Abstract
Fluorescent in situ hybridization (FISH) remains a key technique in microbial ecology. Molecular beacons (MBs) are self-reporting probes that have potential advantages over linear probes for FISH. MB-FISH strategies have been described using both DNA-based and peptide nucleic acid (PNA)-based approaches. Although recent reports have suggested that PNA MBs are superior, DNA MBs have some advantages, most notably cost. The data presented here demonstrate that DNA MBs are suitable for at least some FISH applications in complex samples, providing superior discriminatory power compared to that of corresponding linear DNA-FISH probes. The use of DNA MBs for flow cytometric detection of Pseudomonas putida resulted in approximately double the signal-to-noise ratio of standard linear DNA probes when using laboratory-grown cultures and yielded improved discrimination of target cells in spiked environmental samples, without a need for separate washing steps. DNA MBs were also effective for the detection and cell sorting of both spiked and indigenous P. putida from activated sludge and river water samples. The use of DNA MB-FISH presents another increase in sensitivity, allowing the detection of bacteria in environmental samples without the expense of PNA MBs or multilaser flow cytometry.
Fluorescent in situ hybridization (FISH) with rRNA-targeted probes is a widely used technique (1, 3, 7) that enables phylogenetic identification of individual cells without the need for prior cultivation. Analysis of samples is often achieved using fluorescence microscopy (3, 7) and flow cytometry (2, 16, 17) to enumerate and further study cells of interest. In most environmental samples, bacteria will be slow growing and, hence, will have low ribosome numbers. Consequently, the signal obtained from target cells after FISH will be low, and target cells may be difficult to distinguish from debris and nontarget cells. Accordingly, there has been much effort to increase the fluorescent signal from target cells (2, 6, 10, 12, 13) to improve the signal-to-noise ratio (S:N).
A recent advance in FISH technologies is the replacement of standard linear oligonucleotide probes with molecular beacons. Peptide nucleic acid (PNA) and DNA molecular beacons have been used successfully for the detection of in situ rRNA (15, 20). Molecular beacons are a type of nucleic acid probe used to detect specific nucleic acids in homogenous solutions (14). Designed as probes for reporting the presence of specific amplicons in real-time PCR, molecular beacons substantially increase in fluorescence in the presence of target nucleic acid sequences (reviewed in reference 5). One key advantage in using molecular beacons would be to help minimize noise, thereby also improving the S:N and subsequent detection of cells using FISH.
This study directly compared the discriminatory powers of FISH using linear probes and molecular beacons when samples were analyzed using single-laser flow cytometry. A standard monolabeled fluorescent linear probe and a DNA beacon that targeted the same region on the 16S rRNA molecule (specific for the majority of pseudomonads) were designed. The pseudomonads were targeted in this study due to their ubiquity and importance in many areas of environmental microbiology, especially applications to environmental problems. After optimization of hybridization conditions for the linear probe and beacon, the S:N was directly compared for the two types of probe in pure culture using single-laser flow cytometry. Both probes were then used to analyze environmental samples of water and activated sludge, with and without the artificial addition of target cells.
Comparison of molecular beacons and linear probes with laboratory cultures.
Pseudomonas putida (PaW340) (19) and Escherichia coli (NCIMB12210) were grown aerobically on a rotary shaker in buffered peptone water at 28°C and 37°C, respectively. Cells were fixed with 3 volumes 4% fresh paraformaldehyde in phosphate-buffered saline (PBS) (pH 8.0) for 1 h at room temperature (9) before being washed with PBS and resuspended in PBS/absolute ethanol (1:1; vol/vol). Fixed cells were stored at −20°C.
The oligonucleotide probe Ps440 (Ps440LP; CCCTTCCTCCCAACTT), a modification of a previous oligonucleotide probe (4), is complementary to a 16S rRNA region present in many Pseudomonas species but not in E. coli. The Ps440LP sequence was used as the basis for the molecular beacon (Ps440MB). Stems were designed using an algorithm run in Microsoft Excel (11) (ACGGGCCCTTCCTCCCAACTTCCCGT; stems are in bold). All probes used were labeled with 5′ 6-carboxyfluorescein; the beacons were also labeled with 3′ DABCYL.
Flow cytometric analysis of cells was performed using a FACStar Plus flow cytometer (Becton Dickinson, Cowley, United Kingdom) using a single 488-nm laser at 25 mW. The forward scatter was set to the logarithmic gain and used to trigger events. 6-Carboxyfluorescein fluorescence at 520 nm was analyzed through a fluorescence detector 1 (photo multiplier tube, 750 V) with logarithmic gain.
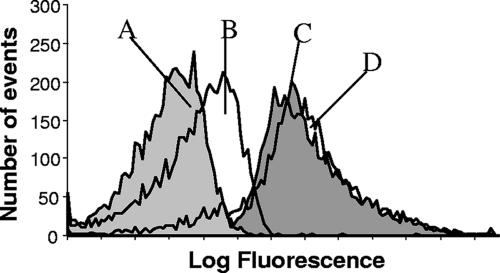
The hybridization of both Ps440LP and Ps440MB was optimized using P. putida as a positive control and E. coli as a negative control. The S:N was calculated by dividing the mean fluorescent intensity (MFI) of P. putida cells by that of E. coli cells. Approximately 2 × 105 cells were added to 50 μl hybridization buffer containing between 0.02 μM and 0.66 μM probe for up to 5 h at temperatures between room temperature and 65°C. The optimal hybridization buffer contained 0.9 M NaCl, 20 mM Tris-HCl (pH 7.2), 0.1% sodium dodecyl sulfate (SDS) (16). Other hybridization buffers tested contained NaCl and MgCl2, and MgCl2 at various concentrations with Tris-HCl and SDS. The optimal hybridization conditions were determined to be 0.1 μM probe for 1 h at 55°C for both probes. Ps440MB and Ps440LP gave S:N of 14.2 (MFI: P. putida, 133.4, and E. coli, 9.4) and 7.9 (MFI: P. putida, 147.3, and E. coli, 18.7), respectively. The increase in the S:N was mainly a result of the decrease in background fluorescence (Fig. 1). The increase in the S:N may be due to closed molecular beacons being “dark” unless hybridized to a target, whereas linear probes are of constant fluorescence whether hybridized or in solution.
