Abstract
Escherichia coli strains isolated from commercial broilers and an experimental flock of chickens were screened to determine phenotypic expression of antimicrobial resistance and carriage of drug resistance determinants. The goal of this study was to investigate the influence of oxytetracycline, sarafloxacin, and enrofloxacin administration on the distribution of resistance determinants and strain types among intestinal commensal E. coli strains isolated from broiler chickens. We detected a high prevalence of resistance to drugs such as tetracycline (36 to 97%), sulfonamides (50 to 100%), and streptomycin (53 to 100%) in E. coli isolates from treated and untreated flocks. These isolates also had a high prevalence of class 1 integron carriage, and most of them possessed the streptomycin resistance cassette, aadA1. In order to investigate the contribution of E. coli strain distribution to the prevalence of antimicrobial resistance and the resistance determinants, isolates from each flock were DNA fingerprinted by enterobacterial repetitive intergenic consensus sequence (ERIC) PCR. Although very diverse E. coli strain types were detected, four ERIC strain types were present on all of the commercial broiler farms, and two of the strains were also found in the experimental flocks. Each E. coli strain consisted of both susceptible and antimicrobial agent-resistant isolates. In some instances, isolates of the same E. coli strain expressed the same drug resistance patterns although they harbored different tet determinants or streptomycin resistance genes. Therefore, drug resistance patterns could not be explained solely by strain prevalence, indicating that mobile elements contributed significantly to the prevalence of resistance.
The poultry industry is a significant economic force in the United States; the National Agricultural Statistics Service estimates that the average value of poultry production is more than 50 billion dollars per year (http://www.nass.usda.gov/Census_of_Agriculture/index.asp; accessed 9 May 2006). The demand for chicken has increased over the past 50 years from 1 million pounds in 1950 to more than 40 billion pounds in 2000, a 4-log10 increase in production. However, the economic losses due to cellulitis and airsacculitis infections in broiler chickens were more than $80 million in 2002 (http://usda.mannlib.cornell.edu/reports/nassr/poultry/ppy-bb; accessed 9 May 2006). Escherichia coli is the primary causative agent of cellulitis, septicemia, and airsacculitis in poultry; therefore, it is the most significant bacterial pathogen of broiler chickens (7, 12). There are several antimicrobials that have been approved for treatment of E. coli infections in broiler chickens, including tetracycline and streptomycin (40, 41). However, some of these antimicrobials are not cost-effective, while others are ineffective due to acquired resistance (3).
The use of antimicrobials in food production is controversial because of data that suggest that usage may lead to an increase in drug resistance in human pathogens. These human food safety concerns have been influential in triggering the European Union to ban the use of antimicrobials as growth promotants in food production (21) and to increase their surveillance for bacterial resistance in food-borne pathogens and indicator organisms (http://www.dfvf.dk/default.asp?ID = 9604; accessed 9 May 2006). Similarly, the U.S. Food and Drug Administration has reevaluated its approach to approving drugs for food animal production and encouraged the withdrawal of the fluoroquinolones sarafloxacin (http://www.fda.gov/cvm/CVM_Updates/PATHLOAD.HTM; accessed 9 May 2006) and enrofloxacin (http://www.fda.gov/cvm/FQWithdrawal.html; accessed 9 May 2006) used to treat E. coli infections in poultry.
The long-term use of antimicrobials for therapy and growth promotion in animals selects for drug resistance in gram-negative pathogens (21). In farm environments, commensal and environmental bacteria may be a reservoir for the transfer of antimicrobial resistance genes to pathogenic bacteria (20, 23, 32). In previous studies, we described the high gene load of resistance determinants in the bacterial community in chicken litter (23, 32). Since bacteria acquire most resistance genes through horizontal transfer, conjugative genetic elements such as plasmids and transposons are common vectors for the dissemination of antimicrobial resistance genes to diverse microorganisms. The purpose of this study was to investigate the influence of antimicrobial administration on the distribution of resistance determinants and strain types among commensal E. coli strains isolated from the broiler chicken intestine. The information obtained in this study should help to elucidate the ecology of resistance in commensal bacteria in a farm environment.
MATERIALS AND METHODS
Sample collection.
Fresh cecal droppings were collected from the surface of the litter of flocks raised on three commercial broiler chicken farms in northeast Georgia. All three chicken farms were contracted to raise broiler chickens for the same poultry company. Samples were obtained during the period when the birds were 3 to 7 weeks old, and the history of antimicrobial usage was known for these commercial farms. This period was chosen because airsacculitis-related mortality is most likely to occur in broiler chickens during this time (1) and we had access to flocks that were given therapeutic antimicrobials. Samples were obtained from three flocks on farm A, which had not used therapeutic antimicrobials for at least 1 year prior to sampling and did not use them during our study. Samples were obtained from two flocks each on farms B and C. Flock 1 on farm B was treated with oxytetracycline, and flock 1 on farm C was treated with the fluoroquinolone sarafloxacin. Samples were obtained from these flocks immediately after antimicrobial administration.
In addition, broiler chicken flocks were raised in a research facility on fresh bedding consisting of pine shavings placed on VirkonS (Dupont, Wilmingon, DE)-disinfected concrete floor pens. One hundred twenty broiler chickens, acquired on the day of hatching from a local commercial hatchery, were raised in one pen and then were split into three treatment groups and one control group, each containing 30 birds, when they were 4 weeks old. All groups were fed a common commercial corn-soy meal broiler diet containing monensin (90 g/ton) and bacitracin methylene disalicylate (50 g/ton). The birds in the treatment groups were given therapeutic concentrations of antimicrobials in their drinking water when they were 4 weeks old. The antimicrobial doses for the treatment groups were as follows: sarafloxacin, 20 ppm for 5 days; enrofloxacin, 25 ppm for 3 days; and oxytetracycline, 25 mg/lb for 5 days. Groups of 10 birds were euthanized by carbon dioxide asphyxiation when they were 3, 5, and 7 weeks old, and the cecal contents were collected for bacterial isolation.
Bacterial isolation and identification.
During each commercial farm sampling approximately 100 cecal droppings were collected using sterile wood applicators and pooled in 30 tubes containing 1 ml of brain heart infusion broth. The contents of the 30 tubes were combined into 10 tubes, diluted with saline (10−3 and 10−5), and plated on MacConkey agar (purchased from Difco Laboratories prior to the merger with BD). The plates were incubated overnight at 37°C. In order to detect phenotypes and genes with a flock prevalence of 5% or greater, 30 isolated colonies were selected from the 10 plates and then streaked for isolation on MacConkey agar, placed in freezer medium (15% glycerol, 1% peptone), and stored at −80°C. Similarly, samples of cecal contents from the experimental birds were plated on MacConkey agar, and 30 isolated colonies were collected and stored frozen. All isolates were identified by using a panel of biochemical tests that included gas production and the sugar fermentation reaction on triple sugar iron agar, indole production, citrate fermentation, ornithine decarboxylase fermentation, and the oxidase reaction in order to determine which isolates were E. coli strains (2).
Antimicrobial susceptibility profiles.
