of the undecaprenyl diphosphate acid pathway in L. lactis, focused on the formation of GPP and FPP, and the reaction catalyzed by FaNES leading to the production of the monoterpene linalool, the major terpene produced in L. lactis, and the sequiterpene nerolidol (30).
Esters are key components of the flavors of strawberry, apple, mango, and other fruits and vegetables. In plants they are typically produced by the enzyme-mediated transfer of the acyl chain from an acyl-CoA ester to an alcohol, and a wide variety of such enzymes in many plant species have been characterized (3, 9, 18, 57, 59).
Here we report the cloning and expression of a linalool/nerolidol synthase and an alcohol acyltransferase from strawberry in L. lactis as examples of the suitability of L. lactis as an expression platform for plant genes. Functional expression was analyzed, and the production of monoterpenes, sesquiterpenes, and long-chain alcohol esters during L. lactis fermentation is reported.
MATERIALS AND METHODS
L. lactis strains and growth conditions.
Strains and plasmids used are listed in Table 1. L. lactis strain NZ9000, an MG1363-derived strain with the nisR and nisK genes integrated into the chromosome, was used for cloning and expression purposes. Strain NZ9000 was grown in M17 medium (61) supplemented with 1% glucose (GM17) at 30°C unless indicated otherwise. The following antibiotics were added when appropriate for the selection of plasmid-containing clones: chloramphenicol (10 μg ml−1) and erythromycin (10 μg ml−1). Growth experiments with milk were carried out using skim milk after sterilization (10 min at 110°C) supplemented with Casitone (0.5%) and glucose (1%) prior to inoculation as a protein and carbon source for the nonproteolytic and Lac− strain L. lactis NZ9000. Milk fermentation tests were carried out at 30°C, without the addition of antibiotics to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strain | ||
| L. lactis subsp. cremoris NZ9000 | MG1363 pepN::nisRK; host strain for the NICE system | 36 |
| Plasmids | ||
| pIL253 | Eryr | 58 |
| pNZ8150 | Cmr; nisA transcriptional fusion vector | 46 |
| pNZ7601 | Cmr; pNZ8150 derivative carrying the SAAT gene | This work |
| pNZ7610 | Eryr; pIL253 derivative carrying the nisF promoter fused to the tRNA1Arg gene of L. lactis strain IL-1403 | This work |
| pNZ7630 | Cmr; pNZ8150 derivative carrying the codon-optimized SAAT gene | This work |
| pNZ7640 | Cmr; pNZ8150 derivative carrying the FaNES gene | This work |
Cmr and Eryr, resistance to chloramphenicol and erythromycin, respectively.
DNA and plasmids.
The genes expressed in L. lactis were originally isolated from Fragassia x ananassa (strawberry). The SAAT gene encodes an alcohol acyltransferase (SAAT) that was previously described by Aharoni et al. (3) (GenBank accession number AF193789). The FaNES gene encodes the Fragassia x ananassa enzyme linalool/nerolidol synthase (FaNES), a monoterpene-sesquiterpene synthase that was initially described by Aharoni and O'Connell (4) and subsequently characterized by Aharoni et al. (1, 4) (GenBank accession number AX529025).
SAAT was previously cloned into the pRSET-B vector (3), designed for expression in E. coli. It was amplified with Pwo DNA polymerase (30 cycles of 15 s at 94°C, 30 s at 47°C, and 90 s at 72°C), using the SAAT forward primer 5′-ATTGGAGAAAATTGAGGTCAG-3′ and SAAT reverse primer 5′-CGCCGCATGCGCCACATAATCTTTCTTAATC-3′. The PCR product was digested with SphI, and the resulting fragment was introduced into the pNZ8150 vector using ScaI and SphI sites, producing a 4,581-bp plasmid designated pNZ7601.
For the pNZ7610 insert, the nucleotide sequence described in the supplemental material was purchased in the pPCR-Script vector from Geneart (Regensburg, Germany). This vector was digested with DraI and KpnI, and the resulting 1,339-bp fragment was cloned into pIL253 using the HaeIII and KpnI sites. Hence, this construct was a fusion between the nisF promoter (15), a spacer region usually preceding L. lactis tRNA operons (49), and the sequence predicted for tRNA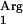 in L. lactis strain IL-1403 (GenBank locus L200111). In this cloning step, the ColE1 origin of replication for E. coli was introduced with the synthetic sequence into the pIL253 vector, producing pNZ7610, an E. coli and L. lactis shuttle vector. The synthetic codon-optimized variant of the SAAT gene (see the nucleotide sequence described in the supplemental material) was purchased from Geneart. For cloning purposes, a PstI site was introduced between the nisA promoter and the ATG start codon of SAAT. The insert was received in the pPCR-script vector and was introduced into pNZ8150 using the PstI and KpnI sites in the vector and in the insert. The resulting vector was designated pNZ7630 and maintains the main characteristics of pNZ8150.
in L. lactis strain IL-1403 (GenBank locus L200111). In this cloning step, the ColE1 origin of replication for E. coli was introduced with the synthetic sequence into the pIL253 vector, producing pNZ7610, an E. coli and L. lactis shuttle vector. The synthetic codon-optimized variant of the SAAT gene (see the nucleotide sequence described in the supplemental material) was purchased from Geneart. For cloning purposes, a PstI site was introduced between the nisA promoter and the ATG start codon of SAAT. The insert was received in the pPCR-script vector and was introduced into pNZ8150 using the PstI and KpnI sites in the vector and in the insert. The resulting vector was designated pNZ7630 and maintains the main characteristics of pNZ8150.
FaNES was amplified using Pwo DNA polymerase, and the resulting fragment was digested with XbaI and introduced into pNZ8150 using ScaI and XbaI sites. The resulting vector carries the FaNES gene under the control of the nisA promoter and was designated pNZ7640. All the plasmids were sequenced to confirm that the sequences obtained were correct. Standard molecular biology techniques and protocols were carried out as described by Sambrook et al. (56).
Enzymatic assays.
For determination of enzyme activities, an aliquot from an overnight culture was used to inoculate (5%, vol/vol) fresh medium, and subsequently, growth was monitored until the optical density at 600 nm (OD600) reached between 0.4 and 0.5 (early exponential phase). The culture was split in two equal subsamples, and nisin (final concentration, 1 ng ml−1) was added to one of these, whereas the other subsample was used as a noninduced control. Aliquots were taken every hour to monitor bacterial growth. At sampling times, 40- to 50-ml aliquots were taken and centrifuged (6,000 rpm, 15 min, 4°C). The pellet was resuspended in 1 ml of cold reaction buffer (described below for each determination), and crude extracts (CE) were prepared by disrupting cells by bead beating (FastprepTM FP120 beater; twice for 30 s each time) using 1 mg of silica sand. An amount of 0.5 ml of buffer was added to wash the sand, and the liquid phase was transferred to an Eppendorf vial, which was centrifuged again (14,000 rpm, 10 min, 4°C). The soluble fractions (cell extracts [CFE]) were used immediately for enzymatic activity assays or stored (−20°C) until the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Acyltransferase activities were determined essentially as described by Aharoni et al. (3). Just before the reaction was started, 70 μl of 1-octanol stock solution (160 mM in reaction buffer) and 50 μl of acetyl-CoA stock solution (4 mM in water) were mixed with 50 μl of the reaction buffer (50 mM Tris-HCl, 1 mM dithiothreitol, pH 8.0) and 30 μl of the sample (CE or CFE) to a final volume of 200 μl in 10-ml vials, and each vial was closed with a Teflon septum. Reactions were carried out at 35°C with vigorous shaking for 5, 10, 15, 20, or 30 min, after which the reaction was stopped by injecting 200 μl CaCl2 (4 M in reaction buffer). Samples were stored at 4°C until injection into a gas chromatograph-mass spectrometer (GC-MS).
