Abstract
1,2,3-Trichloropropane (TCP) is a highly toxic and recalcitrant compound. Haloalkane dehalogenases are bacterial enzymes that catalyze the cleavage of a carbon-halogen bond in a wide range of organic halogenated compounds. Haloalkane dehalogenase LinB from Sphingobium japonicum UT26 has, for a long time, been considered inactive with TCP, since the reaction cannot be easily detected by conventional analytical methods. Here we demonstrate detection of the weak activity (kcat = 0.005 s−1) of LinB with TCP using X-ray crystallography and microcalorimetry. This observation makes LinB a useful starting material for the development of a new biocatalyst toward TCP by protein engineering. Microcalorimetry is proposed to be a universal method for the detection of weak enzymatic activities. Detection of these activities is becoming increasingly important for engineering novel biocatalysts using the scaffolds of proteins with promiscuous activities.
1,2,3-Trichloropropane (TCP) is a xenobiotic compound generated from the manufacture of epichlorohydrin and is used as an industrial solvent, a paint and varnish remover, and a cleaning and degreasing agent (1). TCP can be degraded by haloalkane dehalogenase (DhaA) from Rhodococcus rhodochrous NCIMB 13064 with low catalytic efficiency (kcat = 0.08 s−1; Km = 2.2 mM) (4). DhaA shares a 47.7% protein sequence identity with the haloalkane dehalogenase LinB from Sphingobium japonicum UT26. LinB is an enzyme involved in a biochemical pathway for the degradation of the pesticide lindane (γ-hexachlorocyclohexane), where it catalyzes the conversion of 1,3,4,6-tetrachloro-1,4-cyclohexadiene to 2,4,5-trichloro-2,5-cyclohexadiene-1-ol (16). A wide spectrum of haloalkanes can be converted by LinB; however, TCP was estimated to be a haloalkane that did not serve as a substrate for LinB when conventional analytical methods were used (6, 12). We decided to apply X-ray crystallography to determine the LinB-TCP complex structure and reveal the structural basis for inactivity.
LinB was expressed and purified as described previously (16). The enzyme was crystallized at 278 K using hanging drop vapor diffusion in 0.1 M Tris buffer (pH 8.9), 18 to 20% (wt/vol) polyethylene glycol (PEG) 6000, and 0.2 M calcium acetate. Crystals of approximately 0.4 by 0.3 by 0.04 mm3 were transferred to the mother liquor constituents with 12.5 mM TCP and immersed for 5.5 h. Complete diffraction data were collected at European Synchotron Radiation Facility beam line ID14-2 at 100 K. The cryoprotectant was composed of 20% (vol/vol) sucrose and 20% (wt/vol) PEG 400. The data collected (to 1.6 Å) were processed using HKL (Miller index) programs DENZO and SCALEPACK (18). The structure of the complex of LinB with a halogenated substrate was solved using the coordinates of native LinB (Protein Data Bank entry 1CV2). All calculations were performed using a CCP4 suite of programs (7). The model was refined initially by the rigid-body approach and subsequently by restrained maximum-likelihood optimization. Density modification was applied to improve the quality of the density map. Visual checking of the 2Fo-Fc and Fo-Fc electron density maps (Fo and Fc are the observed and calculated structure factor amplitude, respectively) and manual rebuilding of the model were carried out using the program Xfit (15). Finally, water molecules were added to the model by using the solvent building regime Arp/Warp (13). The refinement converged to Rcryst and Rfree values of 0.140 and 0.162, respectively. The final model contained 10 heteroatoms, including ligand atoms, 3 Ca2+ cations, and 1 Cl− anion. Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Summary of data collection and refinement statistics of crystallographic analysis
| Parametera | Value | |
|---|---|---|
| Wavelength (Å) | 0.933 | |
| Space group | P212121 | |
| Cell parameters (Å, °) | a = 50.57, b = 72.14, | |
| c = 73.31, α = β = γ = 90 | ||
| Total no. of reflections | 325,309 | |
| No. of unique reflections | 36,132 | |
| Resolution range (last | ||
| resolution shell) (Å) | 30-1.6 (1.66-1.6) | |
| Rmerge | 0.052 (0.174) | |
| Completeness (%) | 99.8 (99.5) | |
| Mosaicity | 0.57 | |
| Rcryst | 0.148 | |
| Rfree | 0.162 | |
| No. of protein atoms | 2,735 | |
| No. of water molecules | 337 | |
| No. of heteroatoms | 10 | |
| RMS deviation from ideality
|
||
| Bonds (Å) | 0.012 | |
| Angles (°) | 1.462 | |
| Mean B value (%) | 8.21 | |
Rmerge, reliability factor; Rcryst, crystallographic R factor; Rfree, free R factor, calculated using a random set (5%) of reflections that were omitted during refinement; RMS, root mean square.