The MICs were determined using the Sensititre automated antimicrobial susceptibility system (Trek Diagnostic Systems, Westlake, OH). The antimicrobials tested, using Sensititre avian plates, included amoxicillin, ceftiofur, gentamicin, neomycin, tetracycline, oxytetracycline, spectinomycin, streptomycin, sulfadimethoxine, sulfathiazole, sarafloxacin, and enrofloxacin. The results were interpreted by using Clinical Laboratory Standards Institute (CLSI) guidelines for broth microdilution methods for veterinary E. coli (33). E. coli ATTC 25922 was used as the quality control strain.
Strain typing by ERIC-PCR.
In order to reduce fingerprint variation associated with variation in the quality of genomic DNA preparations, whole bacterial cells (47) were used as enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR) templates. ERIC-PCR was performed with a Rapidcycler (Idaho Technologies, Idaho Falls, ID) using the following program parameters: denaturation at 94°C for 1 s, annealing at 52°C for 10 s, and extension at 72°C for 35 s for 30 cycles, followed by a final extension at 72°C for 4 min. A master mixture containing all of the reagents was prepared for each PCR in order to reduce the variation within trials. One microliter of template was added to 9 μl of the master mixture; each 10-μl PCR mixture contained deoxynucleotides at a concentration of 1 mM, 3 mM MgCl2, PCR buffer (50 mM Tris), 50 pmol of each primer (44), and 0.5 U of Taq DNA polymerase (Roche Molecular Biochemicals, Indianapolis, IN). For standardization, each gel contained a molecular weight size standard and each trial included an E. coli HB101 ERIC-PCR as an internal control. Amplicons were separated on a 1.5% agarose gel containing ethidium bromide (5 μg/ml) at 65 V for 2.3 h. The gels were photographed and digitized, and the bands were detected using the densitometry feature of RFLP Scan, version 3.0 (Scanalytics Inc., Fairfax, VA). To identify related E. coli strains, band patterns (presence or absence) were compared with a 2% molecular weight match tolerance using RFLP Scan, version 3.0. Phylogenetic trees were constructed using TreeCon, version 1.3b (43) with the stepwise clustering algorithm described by Liu and Wang (22). Tree branches were confirmed to contain related strains by visually inspecting the densitometry scans of strains in each branch.
Detection of antimicrobial resistance genes.
The DNA probes for detecting drug resistance genes were generated by PCR with digoxigenin-labeled nucleotides using primers specific for the tetracycline determinants tetA, tetB, tetC, tetD, and tetE (34) and intI1 and intI2 (11). DNA-DNA hybridization was performed as described by Sambrook et al. (38) with hybridization washes at 60°C for intI2 detection (11) and at 68°C for the other DNA probes. Positive isolates were visualized with anti-digoxigenin alkaline phosphatase conjugate and the nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate (BCIP) substrate as described by the manufacturer (Roche Molecular Biochemicals, Indianapolis, IN). The intI1-associated antimicrobial resistance gene cassettes were detected by a PCR-enzyme-linked immunosorbent assay (ELISA) as previously described by Lu et al. (23). The class 2 integron encoded by Tn7 confers drug resistance to streptomycin (aadA1) and trimethoprim (dfrI) (15), and a PCR-ELISA was developed to detect the presence of these genes in some intI2-positive E. coli strains. intI2-associated cassettes were amplified with primer 59be (32) and the 3′intI2 forward primer (10) by performing PCR with the following parameters: initial incubation at 96°C for 5 min, followed by denaturation at 94°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 2.5 min for 30 cycles. Each 10-μl PCR mixture contained digoxigenin-labeled deoxynucleotides at a concentration of 1 mM, 2 mM MgCl2, PCR buffer (50 mM Tris), 50 pmol of each primer, and 1 U of Taq DNA polymerase (Roche Molecular Biochemicals). After amplification, biotinylated oligonucleotide probes were added at a concentration of 0.1 fM, the amplicons were denatured by incubation for 60 s at 96°C, and hybridization was performed at 50°C for 15 min. Positive amplicons were detected by an ELISA using the appropriate probes and controls, as described by Lu et al. (23).
gyrA mutation analysis.
The quinolone resistance-determining region of gyrA was amplified as previously described by Weigel et al. (46). PCR was performed with a Rapidcycler (Idaho Technologies, Idaho Falls, ID) using the following program parameters: initial denaturation at 94°C for 2 min, followed by denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1 min and then a final extension at 72°C for 5 min. The 50-μl mixture contained deoxynucleotides at a concentration of 250 μM, 1.5 mM MgCl2, PCR buffer (50 mM Tris), 100 pmol of each primer, and 2.5 U of Taq DNA polymerase (Roche Molecular Biochemicals). DNA products were separated on a 1.5% agarose gel containing ethidium bromide (5 μg/ml). In order to detect gyrA mutations at Ser-83 and Asp-87 in the PCR amplicon, a restriction fragment length polymorphism assay was performed as described by Ozeki et al. (37).
Statistical analysis.
In order to detect differences in antimicrobial resistance among E. coli strains isolated from different farms and flocks and among birds that were different ages or belonged to different treatment groups, Fisher's exact method (for comparison of two groups) and the Cochran-Mantel-Haenszel method (for comparison of three groups) were used to determine whether isolates from one group were significantly more resistant than other isolates. To detect whether there was a difference in the mean MICs among the treatment and age groups in the research flock, a Kruskal-Wallis test was used to determine whether at least one group had significantly higher mean MICs than other groups. The Wilcoxon-Mann-Whitney test was used to identify a group for which the mean MICs were significantly higher. To analyze antimicrobial resistance gene and integron carriage rates, we fitted logistic models to determine whether one group (defined by age, farm, flock, heat stress, or antimicrobial usage) was more likely to have an antimicrobial resistance gene or integron than another group. For comparison in the logistic model, the P value was determined by the Wald chi-square method. Fisher's exact method was used to determine whether there was a significant difference in the prevalence of carriage of antimicrobial resistance genes between two groups when it was appropriate for the data.
RESULTS
Prevalence of antimicrobial resistance on commercial farms.
A total of 180 isolates from farm A, 150 isolates from farm B, and 120 isolates from farm C were screened for susceptibility to a panel of 12 antimicrobials. Table 1 shows the antimicrobial susceptibilities of E. coli isolates cultured from three flocks of healthy chickens on a farm that had not used therapeutic antimicrobials in the flock house for more than 1 year. We detected a high prevalence of tetracycline (36 to 97%), sulfathiazole (50 to 100%), and streptomycin (53 to 100%) resistance at all sampling times. However, we detected a low prevalence of resistance to fluoroquinolones and the β-lactam amoxicillin. Significantly higher gentamicin MICs were detected for E. coli isolates collected from young birds (MIC50, >8 μg/ml) than for E. coli isolates collected from birds 5 weeks old or older (MIC50, 8 μg/ml; P = 0.0064). Higher oxytetracycline MICs were also detected for isolates cultured from 3-week-old birds.
TABLE 1.