Linalool synthase activity was determined as described previously (1). Briefly, in a 10-ml vial, 500 μl of the CFE in reaction buffer (15 mM 3-morpholino-2-hydroxypropanesulfonic acid, pH 7.0, 10% [vol/vol] glycerol, 10 mM MgCl2, 1 mM MnCl2, 1 mM sodium ascorbate, and 2 mM dithiothreitol) was mixed with 480 μl reaction buffer and with 20 μl GPP (from a 2.1 mM solution). The vial was closed with a Teflon septum and incubated with shaking for 60 min at 30°C. Then, 1 ml CaCl2 (4 M in reaction buffer) was added to stop the reaction. Samples were stored at 4°C until injection into the GC-MS.
Reaction products were detected with a GC-MS (ThermoFinigan) operating in selective ion mode (m/z = 61 for octyl acetate and m/z = 93 for linalool, with an ionization potential of 70 eV). Injection was done by an automatic injector (PAL system) equipped with a solid-phase dynamic extraction fiber (polydiemethylsiloxane, 50 μm by 76 mm; Hamilton) when a low level of product was expected. Before injection, samples were shaken for 15 min at 60°C. GC oven conditions were from 40°C (1-min hold) to 250°C (2-min hold) with a 35°C increase per min. At sampling time, chlorohexane was added as an internal standard. Retention times and spectra were compared with those of authentic standards.
Monitoring of product formation during fermentation.
Fermentation conditions were similar to those used for enzymatic assay sampling. In this case, every 60 min, 2 ml of medium was transferred to a 10-ml vial. Samples were quenched by adding 250 μl of a solution consisting of HgSO4 (2.97 mM) and NaCl (6.8 M) in water. The vial was closed with a Teflon septum and stored at 4°C until GC analysis as described above. For monitoring ester production, SAAT-producing cultures were grown in GM17 medium until the OD600 was 0.4 to 0.5. Subsequently, cells were removed by centrifugation (10 min at 5,000 rpm) and resuspended in the same volume of fresh GM17 or skim milk supplemented with 100 μM 1-octanol and prewarmed at 30°C and, when necessary, nisin (final concentration, 1 ng ml−1) was added. To reduce the possible differences in the media between experiments, 1-octanol was sonicated in GM17 (six times at 10-s intervals) prior to inoculation.
Protein analysis and molecular mass estimation.
The apparent molecular mass of the protein was analyzed by SDS-PAGE on 10% (wt/vol) polyacrylamide gel as described by Sambrook et al. (56). Molecular masses were estimated using the Benchmark protein ladder (Invitrogen).
Protein concentrations in cellular extracts were determined using the BCA protein assay kit (Pierce, Rockford, IL) according to the protocols of the manufacturer, using pure bovine serum albumin (Sigma) as the standard. SDS-PAGE gels were analyzed with an ImaGo compact imaging system (B&L Systems, The Netherlands). Analysis of band patterns and quantification were performed with ImageMaster 1D version 3.0 software (Amersham Pharmacia Biotech, The Netherlands).
RNA isolation and Northern blotting.
Total RNA was isolated by the Macaloid method (35) from exponentially growing cultures. For Northern blot analysis, RNA was separated on 1% formaldehyde agarose gel and blotted and hybridized as described by van Rooijen and de Vos (62). Hybridization probes were radiolabeled with [α-32P]dATP by nick translation. The blots were washed with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 sodium citrate) at 65°C, and hybridizing bands were visualized by autoradiography.
RSCU.
Relative synonymous codon usage values (RSCU) were calculated as described by Fuglsang (21) to compare codon usage in SAAT and FaNES with the codon usage in the entire genome of L. lactis IL-1403 and in highly expressed proteins in L. lactis IL-1403. Total codon usage data were collected from the KEGG database (30), and most of the codon usage of highly expressed proteins was derived from the work of Fuglsang (21).
Statistical analysis.
SPSS software (version 14.0; SPSS, Chicago, IL) was used for the statistical analysis. Two-way analysis of variance, one-way analysis of variance, and Student's t test were used when necessary to establish the presence or absence of significant differences (P ≤ 0.05) in enzymatic activity according to the factors “induction” and “recombinant strain.”
RESULTS
Stability of SAAT- and FaNES-recombinant strains.
The standard plasmid pNZ8150 was used for the cloning of SAAT and FaNES, resulting in pNZ7601 and pNZ7640, respectively. In order to demonstrate that pNZ7601 and pNZ7640 were stable in L. lactis NZ9000, plasmid-containing L. lactis NZ9000 clones were incubated for 100 generations without antibiotic in the medium and the retention of the plasmid was confirmed by comparing numbers of CFU per milliliter on M17 medium with or without chloramphenicol for plasmid selection. Moreover, the intactness of the plasmids in 10 colonies isolated from plates without chloramphenicol every 20 generations was confirmed by PCR and restriction analysis (ScaI and SphI).
Expression of FaNES in L. lactis NZ9000(pNZ7640).
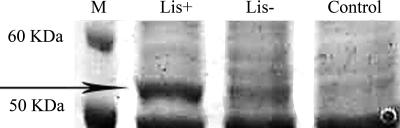
Protein production was evaluated by SDS-PAGE analysis. In crude extracts of induced cultures of L. lactis NZ9000(pNZ7640), an extra band with an apparent molecular mass of 54 kDa was detected, similar to the predicted molecular mass of FaNES (59.2 kDa) (1). This band was observed in samples after 2 h of induction and did not appear in samples of L. lactis NZ9000 with pNZ8150 or in L. lactis NZ9000(pNZ7640) cultures that were not induced with nisin (Fig. 2). According to the densitometry analysis, this protein represented approximately 10% of the total cellular protein. Linalool synthase activity was measured with CFE of nisin-induced cultures of L. lactis NZ9000(pNZ7640). The activity was 0.4 ± 0.1 pmol linalool h−1 μg total protein−1, and no linalool production was detected in the noninduced samples or in the control strain [L. lactis NZ9000(pNZ8150)]. Using purified protein, the specific activity reported by Aharoni et al. (1) was 2.3 nmol h−1 μg protein−1. Hence, it can be calculated that recombinant extracts contained 0.017% of active soluble FaNES. Considering that SDS-PAGE analysis showed that that enzyme accounted for approximately 10% of the soluble protein, these results indicate that the majority of the soluble FaNES in L. lactis is inactive or that other compounds in CFE negatively affected protein activity.
FIG. 2.