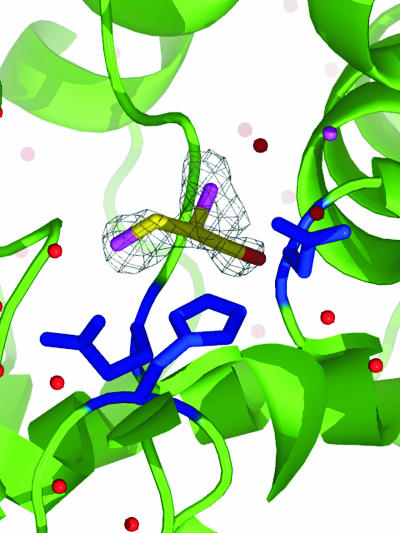
Despite the fact that the activity of LinB with TCP has never been detected in an activity assay in solution under standard conditions, the product of dehalogenation was identified in the crystal structure. Figure 1 depicts (R)-2,3-dichloropropane-1-ol, the product formed during the dehalogenation of TCP. The product molecule is located near the entrance tunnel and makes close contact with several active site residues. The most striking of these are contacts with the catalytic histidine (His272), which creates a strong van der Waals interaction with the ligand molecule. Similar binding of various ligands has been noted previously in other structures of LinB-ligand complexes (14, 17, 21).
FIG. 1.
Product of TCP dehalogenation bound to the active site of LinB. The atoms of (R)-2,3-dichloropropane-1-ol are yellow (carbon), red (oxygen), and purple (chlorine). The residues that make up the catalytic triad are in blue.
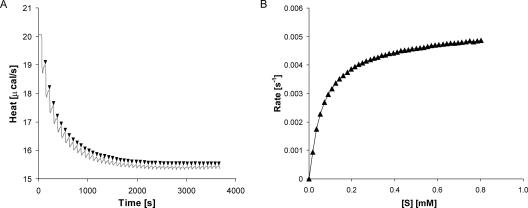
Enzymatic conversion of TCP by LinB was confirmed by the experiment, lasting for 3 days. The reaction mixture was composed of reaction buffer (50 mM Tris and 150 mM NaCl [pH 8.75] at 20°C), 3.3 mM TCP, and 1 mg/ml (0.045 mM) LinB. After 3 days of incubation, the sample was analyzed by gas chromatography-mass spectrometry (Trace MS 2000; Thermo Finnigan) using a CHIRALDEX G-TA capillary gas chromatography column (30 m by 0.25 mm by 0.25 mm) (Alltech Associates, Inc.). The product of the enzymatic conversion was identified in the mixture as 2,3-dichloropropan-1-ol by using National Institute of Standards and Technology mass spectral search program version 1.6d. Gas chromatography was not sensitive enough for quantitative evaluation, and therefore we designed a microcalorimetric procedure to determine the kinetics of the LinB reaction with TCP. A reaction mixture vessel of isothermal titration microcalorimeter model VP-ITC (MicroCal, Northampton, MA) was filled with 1.4 ml of LinB solution at a concentration of 1 mg/ml (50 mM Tris and 150 mM NaCl [pH 8.75] at 37°C). The substrate solution was prepared in the same buffer by the addition of TCP to a final concentration of 3.3 mM. Substrate concentration was verified by gas chromatography before the kinetic experiment was carried out. The gas chromatography Trace GC 2000 model (Thermo Finnigan) was equipped with a flame ionization detector and a capillary column, model DB-FFAP (30 m by 0.25 mm by 0.25 μm; J&W Scientific). The reference power of the microcalorimeter, VP-ITC, was set to 20 μcal·s−1. In the kinetic experiment, the LinB enzyme was titrated in 60-s intervals in the reaction mixture vessel with increasing amounts of a substrate, while pseudo-first-order conditions were maintained. After every injection, the peak of dilution heat was observed, which was followed by a relaxation of the signal to a level corresponding to that of heat produced by the enzymatic reaction. The rate of the heat generated by the enzymatic reaction is equivalent to the decrease in instrumental thermal power (Fig. 2A). The negative deflection, as shown in Fig. 2A, indicated that this reaction is exothermic. Every injection increased the substrate concentration, leading to a further increase in enzyme reaction rate (an increase of heat generated) until the enzymatic reaction was saturated. A total of 45 injections were carried out during titration. The reaction rates reached after every injection (in units of thermal power) were converted to enzyme turnover by using apparent molar enthalpy (ΔHapp), as shown in equation 1, where [P] is the molar concentration of product generated and Q is enzyme-generated thermal power (22).