Antimicrobial susceptibility patterns for E. coli isolates cultured from cecal droppings of untreated commercial broiler chickens on farm Aa
| Antimicrobial | % of susceptible isolates
|
|||||
|---|---|---|---|---|---|---|
| Flock 1
|
Flock 2
|
Flock 3
|
||||
| 3 wk | 6 wk | 3 wk | 6 wk | 3 wk | 6 wk | |
| Amoxicillin | 92 | 96 | 90 | 77 | 100 | 90 |
| Ceftiofur | 100 | 100 | 100 | 100 | 100 | 100 |
| Gentamicinb | 8 | 27 | 13 | 32 | 20 | 70 |
| Neomycin | 100 | 97c | 97 | 71 | 97 | 100 |
| Oxytetracyclineb | 27 | 3c | 58 | 36 | 64 | 44 |
| Streptomycin | 0 | 0 | 0 | 0 | 17 | 47 |
| Sulfathiazole | 0 | 0 | 0 | 0 | 0 | 50 |
| Enrofloxacin | 100 | 100 | 100 | 100 | 100 | 96 |
| Nalidixic acid | 96 | 96 | 94 | 97 | 93 | 62 |
| Sarafloxacin | 96 | 97 | 94 | 97 | 97 | 75 |
The flock houses contained approximately 22,000 birds. Samples of fresh cecal droppings were obtained from the top of the litter bed of broilers when they were 3 and 6 weeks old (30 isolates from each sampling were tested; n = 180). MICs were determined using the Sensititre broth microdilution method with avian plates. Susceptibility was determined using CLSI criteria. No antimicrobials had been used in the flock houses for more than 1 year.
The MICs of gentamicin (P = .0064) and oxytetracycline (P < .0001) for E. coli isolates cultured from young birds (3 weeks old) were significantly higher. Gentamicin is approved for use in day-of-hatch chicks (39) and was used by the companies cooperating in this study.
The MICs of neomycin (P = 0.0035) and oxytetracycline (P = 0.0175) for isolates from flock 1 chickens that were 6 weeks old were higher than the MICs of these antimicrobials for isolates from flock 2 chickens that were 6 weeks old.
Two of the farms in our study had used therapeutic antimicrobials with several flocks of chickens prior to sampling and with two flocks that were sampled in our study. Table 2 shows the susceptibilities of the isolates collected from the treated flocks. We detected a high prevalence of tetracycline (58 to 90%), sulfathiazole (77 to 100%), and streptomycin (74 to 100%) resistance in the E. coli isolates cultured from these farms. Both of these poultry farms had a past history of fluoroquinolone usage (enrofloxacin and sarafloxacin). The sarafloxacin MICs were higher for E. coli isolates that were cultured from a flock that was given sarafloxacin (MIC50, 0.5 μg/ml) than for isolates from the farm with no recent history of fluoroquinolone usage (MIC50, 0.15 μg/ml; P = 0.001). However, we did not observe a significant increase in the prevalence of tetracycline resistance in E. coli isolates after oxytetracycline administration to the commercial broiler flock. One of the difficulties in demonstrating a statistically significant change in the prevalence of tetracycline resistance was related to the high prevalence of resistance in isolates from untreated flocks and the variation in the prevalence of tetracycline resistance due to differences in the ages of the birds and the farms. We detected significant differences in gentamicin MICs (P = 0.0064) and oxytetracycline MICs (P < 0.0001) that were age related, but the pattern of increasing or decreasing resistance related to age varied by farm and flock.
TABLE 2.
Antimicrobial susceptibility patterns for E. coli isolates cultured from the cecal droppings of commercial broiler chickens that were given antimicrobialsa
| Antimicrobial | % of susceptible isolates
|
||||
|---|---|---|---|---|---|
| Farm Bb
|
Farm Cc
|
||||
| 3 wk | 6 wk | 7 wk | 3 wk | 6 wk | |
| Amoxicillind | 76 | 100 | 90 | 93 | 100 |
| Ceftiofur | 100 | 100 | 100 | 100 | 100 |
| Gentamicin | 0 | 3e | 23 | 16 | 16 |
| Neomycin | 100 | 100 | 100 | 100 | 100 |
| Oxytetracyclinef | 10 | 42 | 23 | 33 | 26 |
| Streptomycin | 16 | 0 | 0 | 3 | 26 |
| Sulfathiazole | 0 | 0 | 23 | 6 | 16 |
| Enrofloxacin | 100 | 100 | 100 | 100 | 96 |
| Nalidixic acid | 16 | 28 | 53 | 33 | 20 |
| Sarafloxacing | 23 | 3e | 53 | 16 | 0 |
The flock houses contained approximately 22,000 birds. Samples of fresh cecal droppings were obtained from the top of the litter bed of broilers when they were 3 and 6 weeks old (30 isolates from each sampling were tested; n = 150). Samples were also obtained from the flock on farm B when the chickens were 7 weeks old, after oxytetracycline treatment. MICs were determined using the Sensititre broth microdilution method with avian plates. Susceptibility was determined using CLSI criteria.
Farm B had a history of enrofloxacin and sarafloxacin usage; oxytetracycline was administered to the birds in their water when they were 6.5 weeks old.
The flock house had a history of sarafloxacin and enrofloxacin usage; sarafloxacin was administered to the birds in their water when they were 6 weeks old. Isolates were collected the day after the treatment.
Isolates from farm A (Table 1) were significantly more resistant to amoxicillin than isolates from farm B (P = 0.021) when the birds were 3 weeks old.
The MICs of sarafloxacin (P < 0.0001) and gentamicin (P = 0.0399) for isolates from farm B were significantly higher when the chickens were 6 weeks old than when they were the other ages.
Isolates from farm A were significantly more resistant to oxytetracycline (P = 0.0002) than isolates from farm B when the chickens were 6 weeks old.
Isolates from farm A were significantly more susceptible to sarafloxacin (P < 0.0001) than isolates from farm B and farm C when the chickens were 3 and 6 weeks old.
Prevalence of antimicrobial resistance in broiler chickens in a controlled, environmental setting.
We raised a group of experimental broiler chickens under controlled conditions at our animal facility in order to reduce the contribution of different farm management practices associated with commercial broiler chicken production. These birds were given a therapeutic dose of fluoroquinolones and oxytetracycline; 270 E. coli isolates were screened to determine their susceptibilities to a panel of 12 antimicrobials. Table 3 shows the antimicrobial susceptibilities of E. coli isolates cultured from the experimental birds. As on the commercial farms, there was a low prevalence of susceptibility to antimicrobials such as tetracycline (17 to 57%), streptomycin (20 to 43%), and sulfathiazole (23 to 43%) in the untreated group. Similar to the findings obtained using commercial birds, for the E. coli isolates from the untreated research birds the MICs of neomycin (MIC90, 32 μg/ml versus 2 μg/ml; P = 0.05), tetracycline (MIC50, >8 μg/ ml versus 0.5 μg/ml; P = 0.05), streptomycin (MIC50, 64 μg/ml versus 16 μg/ml; P = 0.05), nalidixic acid (MIC90, 64 μg/ml versus 16 μg/ml; P = 0.05), and sulfathiazole (MIC50, >256 μg/ml versus 256 μg/ml; P = 0.05) were higher when the chickens were 3 weeks old than when the chickens were 5 weeks old. These data confirm that age remains a confounding factor when resistance prevalence data are interpreted. However, if the treatment groups were compared when the chickens were the same ages, the quinolone MICs were significantly more likely (P < 0.05) to be higher for isolates obtained from the enrofloxacin-treated group when the chickens were 5 and 7 weeks old than for isolates obtained from the other treatment groups. But we also found that the MICs of streptomycin were less likely to be higher for E. coli isolates from the enrofloxacin-treated group than for E. coli isolates from the other groups when the chickens were 5 weeks old. For example, the streptomycin MIC90 for isolates in the untreated group was 128 μg/ml, while for 86% of the isolates in the enrofloxacin-treated group the MIC was 8 μg/ml. The enrofloxacin-treated group also exhibited a lower prevalence of resistance to streptomycin, tetracycline, and sulfathiazole after treatment.