Production of FaNES by L. lactis NZ9000 clones. SDS-PAGE analysis of CFE of L. lactis NZ9000(pNZ7640) induced with 1 ng ml−1 nisin for 2 h (Lis+ lane), noninduced L. lactis NZ9000(pNZ7640) (Lis− lane), and L. lactis NZ9000(pNZ8150) induced with 1 ng ml−1 nisin (Control). Lane M contains protein size markers.
Expression of SAAT in L. lactis NZ9000(pNZ7601).
When CE or CFE of nisin-induced L. lactis NZ9000(pNZ7601) were analyzed by SDS-PAGE gels and stained with Coomassie brilliant blue, no extra bands were detected. The specific SAAT activity detected in these extracts was 192 ± 1 pmol of octyl acetate h−1 mg of total protein−1 (Table 2). No detectable octyl acetate production was observed in extracts of L. lactis NZ9000(pNZ8150) or in extracts of noninduced L. lactis NZ9000(pNZ7601) cultures, demonstrating that ester production was due to expression of the SAAT gene (Table 2). Using purified SAAT protein carrying a His tag isolated from E. coli, Aharoni et al. reported a specific SAAT activity for acetyl-CoA and 1-octanol of 4.45 μmol h−1 mg protein−1 (3). According to these data, SAAT protein represents 0.004% of total soluble protein in CFE of L. lactis NZ9000(pNZ7601). This amount of SAAT protein in our L. lactis is, indeed, too low to be detected on a protein gel.
TABLE 2.
Specific SAAT activities in cell extracts of L. lactis NZ9000 strains transformed with different plasmids after the induction of expression with 1 ng ml−1 nisin
| Plasmid(s) in the clone | Description | Activitya |
|---|---|---|
| pNZ7601 | “Wild-type” SAAT in pNZ8150 | 192 ± 1A |
| pNZ7630 | “Codon-optimized” SAAT in pNZ8150 | 225 ± 7A |
| pNZ7601 + pIL253 | PNZ7601 with the empty pIL253 vector | 257 ± 49A |
| pNZ7601 + pNZ7610 | pNZ7601 and pIL253 with tRNA1Arg | 564 ± 14B |
| PNZ7630 + pNZ7610 | “Codon-optimized” gene and pIL253 with tRNA1Arg | <1C |
| pNZ7610 | pIL253 with tRNA1Arg | Not detectableD |
| PNZ8150 | Empty vector | Not detectableD |
Activity was measured in picomoles of octyl acetate per hour per milligram of protein. Superior letters (A to D) placed next to enzymatic activities indicate values that were significantly different (P < 0.05) from values with a different letter.
Transcription analysis.
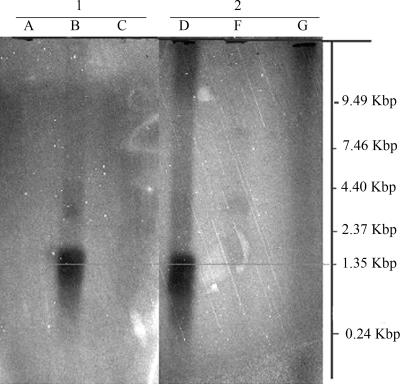
Because the SAAT protein levels detected by SDS-PAGE were much lower than FaNES levels, further research focused on increasing the expression of SAAT protein. As described above, plasmid instability due to the toxicity of the induced protein was not observed. Hence, we hypothesized that limited protein production may be due to low mRNA levels as a result of improper transcription or mRNA instability. Therefore, SAAT mRNA levels in nisin-induced L. lactis NZ9000(pNZ7601) cultures were analyzed and compared with FaNES mRNA concentrations in nisin-induced L. lactis NZ9000(pNZ7640) cultures. Northern blots of mRNA samples form L. lactis NZ9000(pNZ7601) and L. lactis NZ9000(pNZ7640) showed the presence of two clear bands that were around 1.5 kb in induced samples, with similar intensities in both samples (Fig. 3). This size is in agreement with the predicted mRNA size (1,357 bp for SAAT and 1,559 bp for FaNES). No signal was detected with strain NZ9000(pNZ8150) or in noninduced samples, demonstrating that mRNA was produced exclusively in induced recombinant strains carrying plant genes. Therefore, a reduced transcription level or mRNA instability was unlikely to be the major cause of the lower protein level of SAAT produced in recombinant L. lactis NZ9000 strains.
FIG. 3.
Northern blot analysis of L. lactis NZ9000 clones expressing the FaNES gene and SAAT gene blotted with the FaNES probe (lanes 1) and the SAAT probe (lanes 2). Lanes: A, L. lactis NZ9000(pNZ8150) induced with 1 ng ml−1 nisin; B, L. lactis NZ9000(pNZ7640) induced with nisin; C, noninduced L. lactis NZ9000(pNZ7640); D, L. lactis NZ9000(pNZ7601) induced with nisin; E, noninduced L. lactis NZ9000(pNZ7601); F, L. lactis NZ9000(pNZ8150) induced with nisin.
Codon usage analysis.
It is well documented that codon usage is one of the main factors interfering with the efficient production of eukaryotic proteins in microorganisms (46). There are many examples of low translation efficiencies caused by the accumulation of rare codons in bacteria (25, 55) or yeast (8). Accumulations of rare Arg codons can inhibit bacterial growth (23, 51), and in other cases where a protein is highly expressed, aberrant incorporation of amino acids resulted in a high fraction of inactive protein (11).
RSCU were calculated for SAAT and FaNES and plotted in Fig. 4. Frequently used codons in wild-type SAAT or FaNES are rarely used in the highly expressed gene set in L. lactis. These appear as high values in Fig. 4, and the results show that several “rare” codons occur in the SAAT and FaNES (Fig. 4B) sequences. In the SAAT sequence, 35 “rare” codons occur and major “rare” codon accumulations occur at positions 108 to 109 (TTA CGT), 241 to 242 (GAA TCA), 309 to 312 (ATT TTA GAA TTA), and 411 to 412 (ATC ATT). The FaNES gene contains only six rare codons in a total of 520 codons, and there are no rare-codon accumulations in this well-expressed protein. Therefore, we hypothesized that frequencies and the distribution of rare codons are responsible for low protein expression levels due to inefficient translation.
FIG. 4.
Codon usage analysis of SAAT and FaNES. (A) Codon usage in “wild-type” SAAT (▪) and codon usage in “codon-optimized” SAAT (⋄) relative to the codon usage of highly expressed genes in L. lactis. (B) Codon usage in FaNES (⧫) relative to the codon usage of highly expressed genes in L. lactis. The x axis represents the codon position in the protein sequence. Rare codons are defined as codons with a value larger than 40 in the y axis. In both graphs, for representation purposes, the codon usage of AGG, CGG, and CTA in L. lactis is considered to be 0.5 instead of 0.
Optimization of protein expression through codon usage optimization.