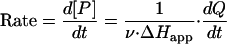 |
(1) |
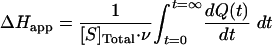 |
(2) |
Apparent molar enthalpy of the TCP conversion by LinB (ΔHapp = −0.0374 kcal·mol−1) was determined in a separate experiment that allowed the reaction to proceed to completion. In the experiment, 3.3 nmol of TCP was fully converted by LinB, and the total heat of conversion was obtained by integration of the VP-ITC signal (22) (equation 2, where [S] is the molar concentration of substrate converted). The calculated enzyme turnover was plotted against the actual concentration of the substrate after every injection (Fig. 2B). Kinetic constants (Table 2) were calculated from the dependence of enzyme turnover on substrate concentration by using Origin 6.1 (OriginLab Corporation).
FIG. 2.
(A) Signal from VP-ITC titration of LinB with TCP (gray solid line). The reaction rates (in units of thermal power) increase with increasing amounts of TCP (black arrows). (B) Dependence of enzyme turnover on the substrate concentration. The enzyme turnover was calculated from reaction rates by using apparent enthalpy (ΔHapp = −0.0374 kcal·mol−1).
TABLE 2.
Kinetic parameters for dehalogenation of TCP by LinB
| Parameter | Value | Error |
|---|---|---|
| Km (mM) | 0.073 | ± 0.001 |
| kcat (s−1) | 0.005 | ± 0.00002 |
| kcat/Km (s−1/mM−1) | 0.068 |
Microcalorimetry is typically used for thermodynamic analysis of macromolecule-ligand binding when heat changes during titration experiments. These data quantify the strength of the interaction. Heat produced or consumed by enzymatic reaction can also be monitored by microcalorimetry (5, 8, 9, 11, 19, 20, 22). Todd and Gomez (22), in their pioneering paper on the application of calorimetry to enzymatic kinetics, summarized the benefits of microcalorimetric assays over conventional methods as universality, nondestructiveness, and precision. In our study, we benefited from another strong feature of the microcalorimetry: sensitivity to the detection of extremely slow enzymatic reactions.
To our knowledge, the activity of LinB with TCP, described by the catalytic constant, kcat = 0.005 s−1, is the slowest ever determined by microcalorimetric assay and published in the scientific literature. The conversion of TCP by LinB is 3,200-fold less effective than the conversion of a good substrate for LinB 1-iodohexane, yet LinB enzyme could become a good starting material for the development of a new biocatalyst for TCP by protein engineering. Previous attempts to improve the activity of DhaA toward TCP by directed evolution resulted in the enzymes showing 3.5-fold (4) and 4-fold (10) more enzyme activity than that of the wild type, while the combination of rational design (3) and directed evolution led to a 36-fold more active protein variant (M. Pavlova et al., unpublished data). Improving LinB activity toward TCP using similar approaches should be feasible. LinB could become a very useful catalyst considering its high affinity toward TCP, with Km equal to 0.073 mM, which is 2 orders of magnitude lower than the value reported for DhaA (Km = 2.2 mM) (4). We further propose that a much broader use for microcalorimetry could be found in enzymology for the detection of weak activities. These activity levels are becoming increasingly important especially for the detection and quantification of enzymes with promiscuous activities (2), since it is generally much easier to improve existing, even very weak, activities than to develop completely new activity within a protein scaffold.
Acknowledgments
This research was financially supported by a grant from INCHEMBIOL (MSM0021622412) and by the Centre for Biocatalysis and Biotransformation (LC06010, from the Czech Ministry of Education).