TABLE 3.
Antimicrobial susceptibility patterns of E. coli isolates cultured from the cecal contents of experimental broiler chickens that were given antimicrobialsa
| Antimicrobial | % of susceptible strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grouped flock (3 wk)f | Group 1 (no treatment)e
|
Group 2 (sarafloxacin treatment)b
|
Group 3 (enrofloxacin treatment)c
|
Group 4 (tetracycline treatment)d
|
|||||
| 5 wk | 7 wk | 5 wk | 7 wk | 5 wk | 7 wk | 5 wk | 7 wk | ||
| Amoxicillin | 100 | 100 | 93 | 100 | 100 | 100 | 97 | 93 | 93 |
| Ceftiofur | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Gentamicin | 23 | 90 | 63 | 87 | 100 | 100 | 93 | 97 | 97 |
| Neomycin | 50 | 93 | 100 | 97 | 97 | 100 | 97 | 77 | 100 |
| Oxytetracycline | 16 | 53 | 57 | 53 | 60 | 70 | 60 | 0 | 3 |
| Streptomycin | 20 | 43 | 20 | 43 | 60 | 87 | 80 | 50 | 60 |
| Sulfathiazole | 23 | 43 | 27 | 43 | 80 | 50 | 83 | 30 | 97 |
| Enrofloxacin | 100 | 100 | 100 | 100 | 100 | 60 | 70 | 100 | 100 |
| Nalidixic acid | 73 | 90 | 53 | 60 | 70 | 46 | 50 | 70 | 96 |
| Sarafloxacin | 83 | 97 | 67 | 83 | 67 | 50 | 53 | 100 | 100 |
Samples of fresh cecal contents were obtained from euthanized birds when they were 3, 5, and 7 weeks old (30 isolates from each sampling were collected; n = 270). MICs were determined using the Sensititre broth microdilution method with avian plates. Susceptibility was determined using CLSI criteria. The research flock contained 120 birds that were separated into four treatment groups when the birds were 4 weeks old.
Sarafloxacin (20 ppm) was administered in the water given to the birds for 5 days when they were 4 weeks old.
Enrofloxacin (25 ppm) was administered in the water given to the birds for 3 days when they were 4 weeks old. The MICs of sarafloxacin (P = 0.0156), enrofloxacin (P = 0.0125), and nalidixic acid (P < 0.0342) were more likely to be higher for the enrofloxacin-treated group than for other treatment groups when the chickens were 5 and 7 weeks old. The isolates from the enrofloxacin-treated group were more susceptible to streptomycin than the isolates from the other groups when the chickens were 5 weeks old (P < 0.001).
Oxytetracycline (25 ppm) was administered in the water of the birds for 5 days when they were 4 weeks old. The isolates from the oxytetracycline group were more likely than the isolates from the untreated group and the fluoroquinolone-treated groups to be resistant to oxytetracycline when the chickens were 5 and 7 weeks old (P < 0.001) and less likely to be resistant to sarafloxacin and enrofloxacin when the chickens were 7 weeks old (P < 0.0002).
The isolates from the untreated group were more likely than the isolates from the other groups to be resistant to streptomycin (P = 0.001) and sulfathiazole (P < 0.003) when the chickens were 7 weeks old.
E. coli strains isolated from 3-week-old birds were more likely to be resistant than the strains isolated from 5-week-old birds (P < 0.05).
Poultry E. coli strain distribution and antimicrobial resistance.
In order to investigate the effects of E. coli strain distribution on the prevalence of antimicrobial resistance, 30 to 50% of the isolates from each flock were DNA fingerprinted, and the results were used to construct phylogenetic trees. Isolates were selected to represent all of the resistance phenotypes and genotypes cultured from each sampling time. Four common ERIC-PCR strain types were detected on the different commercial poultry farms, and two of these strain types were detected among the isolates cultured from research broiler flocks raised in our facilities (Fig. 1).
FIG. 1.
Genetic relatedness of E. coli isolates (n = 172) cultured from flocks on commercial broiler farms and in a research facility. (A) E. coli isolates were fingerprinted using ERIC-PCR, and strain types were identified after phylogenetic analysis using RFLP Scan. Four ERIC-PCR types were found on all three commercial farms (ERIC strains A to D), and two strains were also detected in the research flock (RF) (ERIC strains A and D). The distribution of some E. coli strains was limited to single flocks on a single farm. (B) ERIC-PCR fingerprints of some isolates from the research flock and farm C. Not all bands detected by the typing software are visible. Lanes S contained the molecular weight marker; lanes A contained ERIC strain A.
In order to investigate whether resident E. coli strains affected the prevalence of antimicrobial resistance on farms that did not use therapeutic antimicrobials during this study, we evaluated the distribution and genetic relatedness of E. coli isolates from the flocks on farm A (Fig. 2). While multiple strain types were detected, we found several E. coli strains in flock 3 that persisted as the birds matured. These persistent strains included ERIC strain A. Also, genetically related E. coli strains were found in multiple flocks on this farm, and these strain types included both antimicrobial-susceptible and -resistant isolates (Table 4). These results suggested that a resident E. coli strain could colonize new flocks that enter a house but that the prevalence of antimicrobial resistance was not predictable because of variation in drug resistance within a strain type. However, a comparison of strain types detected in the oxytetracycline-treated flock on farm B indicated that different tetracycline-resistant E. coli strains were present in the birds before and after administration of the antimicrobial (Fig. 3). This finding suggests that oxytetracycline induced E. coli strain succession, but succession was also observed in the untreated flocks on farm A (Fig. 2).
FIG. 2.
Genetic relatedness of E. coli isolates (n = 43) cultured from three sequential flocks on farm A. Samples were obtained from the flocks when the birds were 3 and 6 weeks old. E. coli isolates were fingerprinted using ERIC-PCR, and strain types were identified after phylogenetic analysis. Nearly all E. coli strains exhibited succession within and between flocks; however, two strains (indicated by shading) persisted in flock 3. In addition, genetically related strains were detected in multiple flocks. Drug resistance profiles are indicated for branches containing more than one E. coli isolate. The abbreviations for antimicrobial resistance are as follows: A, amoxicillin; G, gentamicin; T, tetracycline; Sp, spectinomycin; S, streptomycin; Su, sulfathiazole; and Sf, sarafloxacin.