Different solutions have been proposed to improve protein expression when there are codon usage problems. In some cases, gene synthesis with optimal codon usage of the complete gene (60) or in the initial part of the gene (8, 63) proved to be an effective means of increasing protein production. Alternatively, rare-codon tRNA may be coexpressed to increase the availability of rare-codon tRNAs (24). To the best of our knowledge, no examples of the last strategy for improving protein production in L. lactis have been reported. Initially, we tried to improve SAAT protein production by introducing a codon usage-optimized gene containing only 32 of a possible 64 codons, potentially solving in a single strategy the translation initiation and translation blockage problems derived by rare-codon accumulation. The codon usage of the resulting gene was similar to that for the set of highly expressed genes in L. lactis (Fig. 4A). The codon-optimized gene was introduced into the vector pNZ8150; the resulting plasmid was designated pNZ7630 and used to transform L. lactis NZ9000. Protein production and enzyme activity were analyzed in induced cultures of L. lactis NZ9000(pNZ7630) as described above. However, no protein band was observed by SDS-PAGE analysis, and enzyme activities in both CE (data not shown) and CFE (Table 2) were comparable to the activities measured with “wild-type” SAAT in extracts of induced L. lactis NZ9000(pNZ7601). From these results, we concluded that codon optimization did not improve protein production. Similar observations have been made by others working on the optimization of protein expression in E. coli (20) and in L. lactis (Igor Mierau, unpublished results).
Optimization of protein expression through tRNA supplementation.
As a next step, we decided to supplement a potentially rare tRNA. Three primary candidates can be recognized based on the rare-codon analysis of SAAT (Fig. 4A). These are tRNA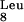 (CUA), tRNA
(CUA), tRNA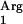 (AGG), and tRNA
(AGG), and tRNA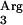 (AGA), and they are frequently reported to interfere with the efficient expression of eukaryotic proteins in bacteria. To our knowledge, no tRNA concentrations have been reported for L. lactis, so we decided to base our selection on the genetic organization of tRNA loci in the genome of strain IL-1403 (10). In this genome, tRNA
(AGA), and they are frequently reported to interfere with the efficient expression of eukaryotic proteins in bacteria. To our knowledge, no tRNA concentrations have been reported for L. lactis, so we decided to base our selection on the genetic organization of tRNA loci in the genome of strain IL-1403 (10). In this genome, tRNA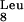 is in a cluster with other tRNA and rRNA genes, and this cluster is under the control of a predicted promoter. The tRNA
is in a cluster with other tRNA and rRNA genes, and this cluster is under the control of a predicted promoter. The tRNA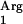 and tRNA
and tRNA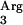 genes, however, are located between two terminators and may not be efficiently expressed. We decided to supplement the tRNA
genes, however, are located between two terminators and may not be efficiently expressed. We decided to supplement the tRNA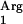 (AGG) gene because it has the lowest RSCU and may also allow the incorporation of arginine at positions corresponding to the AGA codon (21). In the SAAT sequence, AGG appears five times and AGA six times, which represents 58% of Arg codons in the protein.
(AGG) gene because it has the lowest RSCU and may also allow the incorporation of arginine at positions corresponding to the AGA codon (21). In the SAAT sequence, AGG appears five times and AGA six times, which represents 58% of Arg codons in the protein.
Rare codons are used mostly in proteins that are expressed in non-exponential growth phases (22), and in theory, tRNA supplementation should be done carefully to avoid any toxic effect. Therefore, we designed a strategy in which this rare tRNA was added as an extra sequence in an independent plasmid under the control of an inducible promoter, analogously to the strategy successfully applied for E. coli (60). The nisF promoter was selected because it is inducible by nisin, is strictly controlled, and is compatible with the NICE system (15). By this strategy, the pIL253 vector was used to insert a copy of the L. lactis tRNA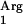 gene under the control of the nisF promoter, generating the plasmid pNZ7610. This plasmid was transformed into L. lactis NZ9000, resulting in L. lactis NZ9000(pNZ7610). Also, the control strain L. lactis NZ9000(pIL253), with unmodified pIL253, was created. Strains with pIL253 or pNZ7610 are erythromycin resistant. The pIL253-derived vectors are compatible with plasmids derived from pNZ8150 carrying a chloramphenicol resistance marker (45). Cotransformation of L. lactis NZ9000 with pNZ7601 and pNZ7610 results in a strain carrying both the SAAT and tRNA
gene under the control of the nisF promoter, generating the plasmid pNZ7610. This plasmid was transformed into L. lactis NZ9000, resulting in L. lactis NZ9000(pNZ7610). Also, the control strain L. lactis NZ9000(pIL253), with unmodified pIL253, was created. Strains with pIL253 or pNZ7610 are erythromycin resistant. The pIL253-derived vectors are compatible with plasmids derived from pNZ8150 carrying a chloramphenicol resistance marker (45). Cotransformation of L. lactis NZ9000 with pNZ7601 and pNZ7610 results in a strain carrying both the SAAT and tRNA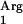 genes under the control of a nisin-inducible promoter. Control strains were constructed by replacing either pNZ7601 or pNZ7610 with the corresponding empty vector pNZ8150 or pIL252, respectively. All of these control strains were able to grow on GM17 supplemented with erythromycin and chloramphenicol.
genes under the control of a nisin-inducible promoter. Control strains were constructed by replacing either pNZ7601 or pNZ7610 with the corresponding empty vector pNZ8150 or pIL252, respectively. All of these control strains were able to grow on GM17 supplemented with erythromycin and chloramphenicol.
SAAT expression in tRNA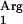 gene-supplemented clones.
gene-supplemented clones.
The supplementation with the tRNA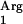 gene was a successful strategy. The coexpression of SAAT and the tRNA
gene was a successful strategy. The coexpression of SAAT and the tRNA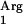 gene resulted in a threefold increase in specific SAAT activity in L. lactis NZ9000(pNZ7630pNZ7610) CFE, compared to activities in “wild-type” and “codon-optimized” SAAT clones (Table 2). The specific octyl acetate production rate with L. lactis NZ9000(pNZ7601pIL253) was similar to that in L. lactis NZ9000(pNZ7601), demonstrating that the increased acyltransferase activity is due the expression of the tRNA
gene resulted in a threefold increase in specific SAAT activity in L. lactis NZ9000(pNZ7630pNZ7610) CFE, compared to activities in “wild-type” and “codon-optimized” SAAT clones (Table 2). The specific octyl acetate production rate with L. lactis NZ9000(pNZ7601pIL253) was similar to that in L. lactis NZ9000(pNZ7601), demonstrating that the increased acyltransferase activity is due the expression of the tRNA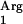 gene (Table 2). No octyl acetate production was detected with L. lactis NZ9000(pNZ7610), demonstrating that the SAAT gene is necessary for the enzymatic activity. Finally, when the tRNA
gene (Table 2). No octyl acetate production was detected with L. lactis NZ9000(pNZ7610), demonstrating that the SAAT gene is necessary for the enzymatic activity. Finally, when the tRNA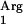 gene was coexpressed with the codon-optimized gene in strain L. lactis NZ9000(pNZ7630pNZ7610), acyltransferase activity was reduced dramatically to below the detection limit (Table 2). This codon-optimized gene no longer contained the rare codon AGG or AGC, and hence the overexpression of tRNA
gene was coexpressed with the codon-optimized gene in strain L. lactis NZ9000(pNZ7630pNZ7610), acyltransferase activity was reduced dramatically to below the detection limit (Table 2). This codon-optimized gene no longer contained the rare codon AGG or AGC, and hence the overexpression of tRNA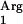 may lead to the accumulation of Arg linked to tRNA
may lead to the accumulation of Arg linked to tRNA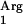 , which cannot be used for protein synthesis and ultimately may result in reduced levels of usable tRNAArg molecules. Analysis of the noninduced clones resulted in no octyl acetate production, which is in agreement with the tight control of the NICE system. A sample of all CFE was subjected to SDS-PAGE analysis. No protein band with the expected molecular mass could be visualized in extracts of nisin-induced clones.