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Agency for Toxic Substances and Disease Registry. 1992. Toxicological profile for 1,2,3-trichloropropane. U.S. Department of Health and Human Services, Public Health Service, Washington, DC. [PubMed]
- 2.Aharoni, A., L. Gaidukov, O. Khersonsky, S. M. Gould, C. Roodveldt, and D. S. Tawfik. 2004. The “evolvability” of promiscuous protein functions. Nat. Genet. 37:73-76. [DOI] [PubMed] [Google Scholar]
- 3.Banas, P., M. Otyepka, P. Jerabek, M. Petrek, and J. Damborsky. 2006. Mechanism of enhanced conversion of 1,2,3-trichloropropane by mutant haloalkane dehalogenase revealed by molecular modeling. J. Comput.-Aided Mol. Des. 20:375-383. [DOI] [PubMed] [Google Scholar]
- 4.Bosma, T., J. Damborsky, G. Stucki, and D. B. Janssen. 2002. Biodegradation of 1,2,3-trichloropropane through directed evolution and heterologous expression of a haloalkane dehalogenase gene. Appl. Environ. Microbiol. 68:3582-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, L., A. Cao, and L. Lai. 2001. An isothermal titration calorimetric method to determine the kinetics parameters of enzyme catalytic reaction by employing the product inhibition as probe. Anal. Biochem. 299:19-23. [DOI] [PubMed] [Google Scholar]
- 6.Chaloupkova, R., J. Sykorova, Z. Prokop, A. Jesenska, M. Monincova, M. Pavlova, Y. Nagata, and J. Damborsky. 2003. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 278:52622-52628. [DOI] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 8.D′Amico, S., J. S. Sohier, and G. Feller. 2006. Kinetics and energetics of ligand binding determined by microcalorimetry: insights into active site mobility in a psychrophilic alpha-amylase. J. Mol. Biol. 358(5):1296-1304. [DOI] [PubMed] [Google Scholar]
- 9.Debord, J., B. Verneuil, J.-C. Bollinger, L. Merle, and T. Dantoine. 2006. Flow microcalorimetry study of butyrylcholinesterase kinetics and inhibition. Anal. Biochem. 354:299-304. [DOI] [PubMed] [Google Scholar]
- 10.Gray, K. A., T. H. Richardson, K. Kretz, J. M. Short, F. Bartnek, R. Knowles, L. Kan, P. E. Swanson, and D. E. Robertson. 2001. Rapid evolution of reversible denaturation and elevated melting temperature in a microbial haloalkane dehalogenase. Adv. Synth. Catal. 343:607-617. [Google Scholar]
- 11.Jeoh, T., J. O. Baker, A. K. Mursheda, M. E. Himmel, and W. S. Adney. 2005. Beta-D-glucosidase reaction kinetics from isothermal titration microcalorimetry. Anal. Biochem. 347:244-253. [DOI] [PubMed] [Google Scholar]
- 12.Kmunicek, J., K. Hynkova, T. Jedlicka, Y. Nagata, A. Negri, F. Gago, R. C. Wade, and J. Damborsky. 2005. Quantitative analysis of substrate specificity of haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26. Biochemistry 44:3390-3401. [DOI] [PubMed] [Google Scholar]
- 13.Lamzin, V. S., and K. S. Wilson. 1997. Automated refinement for protein crystallography. Methods Enzymol. 277:269-305. [DOI] [PubMed] [Google Scholar]
- 14.Marek, J., J. Vevodova, I. Kuta-Smatanova, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi, and J. Damborsky. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39:14082-14086. [DOI] [PubMed] [Google Scholar]
- 15.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 16.Nagata, Y., K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova, and M. Takagi. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63:3707-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oakley, A. J., Z. Prokop, M. Bohac, J. Kmunicek, T. Jedlicka, M. Monincova, I. Kuta-Smatanova, Y. Nagata, J. Damborsky, and M. C. J. Wilce. 2002. Exploring the structure and activity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26: evidence for product and water mediated inhibition. Biochemistry 41:4847-4855. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 19.Saboury, A. A., and A. A. Moosavi-Movahedi. 1997. A simple novel method for determination of an inhibition constant by isothermal titration microcalorimetry. The effect of fluoride ion on urease. J. Enzyme Inhib. 12:273-279. [DOI] [PubMed] [Google Scholar]
- 20.Stodeman, A., and F. P. Schwarz. 2002. Importance of product inhibition in the kinetics of the acylase hydrolysis reaction by differential stopped flow microcalorimetry. Anal. Biochem. 308:285-293. [DOI] [PubMed] [Google Scholar]
- 21.Streltsov, V. A., Z. Prokop, J. Damborsky, Y. Nagata, A. J. Oakley, and M. C. J. Wilce. 2003. Haloalkane dehalogenase LinB from Sphingomonas paucimbilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates. Biochemistry 42:10104-10112. [DOI] [PubMed] [Google Scholar]
- 22.Todd, M. J., and J. Gomez. 2001. Enzyme kinetics determined using calorimetry: a general assay for enzyme activity? Anal. Biochem. 296:179-187. [DOI] [PubMed] [Google Scholar]