TABLE 4.
Distribution of ERIC-PCR strains and their antimicrobial resistance phenotypes and genotypes among E. coli strains isolated from broiler chicken flocksa
| ERIC strain | Antimicrobial resistance phenotype | No. of strains with resistance phenotype on:
|
Resistance genotype on:
|
||||
|---|---|---|---|---|---|---|---|
| Farm A | Farm B | Farm C | Farm A | Farm B | Farm C | ||
| A | G T Sp S Su Sf | 5 | 1 | intI1 (aadA) | intI1 (aadA), tetA | ||
| G T Sp S Su | 9 | 2 | 3 | intI1 (aadA) | intI1 (aadA) | intI1 (aadA) | |
| intI1 (aadA), tetA | intI1 (?)b | ||||||
| intI1 (aadA), tetB | |||||||
| G T Su | 1 | intI1 (aadA), tetA | |||||
| G Sp S Su | 3 | 1 | intI1 (aadA), tetA | intI1 (aadA) | |||
| intI1 (aadA) | |||||||
| G Sp S Su | 1 | intI1 (aadA) | |||||
| T Sp S Su Sf | 1 | intI1 (aadA), intI2 | |||||
| T Su | 1 | tetB | |||||
| T S | 1 | 2 | tetB | intI1 (aadA) | |||
| T | 2 | intI2 | |||||
| Sf | 1 | ||||||
| Susceptible | 1 | 2 | |||||
| B | G T Sp S Su Sf | 1 | 1 | intI1 (aadA) | intI1 (aadA), tetB | ||
| G T Sp S Su | 7 | 3 | 1 | intI1 (aadA), tetA | intI1 (aadA), tetA | intI1 (aadA) | |
| intI1 (aadA), intI2, tetA | intI1 (aadA), tetB | ||||||
| intI1 (aadA, dfr), tetA | intI1 (aadA) | ||||||
| G T Sp Su | 1 | intI1 (aadA), tetA | |||||
| C | G T Sp S Su Sf | 1 | 3 | 3 | intI1 (aadA), tetA | intI1 (aadA) | intI1 (aadA) |
| G T Sp S Su | 11 | 7 | 2 | intI1 (aadA), tetA | intI1 (aadA), tetA | intI1 (aadA), intI2 | |
| intI1 (aadA), tetB | intI1 (aadA) | intI1 (aadA) | |||||
| intI1 (aadA) | |||||||
| G Sp S Su | 1 | intI1 (aadA) | |||||
| G Sp Su Sf | 4 | intI1 (aadA) | |||||
| intI1b | |||||||
| A T | 1 | intI1 | |||||
| T S | 2 | intI1 (bla) | |||||
| Sf | 1 | 1 | intI1 (aadA, bla) | ||||
| D | G T Sp S Su Sf | 2 | intI1 (aadA), tetB | ||||
| intI1 (aadA) | |||||||
| G T Sp S Su | 2 | 1 | intI1 (aadA), tetB | intI1 (aadA) | |||
| G Sp S Su Sf | 1 | intI1 (aadA) | |||||
| Sf | 1 | intI1 (aadA) | |||||
Strain types were determined by ERIC-PCR. Class 1 and class 2 integrase genes were detected using DNA-DNA hybridization, and class 1-associated gene cassettes of intI1-positive isolates were screened using PCR-ELISA. The probes were pooled to screen for the presence of β-lactamase (bla), dihydrofolate reductase (dfr), and aminoglycoside resistance (AG) cassettes. The aminoglycoside resistance pool did not contain the aadA1,2 probe. A gene in parentheses following intI1 is an antimicrobial resistance gene present in the integron. The genes listed in parentheses do not reflect the order in the integron or the number of integrons present in the isolates. The abbreviations for antimicrobial resistance are as follows: A, amoxicillin; G, gentamicin; T, tetracycline; Sp, spectinomycin; S, streptomycin; Su, sulfathiazole; and Sf, sarafloxacin.
PCR-ELISA did not detect β-lactamase (bla), dihydrofolate reductase (dfr), or aminoglycoside resistance cassettes in this integron.
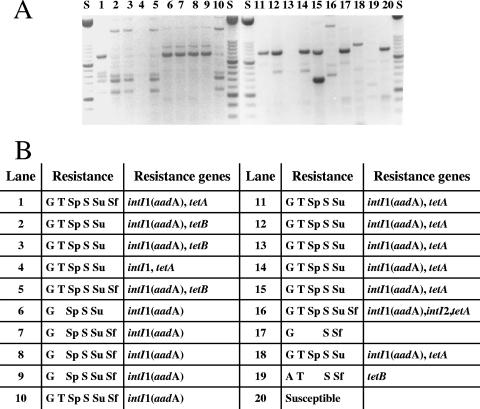
FIG. 3.
Antibiotic resistance phenotypes and genotypes for E. coli strains isolated from an oxytetracycline-treated commercial broiler flock. (A) ERIC-PCR fingerprints. Not all bands detected by the typing software are visible. Lanes S contained a molecular weight marker; lanes 1 to 10 contained isolates from 6-week-old birds in flock 1 on farm B prior to oxytetracycline administration; lanes 11 to 19 contained isolates from 7-week-old birds after treatment; and lane 20 contained E. coli HB101. (B) Antibiotic resistance phenotypes and genotypes of the isolates. The abbreviations are the same as those described in the legend to Fig. 2.
Farm C also possessed antimicrobial-resistant strains that persisted throughout the grow-out period of one flock, including ERIC strains A and C. Because there was treatment-independent variation in the prevalence of antimicrobial resistance, we investigated whether the prevalence of specific strain types could explain the prevalence of fluoroquinolone resistance on this farm, which had a history of usage. Eighty-four percent of the E. coli isolates from 3-week-old birds were sarafloxacin resistant or intermediate, and all isolates from treated 6-week-old birds were resistant (Table 2). ERIC-PCR strain typing revealed three sarafloxacin-resistant strains in 3-week-old birds, including ERIC strains A and C (Fig. 4). Five resistant strains were isolated from the treated birds, including ERIC strains A, B, and D. The other two resistant strains must have been present at levels below the detection level, and these E. coli strains could have persisted in the litter between flocks. Therefore, the data from the research flock could eliminate the confounding factor of litter contamination. However, 10% of the E. coli isolates from the untreated research control group were sarafloxacin resistant. Two sarafloxacin-resistant strains were detected, and one of them was ERIC strain A. Both of these E. coli strains possessed the gyrA (Thr83Ser) mutation, as did the sarafloxacin-resistant strains detected in the sarafloxacin-treated research group. ERIC strain A was also the dominant susceptible strain detected in the birds prior to administration of antimicrobials, indicating that sarafloxacin administration selected for additional fluoroquinolone-resistant strains that were present at a low density in the bacterial community.
FIG. 4.