, which cannot be used for protein synthesis and ultimately may result in reduced levels of usable tRNAArg molecules. Analysis of the noninduced clones resulted in no octyl acetate production, which is in agreement with the tight control of the NICE system. A sample of all CFE was subjected to SDS-PAGE analysis. No protein band with the expected molecular mass could be visualized in extracts of nisin-induced clones.
Production of terpenes by FaNES-producing L. lactis NZ9000 clones during fermentation in GM17 or milk.
L. lactis NZ9000(pNZ7640) carrying the FaNES gene was grown in GM17 medium and induced with nisin at early exponential phase (OD600 = 0.4). Linalool synthase uses GPP as a substrate for linalool production. GPP is an intermediate in the production of lipid II in L. lactis (Fig. 1). In this pathway, GPP is converted to farnesyl-PP (FPP), which is also accepted as a substrate by FaNES and is converted to the sesquiterpene nerolidol. We hypothesized that FaNES-producing L. lactis NZ9000 strains may be able to produce both linalool and nerolidol, and therefore samples were removed from the medium to monitor terpene production.
Initially, we used purge-and-trap analysis to concentrate the volatile compounds formed during L. lactis NZ9000(pNZ7640) fermentation in GM17. Chromatographic conditions were similar to those used by Aharoni et al. (1). Comparative analysis of induced and noninduced samples showed that FaNES expression indeed resulted in the production of two compounds that were identified as linalool and nerolidol, respectively, by comparing their retention times and mass spectra with those of authentic standards. In all samples, the amount of linalool produced was four times higher than the amount of nerolidol, and therefore we focused subsequent experiments on linalool production.
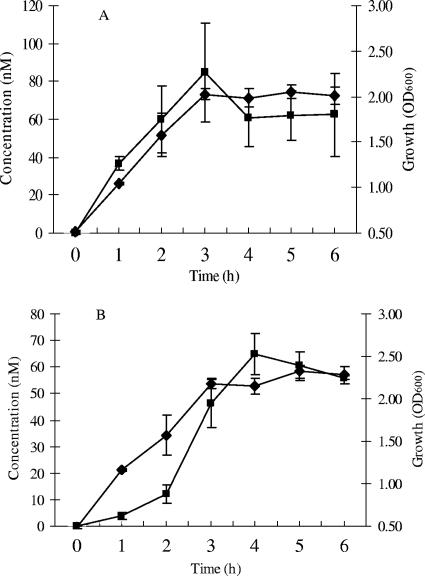
In L. lactis NZ9000(pNZ7640), linalool was rapidly produced upon induction with nisin, and after 3 h, a linalool concentration of 85 ± 26 nM was detected (Fig. 5A). Subsequently, the linalool concentration remained constant during the stationary phase. The results show that there is a clear correlation between bacterial growth and linalool production, and it can be calculated that approximately 1.3 pmol of linalool was produced per mg of cells (with 30 mg ml−1 being produced per OD600 unit) (52). When L. lactis NZ9000(pNZ7640) cell suspensions or CFE were incubated with linalool, no degradation of the monoterpene was observed (data not shown), indicating that the constant linalool levels are not caused by an equilibrium between linalool formation and degradation. When fresh GM17 medium was added to a fully grown culture, linalool levels increased in proportion to the bacterial growth (data not shown). The correlation between linalool and growth may be related to GPP availability. In nongrowing cells, the lipid II synthesis rates and GPP levels may be low. Hence, nongrowing cells may no longer produce linalool due to limiting levels of GPP. Alternatively, FaNES may be sensitive to low intracellular pH levels in the later stages of the fermentation process.
FIG. 5.
Linalool production in GM17 and skim milk by recombinant L. lactis NZ9000(pNZ7640) expressing FaNES. (A) Fermentation in GM17; (B) fermentation in skim milk. ⧫, bacterial growth; ▪, linalool concentration. At time zero, nisin was added (1 ng ml−1) for the induction of protein expression. Data are averages of sample results from three independent cultures, and the standard deviations are depicted.
FaNES productivity during fermentation was high compared with activities determined in CFE. The production rate in the first 3 h of the fermentation process was 146 pmol linalool h−1 mg total protein−1. This corresponds to approximately 40% of the production rate measured in vitro with CFE (400 pmol linalool h−1 mg total protein−1). Differences in the substrate concentrations or intracellular pH values that are suboptimal for FaNES could explain these differences.
Finally, FaNES was expressed during growth in skim milk supplemented with Casitone and glucose, required for proper growth of the expression host L. lactis NZ9000. Linalool was produced to a concentration of 65.0 ± 7.8 nM after 4 h of incubation, and subsequently, the concentration remained constant (Fig. 5B). There was no detectable linalool production in noninduced samples or in the clones with the empty vector (pNZ8150) in all fermentation tests. These results are in agreement with the results obtained with GM17 medium and clearly indicate that L. lactis NZ9000(pNZ7640) can be used for the in situ production of the plant flavor compound linalool in fermented milk.
Octyl acetate production during fermentation in GM17 and milk.
L. lactis NZ9000(pNZ7601) carrying the wild-type SAAT gene was grown in GM17 and skim milk to study ester production. In initial fermentations in GM17, we did not observe significant ester production (data not shown). SAAT catalyzes the production of ethyl esters from acetyl-CoA and terminal alcohols, and we postulated that levels of alcohols in GM17 are too low to allow ester production. Various alcohols can be used as substrates by SAAT with 1-octanol as the preferred substrate (3). Therefore, 1-octanol was added to the medium to a concentration of 100 μM as the alcohol substrate. No growth inhibition of L. lactis NZ9000 occurred at this concentration of 1-octanol (data not shown). For monitoring ester production in the SAAT-producing clone, cultures were grown in GM17 medium until the OD600 was 0.4. The cells were harvested by centrifugation and resuspended in the same volume of fresh GM17 supplemented with 100 μM 1-octanol.
The results clearly demonstrated that alcohol addition was necessary for measurable ester production and that octyl acetate production was linear with time for at least 4 h after induction and transfer to the fresh medium (data not shown). The octyl acetate production rate with an induced culture of L. lactis NZ9000(pNZ7601) was 326 pmol octyl acetate h−1 mg of total protein−1, which is nine times higher than the activity observed with a noninduced culture and with a clone carrying the pNZ8150 vector (Table 3). This background octyl acetate production activity may be caused by the indigenous L. lactis esterase EstA, which is capable of catalyzing ester production through the condensation of acid and alcohols or through transacylation reactions (39, 47) We also included the L. lactis NZ9000(pNZ7601pNZ7610) strain producing tRNA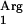 and L. lactis NZ9000(pNZ7630) carrying the codon-optimized SAAT gene in these experiments. The octyl acetate production rates with induced and noninduced cultures of these clones were similar to production rates with their L. lactis NZ9000(pNZ7601) counterparts (Table 3). Finally, we evaluated the possibility of producing octyl acetate during fermentation in skim milk supplemented with glucose and Casitone. Cells were inoculated in skim milk (0.1%), and when necessary, nisin was added at an OD600 of 0.4 to 0.5. The results showed that during skim milk fermentation, L. lactis NZ9000(pNZ7601) can produce 776 pmol octyl acetate h−1 mg total protein−1 after induction with nisin (Table 3), which is almost twofold higher than production rates in GM17. Octyl acetate production was linear during the initial 5 h.
and L. lactis NZ9000(pNZ7630) carrying the codon-optimized SAAT gene in these experiments. The octyl acetate production rates with induced and noninduced cultures of these clones were similar to production rates with their L. lactis NZ9000(pNZ7601) counterparts (Table 3). Finally, we evaluated the possibility of producing octyl acetate during fermentation in skim milk supplemented with glucose and Casitone. Cells were inoculated in skim milk (0.1%), and when necessary, nisin was added at an OD600 of 0.4 to 0.5. The results showed that during skim milk fermentation, L. lactis NZ9000(pNZ7601) can produce 776 pmol octyl acetate h−1 mg total protein−1 after induction with nisin (Table 3), which is almost twofold higher than production rates in GM17. Octyl acetate production was linear during the initial 5 h.
TABLE 3.
Octyl acetate production rates by SAAT-producing L. lactis NZ9000 strains transformed with different plasmidsa
| Fermentation medium and plasmid(s) used | Activity (pmol h−1 mg protein−1)b
|
|
|---|---|---|
| Induced | Noninduced | |
| GM17 | ||
| pNZ7601 | 326 ± 87A | 65 ± 33B |
| pNZ7601 + pNZ7610 | 371 ± 2A | 73 ± 38B |
| pNZ7630 | 355 ± 57A | 40 ± 1B |
| pNZ8150 | 40 ± 9B | 55 ± 7B |
| Skim milk | ||
| pNZ7601 | 776 ± 66C | 80 ± 8D |
| pNZ8150 | 74 ± 13D | 68 ± 9D |
Fermentation was performed with GM17 medium or skim milk, and nisin was added at a concentration of 1 ng ml−1 at early exponential phase for the induction of protein production.
Superior letters (A to D) placed next to enzymatic activities indicate values that were significantly different (P < 0.05) from other values with a different letter.
DISCUSSION
We have cloned and expressed in L. lactis two proteins from the secondary metabolism of strawberry. Both proteins are involved in the production of flavor compounds (1, 3). The proteins could be functionally expressed, but the amount of active protein produced was much lower than that typically observed with bacterial proteins in L. lactis (45). For FaNES, we observed significant protein production, but our results indicate that only a fraction of the protein was active. Protein synthesis in a SAAT-expressing clone was much less efficient, and we were able to demonstrate that the efficiency of expression could be improved almost threefold using a rare-tRNA supplementation approach. Rare-tRNA supplementation has been successfully applied in E. coli and other microbial cell factories (6, 7, 11, 14). To our knowledge, the current study is the first successful example of rare-tRNA supplementation in L. lactis. It is important to notice that low yields, or even the complete absence, of functional expression levels are frequently observed when plant enzymes are expressed in E. coli or other microbial expression platforms. For instance, melon alcohol acyltransferase, an enzyme highly similar to SAAT, could not be functionally expressed in E. coli, whereas in this case, active enzyme could be produced in recombinant yeast (64). Our results and those presented by Martinez-Cuesta (42) represent three successful examples of very different plant proteins that were actively produced in L. lactis and justify a broader exploration of the suitability of this organism as a complementary screening host for elucidating protein function and screening of plant cDNA libraries in a food grade bacterial expression platform.
FaNES-expressing L. lactis NZ9000 was able to produce linalool and nerolidol without substrate addition. FaNES catalyzes the synthesis of these compounds from GPP and FPP, respectively (Fig. 1), and L. lactis has the enzymatic machinery necessary to produce GPP and FPP as intermediates in the undecaprenyl diphosphate acid pathway (30). Using the linalool synthetase activity measured in CFE, it can be calculated that the productivity of growing cells was approximately 40% of the maximal productivity. This indicates that indigenous GPP levels were not severely limiting for linalool production in growing cells, especially when the fact that intracellular conditions may be suboptimal for the enzyme is taken into account. To our knowledge there are no reports of GPP or FPP concentrations in L. lactis, but this example demonstrates the potential of using L. lactis for the characterization of enzymes for the biosynthesis of monoterpenes and sesquiterpenes, classes of compounds containing a wide range of high-value bioactive compounds. Several groups have reported the construction of microbial cell factories for these compounds, as exemplified by the introduction in yeast or in E. coli of the biosynthetic pathways for monoterpenes, sesquiterpenes, and carotenoids (12, 19, 41, 65). The efficient metabolic engineering strategies that are available for L. lactis could enable the construction of efficient cell factories for the production of target terpenes (26).
SAAT-expressing L. lactis NZ9000 required the addition of long-chain alcohols for the production of ethyl esters. These results show that acetyl-CoA levels were sufficient to sustain a base-level ester production, but the equal levels of productivity of L. lactis NZ9000 clones producing different SAAT levels indicates that acetyl-CoA levels may limit ester formation. Glucose, which was used as a carbon source in our experiments, is converted mainly to l-lactate, whereas approximately 2% is converted to acetyl-CoA; ultimately, ethanol, acetate, or biomass formation occurs (16, 50). In our clones, at least three enzymes may compete for acetyl-CoA pools (phosphotransacetylase, acetaldehyde/alcohol dehydrogenase, and SAAT), and the relative amount of acetyl-CoA used for ester production depends on the amounts and kinetic parameters of these enzymes. Lactococcal primary metabolism can be efficiently rerouted, and this offers the potential of increasing acetyl-CoA levels (48) and hence ester productivity.
The octyl acetate and linalool concentrations produced in fermentation tests are higher than the odor threshold values for these molecules. The octyl acetate concentration in GM17 of 1.9 μM is 27 times higher than the reported odor threshold for octyl acetate in water (Flavor-Base, version 2004, Leffinger & Associates, Canton, GA). Analogously, linalool levels of 13 ppb (85 nM) are twofold higher than its odor threshold (6 ppb) in water (37). In a preliminary sensorial analysis done with GM17 cultures, however, neither octyl acetate nor linalool could be detected, probably because of the strong background odor of this medium. The expression plasmids that were used can be easily transferred into a food grade expression system by exchanging the chloramphenicol marker with lacF as the selective marker (45, 53). Hence, L. lactis may be a valuable production host for plant-derived bioactive compounds for food applications.
Supplementary Material
Acknowledgments
Igor Hernández acknowledges Marke Beerthuyzen, Iris van Swam, and Jilbert Bruinsma for excellent assistance. Anders Fuglsang is acknowledged for assistance with the codon usage analysis.