ERIC-PCR fingerprints of fluoroquinolone-resistant E. coli strains isolated from farm C. Not all bands detected by the typing software are visible. Lanes S contained a molecular weight marker; pretreatment isolates were cultured from 3-week-old birds prior to sarafloxacin treatment; posttreatment isolates were cultured from 6-week-old birds. Lanes A, B, and D contained ERIC strains A, B, and D, respectively.
Prevalence of tetracycline resistance genes.
Although tetracycline resistance can be mediated by 36 different genetic elements, the tetA to tetE genes have been found to be the most prevalent such elements in E. coli isolates (8). tetA (10 to 60% of isolates) and tetB (10 to 50%) were the alleles most commonly detected in the flocks regardless of treatment history. On farm C, there was no significant difference in tetA (13% versus 46%), tetB (36% versus 40%), or tetC (10% versus 0%) carriage after sarafloxacin treatment (P > 0.1). However, on farm B, there was a significant decrease in tetB carriage (50% versus 10%) (P = 0.0003) after tetracycline administration, and there was a significant increase in tetA carriage (10% versus 27%) (P < 0.0001). In addition, tetA was more likely to be detected in isolates from farm B than in isolates farm A (P = 0.0002) or farm C (P = 0.0005), suggesting that the tetracycline administration on farm B may have affected tetracycline allele carriage. Past usage of tetracycline on a farm may affect the persistence of a particular tetracycline determinant; however, there were significant age (P < 0.0338) and farm (P = 0.0041) interactions that may have confounded the interpretation of the data. In some cases we found a higher prevalence of tet gene carriage than tetracycline resistance, suggesting that silent carriage was not usual. We also detected a higher prevalence of tetracycline resistance than carriage of the tetA to tetE alleles, suggesting that other tet genes mediated some resistance. This was particularly true on the commercial farms, indicating that there may be great diversity of tet determinants in the community of environmental bacteria.
We also wanted to determine whether carriage of a particular tetracycline resistance determinant correlated with the presence of a particular E. coli strain. In untreated experimental flocks, genetically related E. coli strains carried the same tetracycline resistance gene allele. However, on the commercial farms, similar E. coli strains carried different tetracycline resistance determinants. These data suggest that there may have been a greater abundance and diversity of tetracycline alleles in the bacterial communities of the commercial broiler chicken farms. These data indicate that the selective pressure of tetracycline usage may influence the diversity of resistance genes that persist in an environment.
Prevalence of integron-encoded antimicrobial resistance genes.
Integrons have not been shown to encode tetracycline resistance, but they often encode resistance to aminoglycosides, β-lactams, cephalosporins, chloramphenicol, and trimethoprim. Therefore, we investigated the contribution of integrons to resistance in commensal E. coli strains isolated from treated and untreated birds (Table 5). The most common integrase gene detected in the E. coli isolates was the class 1 integron intI1 gene (20 to 100%), which frequently contained a streptomycin resistance cassette (23.3 to 100%) and/or a trimethoprim resistance cassette (0 to 17.6%). The class 2 integrons (prevalence, 0 to 20%) that were screened by PCR-ELISA contained a dfr cassette (67%) and an aadA1 cassette (83%). The presence of these cassettes in the class 2 integron-positive isolates suggests that Tn7 or related transposons probably encoded the resistance (15).
TABLE 5.
Distribution of integron carriage and class 1 integron-associated cassettes in E. coli strains isolated from commercial broiler chicken flocksa
| Farm | Flock | Age (wk) | % of positive strains
|
|||||
|---|---|---|---|---|---|---|---|---|
| intI1 | intI2 | aadA1,2b | blab | AGb | dfrb | |||
| A | 1 | 3 | 96 | 0 | 94.1 | 0 | 0 | 17.6 |
| 6 | 100 | 3 | 86.7 | 3.3 | 0 | 3.3 | ||
| 2 | 3 | 40 | 6 | 90 | 0 | 10 | 0 | |
| 6 | 50 | 10 | 70 | 20 | 0 | 0 | ||
| 3 | 3 | 51 | 3 | 90 | 3.3 | 10 | 6.7 | |
| 6 | 20 | 0 | 55.2 | 3.4 | 0 | 3.4 | ||
| B | 1 | 3 | 97 | 6 | 100 | 0 | 0 | 6.7 |
| 6 | 100 | 0 | 78.6 | 0 | 0 | 0 | ||
| 7 | 93 | 6 | 76.7 | 0 | 0 | 0 | ||
| 2 | 3 | 60 | 0 | 90 | 0 | 0 | 0 | |
| 6 | 66 | 6 | 83.3 | 0 | 0 | 6.7 | ||
| C | 1 | 3 | 93 | 10 | 92.9 | 0 | 0 | 14.3 |
| 6 | 83 | 0 | 86.7 | 0 | 3.3 | 16.7 | ||
| 2 | 3 | 46 | 0 | 48.3 | 24.1 | 0 | 3.4 | |
| 6 | 23 | 13 | 23.3 | 0 | 0 | 0 | ||
Class 1 and class 2 integrase genes were detected using DNA-DNA hybridization. Class 1-associated gene cassettes of intI1-positive isolates were screened using PCR-ELISA.
The digoxigenin-labeled probes were pooled to screen for the presence of β-lactamase (bla), dihydrofolate reductase (dfr), and aminoglycoside resistance (AG) cassettes in intI1-positive isolates by PCR-ELISA. The aminoglycoside resistance pool did not contain the aadA1,2 probe.
Antimicrobial administration significantly affected the prevalence of integron carriage in the E. coli strains screened in this study. On farm B, there was a significant decrease in intI1 carriage after tetracycline treatment (P = 0.0077), and there was a significant decrease in intI2 carriage (P = 0.0053) after sarafloxacin treatment on farm C. These data suggest that antimicrobial administration can affect integron carriage. However, significant differences between the ages (P = 0.0036), farms (P = 0.0008), and flocks (P = 0.0001) indicated that other factors significantly contributed to the variation in integron carriage.
Prevalence of multidrug resistance in commensal E. coli strains from chickens.
More than 75% of the E. coli isolates from commercial flocks were resistant to three or more antimicrobials. However, there were different E. coli strains that exhibited the same drug resistance patterns, and there were similar strains that displayed different antimicrobial resistance patterns. Table 4 shows the resistance phenotypes and genotypes of the four common E. coli strains found on the commercial broiler farms. In some instances, isolates of the same E. coli strain expressed the same drug resistance patterns although they harbored different tet determinants or streptomycin resistance genes. In addition, we detected sarafloxacin-resistant members of some E. coli strains on each commercial farm and the research farm, indicating that dissemination of sarafloxacin-resistant strains may also be responsible for the presence of fluoroquinolone resistance in the absence of usage.
DISCUSSION
The purpose of this study was to investigate the influence of antimicrobial administration on the prevalence of resistance in commensal E. coli strains isolated from broiler chickens. In this study we cultured isolates from birds with known antimicrobial treatment histories and characterized their antimicrobial resistance profiles and their carriage of specific drug resistance genes. Furthermore, in order to determine whether the prevalence of antimicrobial resistance was due to the presence of persistent resistant E. coli strain types, we evaluated the distribution of strains and their antimicrobial resistance genes. We found a high prevalence of resistance to tetracycline, sulfonamides, and streptomycin in all commercial flocks, although these drugs were not used in most cases. These results were supported by data from the experimental flocks which demonstrated that even in controlled settings with clean pens and fresh bedding, there was high prevalence of resistance to antimicrobials not commonly used in broiler chicken husbandry. These data are similar to data in previously published studies that illustrated that usage patterns may not correlate with resistance prevalence (14, 19, 42).