Igor Hernández acknowledges his postdoctoral fellowship from the Department of Education, Universities, and Research of the Basque Government.
Footnotes
Published ahead of print on 5 January 2007.
Supplemental material for this article may be found at http://aem.asm.org.
REFERENCES
- 1.Aharoni, A., A. P. Giri, F. W. Verstappen, C. M. Bertea, R. Sevenier, Z. Sun, M. A. Jongsma, W. Schwab, and H. J. Bouwmeester. 2004. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16:3110-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aharoni, A., M. A. Jongsma, and H. J. Bouwmeester. 2005. Volatile science? Metabolic engineering of terpenoids in plants. Trends Plant Sci. 10:594-602. Epub 14 November 2005. [DOI] [PubMed] [Google Scholar]
- 3.Aharoni, A., L. C. Keizer, H. J. Bouwmeester, Z. Sun, M. Alvarez-Huerta, H. A. Verhoeven, J. Blaas, A. M. van Houwelingen, R. C. De Vos, H. van der Voet, R. C. Jansen, M. Guis, J. Mol, R. W. Davis, M. Schena, A. J. van Tunen, and A. P. O'Connell. 2000. Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12:647-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aharoni, A., and A. P. O'Connell. 2002. Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J. Exp. Bot. 53:2073-2087. [DOI] [PubMed] [Google Scholar]
- 5.Azodanlou, R., C. Darbellay, J. L. Luisier, J. C. Villettaz, and R. Amado. 2003. Quality assessment of strawberries (Fragaria species). J. Agric. Food Chem. 51:715-721. [DOI] [PubMed] [Google Scholar]
- 6.Baca, A. M., and W. G. J. Hol. 2000. Overcoming codon bias: a method for high-level overexpression of Plasmodium and other AT-rich parasite genes in Escherichia coli. Int. J. Parasitol. 30:113-118. [DOI] [PubMed] [Google Scholar]
- 7.Baden, H. A., S. P. Sarma, R. B. Kapust, R. A. Byrd, and D. S. Waugh. 1998. The amino-terminal domain of human STAT4. Overproduction, purification, and biophysical characterization. J. Biol. Chem. 273:17109-17114. [DOI] [PubMed] [Google Scholar]
- 8.Batard, Y., A. Hehn, S. Nedelkina, M. Schalk, K. Pallett, H. Schaller, and D. Werck-Reichhart. 2000. Increasing expression of P450 and P450-reductase proteins from monocots in heterologous systems. Arch. Biochem. Biophys. 379:161-169. [DOI] [PubMed] [Google Scholar]
- 9.Beekwilder, J., M. Alvarez-Huerta, E. Neef, F. W. A. Verstappen, H. J. Bouwmeester, and A. Aharoni. 2004. Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 135:1865-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calderone, T. L., R. D. Stevens, and T. G. Oas. 1996. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 262:407-412. [DOI] [PubMed] [Google Scholar]
- 12.Crock, J., M. Wildung, and R. Croteau. 1997. Isolation and bacterial expression of a sesquiterpene synthase cDNA clone from peppermint (Mentha x piperita, L.) that produces the aphid alarm pheromone (E)-beta-farnesene. Proc. Natl. Acad. Sci. USA 94:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dejong, J. M., Y. Liu, A. P. Bollon, R. M. Long, S. Jennewein, D. Williams, and R. B. Croteau. 2006. Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol. Bioeng. 93:212-224. [DOI] [PubMed] [Google Scholar]
- 14.Del Tito, B. J., Jr., J. M. Ward, J. Hodgson, C. J. Gershater, H. Edwards, L. A. Wysocki, F. A. Watson, G. Sathe, and J. F. Kane. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 177:7086-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vos, W. M., and J. Hugenholtz. 2004. Engineering metabolic highways in lactococci and other lactic acid bacteria. Trends Biotechnol. 22:72-79. [DOI] [PubMed] [Google Scholar]
- 17.Dudareva, N., L. Cseke, V. M. Blanc, and E. Pichersky. 1996. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8:1137-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudareva, N., J. C. D'Auria, K. H. Nam, R. A. Raguso, and E. Pichersky. 1998. Acetyl-CoA:benzylalcohol acetyltransferase—an enzyme involved in floral scent production in Clarkia breweri. Plant J. 14:297-304. [DOI] [PubMed] [Google Scholar]
- 19.Farmer, W. R., and J. C. Liao. 2000. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18:533-537. [DOI] [PubMed] [Google Scholar]
- 20.Flick, K., S. Ahuja, A. Chene, M. T. Bejarano, and Q. Chen. 2004. Optimized expression of Plasmodium falciparum erythrocyte membrane protein 1 domains in Escherichia coli. Malar. J. 3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuglsang, A. 2003. Lactic acid bacteria as prime candidates for codon optimization. Biochem. Biophys. Res. Commun. 312:285-291. [DOI] [PubMed] [Google Scholar]
- 22.Fuglsang, A. 2003. Strong associations between gene function and codon usage. APMIS 111:843-847. [DOI] [PubMed] [Google Scholar]
- 23.Gao, W., S. Tyagi, F. R. Kramer, and E. Goldman. 1997. Messenger RNA release from ribosomes during 5′-translational blockage by consecutive low-usage arginine but not leucine codons in Escherichia coli. Mol. Microbiol. 25:707-716. [DOI] [PubMed] [Google Scholar]
- 24.Gustafsson, C., S. Govindarajan, and J. Minshull. 2004. Codon bias and heterologous protein expression. Trends Biotechnol. 22:346-353. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, C. S., B. Bose, and R. T. Sauer. 2002. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:3440-3445. Epub 12 March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz, J., M. Kleerebezem, M. Starrenburg, J. Delcour, W. de Vos, and P. Hols. 2000. Lactococcus lactis as a cell factory for high-level diacetyl production. Appl. Environ. Microbiol. 66:4112-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, E. I., M. Kaneko, Y. Ohnishi, and S. Horinouchi. 2003. Production of plant-specific flavanones by Escherichia coli containing an artificial gene cluster. Appl. Environ. Microbiol. 69:2699-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson, B. E., E. A. Hart-Wells, and S. P. Matsuda. 2003. Metabolic engineering to produce sesquiterpenes in yeast. Org. Lett. 5:1629-1632. [DOI] [PubMed] [Google Scholar]
- 29.Jia, J. W., J. Crock, S. Lu, R. Croteau, and X. Y. Chen. 1999. (3R)-linalool synthase from Artemisia annua L.: cDNA isolation, characterization, and wound induction. Arch. Biochem. Biophys. 372:143-149. [DOI] [PubMed] [Google Scholar]
- 30.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaneko, M., E. I. Hwang, Y. Ohnishi, and S. Horinouchi. 2003. Heterologous production of flavanones in Escherichia coli: potential for combinatorial biosynthesis of flavonoids in bacteria. J. Ind. Microbiol. Biotechnol. 30:456-461. [DOI] [PubMed] [Google Scholar]
- 32.Kappers, I. F., A. Aharoni, T. W. van Herpen, L. L. Luckerhoff, M. Dicke, and H. J. Bouwmeester. 2005. Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 309:2070-2072. [DOI] [PubMed] [Google Scholar]
- 33.Kris-Etherton, P. M., K. D. Hecker, A. Bonanome, S. M. Coval, A. E. Binkoski, K. F. Hilpert, A. E. Griel, and T. D. Etherton. 2002. Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 113(Suppl. 9B):71S-88S. [DOI] [PubMed] [Google Scholar]
- 34.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 35.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 36.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 37.Reference deleted.