However, we also detected broad distribution of several antimicrobial-resistant E. coli strains on all the farms in our study, including birds raised in our research facilities. Previous studies have shown the persistence of clonal pathogenic E. coli strains in poultry houses, but commensal E. coli isolates have been shown to have a wide diversity of genotypes (9, 26, 35). ERIC-PCR was used in our study to strain type the E. coli isolates, and this method has been used by other workers to type E. coli strains from poultry (6, 9, 35) and swine (31, 45) and uropathogenic isolates (48). Although Meacham et al. (27) demonstrated that low-stringency PCR conditions could generate false diversity, a multilaboratory study performed by Grundmann et al. (13) showed that standardization reduced variation. We used a whole-cell template to reduce the variation associated with genomic DNA preparation and standardized reaction conditions by preparing a master mixture, using the same thermocycler, and including a positive control strain in each experiment. ERIC-PCR typing revealed significant diversity among E. coli strains isolated in our study, but there also appeared to be a propensity for some resistant strains to persist in the farm environment and colonize new flocks.
These findings have important implications for poultry flock house and litter management. In several studies workers have described the microbial diversity and activity of organisms that survive in litter (10, 23, 25); therefore, it is important to understand the implications of litter management for the persistence of E. coli and other microorganisms. Some poultry companies remove litter from the house prior to every new flock, while others place fresh bedding on top of used litter and replace the litter a few times a year. Litter management varies because companies have different approaches to developing the intestinal microbiota of young chicks in order to improve resistance to infectious diseases. In addition, the cost of drug resistance plasmid carriage has been shown to be reduced as E. coli strains evolve over successive generations (28). Therefore, the coevolution of E. coli populations and the antimicrobial resistance gene load in litter may have a greater influence on the prevalence of antimicrobial resistance than antimicrobial usage alone has. We have also shown in previous studies that the litter may contain the same antimicrobial resistance genes that were detected in the commensal E. coli strains (23, 32), suggesting that the litter environment can serve as a reservoir for antimicrobial resistance gene carriage and genetic exchange among abundant members of the litter bacterial community (36). This persistent antimicrobial resistance gene reservoir may also explain the limited diversity of tet alleles and integron cassettes that were observed for commensal E. coli strains.
Integron-mediated antimicrobial resistance has been shown to be common in avian E. coli strains and other veterinary pathogens (3, 11). Our study also confirmed that class 1 integrons are very common among commensal poultry E. coli strains. In some cases, antimicrobial administration significantly affected the carriage of integrons, illustrating the influence of usage on antimicrobial resistance genetic carriage. Antimicrobial administration appeared to have more effect on antimicrobial resistance gene distribution than on the phenotypic prevalence of antimicrobial resistance of E. coli isolates in this study. These findings are similar to those of Blake et al. (5), who found that tetracycline administration changed the carriage of particular tet genes in commensal E. coli strains. In our study, changes in carriage also appeared to be due to changes in E. coli community structure.
We encountered difficulty in correlating increased antimicrobial resistance with antimicrobial usage because of the wide variation in prevalence among commensal E. coli strains from the commercial farms. Langlois et al. (18) also found that the location of housing and the age of pigs affected the antimicrobial resistance of the fecal coliforms in an untreated herd; other studies have shown similar effects (29, 30). Likewise, we found that younger birds were more likely to contain E. coli strains that were resistant to gentamicin and oxytetracycline, although antimicrobial resistance also varied among flocks and farms. Similar age-related and farm-to-farm differences in antimicrobial susceptibility of commensal E. coli strains have also been reported for cattle (4, 16, 17). E. coli is most abundant among the intestinal bacterial community when the birds are young and wanes as the birds mature (24). Therefore, poultry production practices that affect the density of drug-resistant genes and affect the abundance of E. coli may be very important for influencing the prevalence of antimicrobial resistance.
In conclusion, the data obtained in this study indicate that many factors contribute to the prevalence of drug resistance in commensal bacterial communities of animals. Understanding the ecology of bacterial communities present in animal environments should allow informed inquiries concerning the degree that specific organisms affect the overall ecology of resistance with respect to genetic exchange and interaction with members of the microbial community. The information gathered from these types of studies may help us manage the evolution of antimicrobial resistance in the future.
Acknowledgments
Funds from the Food and Drug Administration supported work in the laboratory of M.D.L. USDA NRICGP grant 99-35212-8680 supported work in the laboratory of J.J.M.
We thank Linda Purvis for participating in the sample collection on the commercial farms. We also thank Cathy Goldstein and Kim DiCono for contributing to the template preparation and DNA hybridization procedures.
Footnotes
Published ahead of print on 28 December 2006.
REFERENCES
- 1.Barnes, H. J., J. P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-652. In Y. M. Saif (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames.
- 2.Baron, E. J., L. R. Peterson, and S. M. Finegold. 1994. Enterobacteriaceae, p. 370. In Bailey and Scott's diagnostic microbiology, 9th ed. Mosby, St. Louis, MO.
- 3.Bass, L., C. A. Liebert, M. D. Lee, A. O. Summers, D. G. White, S. G. Thayer, and J. J. Maurer. 1999. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43:2925-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge, A. C., E. R. Atwill, and W. M. Sischo. 2005. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev. Vet. Med. 69:25-38. [DOI] [PubMed] [Google Scholar]
- 5.Blake, D. P., R. W. Humphry, K. P. Scott, K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J. Appl. Microbiol. 94:1087-1097. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho de Moura, A. C., K. Irino, and M. C. Vidotto. 2001. Genetic variability of avian Escherichia coli strains evaluated by enterobacterial repetitive intergenic consensus and repetitive extragenic palindromic polymerase chain reaction. Avian Dis. 45:173-181. [PubMed] [Google Scholar]
- 7.Cheville, N. F., and L. H. Arp. 1978. Comparative pathologic findings of Escherichia coli infection in birds. J. Am. Vet. Med. Assoc. 173:584-587. [PubMed] [Google Scholar]
- 8.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Microbiol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias da Silveira, W., A. Ferreira, M. Lancellotti, I. A. Barbosa, D. S. Leite, A. F. de Castro, and M. Brocchi. 2002. Clonal relationships among avian Escherichia coli isolates determined by enterobacterial repetitive intergenic consensus (ERIC)-PCR. Vet. Microbiol. 89:323-328. [DOI] [PubMed] [Google Scholar]
- 10.Dilly, O., J. Bloem, A. Vos, and J. C. Munch. 2004. Bacterial diversity in agriculture soils during litter decomposition. Appl. Environ. Microbiol. 70:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein, C., M. D. Lee, S. Sanchez, C. Hudson, B. Phillips, B. Register, M. Grady, C. Liebert, A. O. Summers, D. G. White, and J. J. Maurer. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45:723-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomis, S. M., R. Goodhope, L. Kumor, N. Caddy, C. Riddell, A. A. Petter, and J. J. Allan. 1997. Experimental reproduction of Escherichia coli cellulitis and septicemia in broiler chickens. Avian Dis. 41:234-240. [PubMed] [Google Scholar]
- 13.Grundmann, H. J., K. J. Towner, L. Dijkshoorn, P. Gerner-Smidt, M. Maher, H. Seifert, and M. Vaneechoutte. 1997. Multicenter study using standardized protocols and reagents for evaluation of reproducibility of PCR-based fingerprinting of Acinetobacter spp. J. Clin. Microbiol. 35:3071-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra, B., E. Junker, A. Schroeter, B. Malorny, S. Lehmann, and R. Helmuth. 2003. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine, and poultry. J. Antimicrob. Chemother. 52:489-492. [DOI] [PubMed] [Google Scholar]
- 15.Hasson, K., L. Sundstrom, A. Pelletier, and P. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyle, D. V., H. I. Knight, D. J. Shaw, K. Hillman, M. C. Pearce, J. C. Low, G. J. Gun, and M. E. Woolhouse. 2004. Acquisition and epidemiology of antibiotic-resistant Escherichia coli in a cohort of newborn calves. J. Antimicrob. Chemother. 53:867-871. [DOI] [PubMed] [Google Scholar]
- 17.Khachatryan, A. R., D. D. Hancock, T. E. Besser, and D. R. Call. 2004. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl. Environ. Microbiol. 70:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langlois, B. E., K. A. Dawson, I. Leak, and D. K. Aaron. 1988. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 54:1341-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanz, R., P. Kuhnert, and P. Boerlin. 2003. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 91:73-84. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. D., S. Sanchez, M. Zimmer, U. Idris, M. E. Berrang, and P. F. McDermott. 2002. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob. Agents Chemother. 46:3660-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy, S. B. 1998. Multidrug resistance: a sign of the times. N. Engl. J. Med. 338:1376-1378. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y. Y., and Y. L. Wang. 1979. Application of stepwise cluster analysis in medical research. Sci. Sin. 22:1082-1094. [PubMed] [Google Scholar]
- 23.Lu, J., S. Sanchez, C. Hofacre, J. J. Maurer, B. G. Harmon, and M. D. Lee. 2003. Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, J., U. Idris, B. Harmon, C. Hofacre, J. J. Maurer, and M. D. Lee. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, S. A., and M. A. McCann. 1998. Microbiological survey of Georgia poultry litter. J. Appl. Poult. Res. 7:90-98. [Google Scholar]
- 26.Maurer, J. J., M. D. Lee, C. Lobsinger, T. Brown, M. Maier, and S. G. Thayer. 1998. Molecular typing of avian Escherichia coli isolates by random amplification of polymorphic DNA. Avian Dis. 42:431-451. [PubMed] [Google Scholar]
- 27.Meacham, K. J., L. Zhang, B. Foxman, R. J. Bauer, and C. F. Marrs. 2003. Evaluation of genotyping large numbers of Escherichia coli isolates by enterobacterial repetitive intergenic consensus-PCR. J. Clin. Microbiol. 41:5224-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Modi, R. I., C. M. Wilke, R. F. Rosenweig, and J. Adam. 1991. Plasmid macro-evolution: selection of deletions during adaptation in a nutrient limited environment. Genetica 84:195-202. [DOI] [PubMed] [Google Scholar]
- 29.Molitoris, E., D. J. Fagerberg, C. L. Quarles, and M. I. Krichevsky. 1987. Changes in antimicrobial resistance in fecal bacteria associated with pig transit and holding times in slaughter plants. Appl. Environ. Microbiol. 53:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moro, M. H., G. W. Beran, L. J. Hoffman, and R. W. Griffith. 1998. Effects of cold stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. Lett. Appl. Microbiol. 27:251-254. [DOI] [PubMed] [Google Scholar]
- 31.Namvar, A., and K. Warriner. 2006. Application of enterobacterial repetitive intergenic consensus-polymerase chain reaction to trace the fate of generic Escherichia coli within a high capacity pork slaughter line. Int. J. Food Microbiol. 108:155-163. [DOI] [PubMed] [Google Scholar]
- 32.Nandi, S., J. J. Maurer, C. L. Hofacre, and A. O. Summers. 2004. Gram positive bacteria, major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 101:7118-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 1999. NCCLS document M31-A. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 34.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 35.Ngeleka, M., J. K. Kwaga, D. G. White, T. S. Whittam, C. Riddell, R. Goodhope, A. A. Potter, and B. Allan. 1996. Escherichia coli cellulitis in broiler chickens: clonal relationships among strains and analysis of virulence-associated factors of isolates from diseased birds. Infect. Immun. 64:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nield, B. S., A. J Holmes, M. R. Gillings, G. D. Recchia, B. C. Mabbutt, K. M. Nevalainen, and H. W. Stokes. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195:59-65. [DOI] [PubMed] [Google Scholar]
- 37.Ozeki, S., T. Deguchi, M. Nakano, M. Kawamura, T. Nishino, and Y. Kawada. 1997. Development of rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J. Clin. Microbiol. 35:2315-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1, p. 1.89-1.100. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 39.United States Department of Health and Human Services. 2001. Code of federal regulations, title 21, vol. 6, part 522, revised on April 1. Implantation or injectable dosage form new animal drugs: gentamicin sulfate injection. Document 21CFR522.1044. U.S. Government Printing Office, Washington, DC.
- 40.United States Department of Health and Human Services. 2001. Code of federal regulations, title 21, vol. 6, part 520, revised on April 1. Oral dosage form new animal drugs: streptomycin sulfate oral solution. Document 21CFR520.2158a. U.S. Government Printing Office, Washington, DC.
- 41.United States Department of Health and Human Services. 2003. Code of federal regulations, title 21, vol. 68, part 520, revised April 1. Oral dosage form new animal drugs: oxytetracycline hydrochloride soluble powder. Document 21CFR520.1660d. U.S. Government Printing Office, Washington, DC.
- 42.van den Bogaard, A. E., M. Hazen, M. Hoyer, P. Osstenbach, and E. E. Stobberingh. 2002. Effects of flavophospholipol on resistance in fecal Escherichia coli and enterococci of fattening pigs. J. Antimicrob. Chemother. 46:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 44.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warriner, K., T. G. Aldsworth, S. Kaur, and C. E. Dodd. 2002. Cross-contamination of carcasses and equipment during pork processing. J. Appl. Microbiol. 93:169-177. [DOI] [PubMed] [Google Scholar]
- 46.Weigel, L. M., C. P. Steward, and F. C. Tenover. 1998. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42:2661-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woods, C. R., J. Versalovic, T. Koeuth, and J. R. Lupski. 1993. Whole-cell repetitive element sequence-based polymerase chain reaction allows rapid assessment of clonal relationships of bacterial isolates. J. Clin. Microbiol. 31:1927-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]