- 38.Lewinsohn, E., F. Schalechet, J. Wilkinson, K. Matsui, Y. Tadmor, K. H. Nam, O. Amar, E. Lastochkin, O. Larkov, U. Ravid, W. Hiatt, S. Gepstein, and E. Pichersky. 2001. Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127:1256-1265. [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, S. Q., R. Holland, and V. L. Crow. 2003. Ester synthesis in an aqueous environment by Streptococcus thermophilus and other dairy lactic acid bacteria. Appl. Microbiol. Biotechnol. 63:81-88. [DOI] [PubMed] [Google Scholar]
- 40.Lucker, J., M. K. El Tamer, W. Schwab, F. W. Verstappen, L. H. van der Plas, H. J. Bouwmeester, and H. A. Verhoeven. 2002. Monoterpene biosynthesis in lemon (Citrus limon). cDNA isolation and functional analysis of four monoterpene synthases. Eur. J. Biochem. 269:3160-3171. [DOI] [PubMed] [Google Scholar]
- 41.Martin, V. J., D. J. Pitera, S. T. Withers, J. D. Newman, and J. D. Keasling. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796-802. Epub 1 June 2003. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Cuesta, M. C., M. J. Gasson, and A. Narbad. 2005. Heterologous expression of the plant coumarate: CoA ligase in Lactococcus lactis. Lett. Appl. Microbiol. 40:44-49. [DOI] [PubMed] [Google Scholar]
- 43.Matthews, P. D., and E. T. Wurtzel. 2000. Metabolic engineering of carotenoid accumulation in Escherichia coli by modulation of the isoprenoid precursor pool with expression of deoxyxylulose phosphate synthase. Appl. Microbiol. Biotechnol. 53:396-400. [DOI] [PubMed] [Google Scholar]
- 44.McGarvey, D. J., and R. Croteau. 1995. Terpenoid metabolism. Plant Cell 7:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mierau, I., and M. Kleerebezem. 2005. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 68:705-717. [DOI] [PubMed] [Google Scholar]
- 46.Mierau, I., K. Olieman, J. Mond, and E. J. Smid. 2005. Optimization of the Lactococcus lactis nisin-controlled gene expression system NICE for industrial applications. Microb. Cell Fact. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nardi, M., C. Fiez-Vandal, P. Tailliez, and V. Monnet. 2002. The EstA esterase is responsible for the main capacity of Lactococcus lactis to synthesize short chain fatty acid esters in vitro. J. Appl. Microbiol. 93:994-1002. [DOI] [PubMed] [Google Scholar]
- 48.Neves, A. R., R. Ventura, N. Mansour, C. Shearman, M. J. Gasson, C. Maycock, A. Ramos, and H. Santos. 2002. Is the glycolytic flux in Lactococcus lactis primarily controlled by the redox charge? Kinetics of NAD(+) and NADH pools determined in vivo by 13C NMR. J. Biol. Chem. 277:28088-28098. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson, D., and E. Johansen. 1994. A conserved sequence in tRNA and rRNA promoters of Lactococcus lactis. Biochim. Biophys. Acta 1219:141-144. [DOI] [PubMed] [Google Scholar]
- 50.Nordkvist, M., N. B. Jensen, and J. Villadsen. 2003. Glucose metabolism in Lactococcus lactis MG1363 under different aeration conditions: requirement of acetate to sustain growth under microaerobic conditions. Appl. Environ. Microbiol. 69:3462-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olivares-Trejo, J. J., J. G. Bueno-Martinez, G. Guarneros, and J. Hernandez-Sanchez. 2003. The pair of arginine codons AGA AGG close to the initiation codon of the lambda int gene inhibits cell growth and protein synthesis by accumulating peptidyl-tRNAArg4. Mol. Microbiol. 49:1043-1049. [DOI] [PubMed] [Google Scholar]
- 52.Pedersen, M. B., B. J. Koebmann, P. R. Jensen, and D. Nilsson. 2002. Increasing acidification of nonreplicating Lactococcus lactis ΔthyA mutants by incorporating ATPase activity. Appl. Environ. Microbiol. 68:5249-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platteeuw, C., I. van Alen-Boerrigter, S. van Schalkwijk, and W. M. de Vos. 1996. Food-grade cloning and expression system for Lactococcus lactis. Appl. Environ. Microbiol. 62:1008-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ro, D. K., E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Chang, S. T. Withers, Y. Shiba, R. Sarpong, and J. D. Keasling. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940-943. [DOI] [PubMed] [Google Scholar]
- 55.Roche, E. D., and R. T. Sauer. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 57.Shalit, M., N. Katzir, Y. Tadmor, O. Larkov, Y. Burger, F. Shalekhet, E. Lastochkin, U. Ravid, O. Amar, M. Edelstein, Z. Karchi, and E. Lewinsohn. 2001. Acetyl-CoA: alcohol acetyltransferase activity and aroma formation in ripening melon fruits. J. Agric. Food Chem. 49:794-799. [DOI] [PubMed] [Google Scholar]
- 58.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 59.Souleyre, E. J. F., D. R. Greenwood, E. N. Friel, S. Karunairetnam, and R. D. Newcomb. 2005. An alcohol acyl transferase from apple (cv. Royal Gala), MpAAT1, produces esters involved in apple fruit flavor. FEBS J. 272:3132-3144. [DOI] [PubMed] [Google Scholar]
- 60.Spanjaard, R. A., K. Chen, J. R. Walker, and J. van Duin. 1990. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNA(Arg). Nucleic Acids Res. 18:5031-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Rooijen, R. J., and W. M. de Vos. 1990. Molecular cloning, transcriptional analysis, and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499-18503. [PubMed] [Google Scholar]
- 63.Vervoort, E. B., A. van Ravestein, N. N. van Peij, J. C. Heikoop, P. J. van Haastert, G. F. Verheijden, and M. H. Linskens. 2000. Optimizing heterologous expression in dictyostelium: importance of 5′ codon adaptation. Nucleic Acids Res. 28:2069-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yahyaoui, F. E., C. Wongs-Aree, A. Latche, R. Hackett, D. Grierson, and J. C. Pech. 2002. Molecular and biochemical characteristics of a gene encoding an alcohol acyl-transferase involved in the generation of aroma volatile esters during melon ripening. Eur. J. Biochem. 269:2359-2366. [DOI] [PubMed] [Google Scholar]
- 65.Yoon, S. H., Y. M. Lee, J. E. Kim, S. H. Lee, J. H. Lee, J. Y. Kim, K. H. Jung, Y. C. Shin, J. D. Keasling, and S. W. Kim. 2006. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol. Bioeng. 17:17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.