FIG. 1.
Flow cytometry histograms showing fluorescence of cells after hybridization with Ps440MB and Ps440LP in optimal conditions. (A) E. coli cells hybridized with Ps440MB. (B) E. coli cells hybridized with Ps440LP. (C) P. putida cells hybridized with Ps440MB. (D) P. putida cells hybridized with Ps440LP.
The effect of posthybridization washing was also investigated, by the centrifugation of hybridized cells followed by resuspension in PBS (16). SDS was included in the hybridization buffer both to increase probe accessibility into the cells and to reduce clumping upon washing. Centrifugation was performed at 2,700 × g or 11,900 × g for 30 seconds to 5 min. While the S:N increased for both probes, the percentage of single cells decreased by up to 30%, presumably because of clumping, and up to 50% of cells were lost. Due to this, cell washing should be avoided when enumeration is important (16) and also since it may cause damage to and clumping of larger cells (13). Concentration, fixation, and washing of cells on filters also cause cell loss (13). Thus, the use of molecular beacons for FISH represents a development that reduces the need for washing and consequently reduces cell clumping and loss.
FISH detection of target bacteria in river water and activated sludge.
Activated sludge samples were taken from Countess Wear sewage treatment works, Exeter, United Kingdom. Countess Wear is a medium-scale municipal plant, serving ∼150,000 population equivalents, taking domestic and industrial wastes. Samples were prepared according to previous protocols (8) and then fixed and stored (as described above) at a concentration of 108 cells per ml. Before fixation, culturable counts were made in duplicate on C-F-C agar (Oxoid, Basingstoke, United Kingdom) to estimate the numbers of fluorescent pseudomonads. To test the system, approximately 500 fixed P. putida cells per microliter of fixed sample were added to a subset of samples. Known concentrations of fluorescent beads were added to samples to measure the volume of sample analyzed and permit the calculation of the number of target cells per volume of original sample.
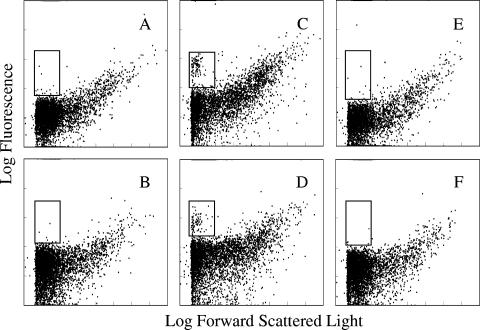
Samples were hybridized with Ps440LP, Eub338′ (2) (to act as a nonspecific binding control for linear probes), Ps440MB, and an entirely separate beacon, xylE MB (CGCTCGCGACCTATGACCTGTTCACCGAGCG) (J. Lenaerts et al., unpublished data) (to act as a nonspecific binding control for beacons). Molecular beacons proved to be more powerful than linear probes at discriminating target cells from debris and nontarget cells (Fig. 2). This increase in discriminatory power is because molecular beacons increase the S:N by reducing background fluorescence. Hence, FISH with molecular beacons allows increased discrimination of target events due to fewer nonspecific events appearing within the same region of the flow cytometry dot plot (Fig. 2).
FIG. 2.
Flow cytometry dot plots of activated sludge samples, with and without fixed exponential-phase P. putida cells added, hybridized with Ps440MB (A and C), Ps440LP (B and D), xylE MB (E), and Eub338′ (F). Samples amended with P. putida cells are shown in panels C and D. The inset rectangles highlight the region where labeled target events should appear. All plots display 5,000 events.
The numbers of target cells were calculated by subtracting the counts of the nonspecific binding controls (Eub338′ and xylE MB) from the total counts (Ps440LP and Ps440MB). Where target cells were added, Ps440MB detected significantly more cells without any posthybridization wash step than Ps440LP (P = 0.047; mean cell count of 418 compared to mean cell count of 326, respectively, per microliter of fixed sample). The number of target cells detected decreased after washing. The numbers of cells detected by Ps440MB decreased significantly (P = 0.010). The decrease in cell numbers detected by Ps440LP was not significant (P = 0.077). There was no significant difference between the numbers of target cells detected with Ps440MB and Ps440LP after washing (P = 0.550). Before washing, the molecular beacon enabled the detection of significantly more target cells than the linear probe, due to its superior S:N. It is proposed that after washing, both probes detected lower but similar numbers of target cells, presumably due to both approaches providing useful S:N values, but with fewer cells detected in total due to losses from the washing steps. Because molecular beacons increase the S:N by reducing background fluorescence, the area on flow cytometry dot plots occupied by cells with low cellular rRNA contents is made more clear, as noise is reduced (Fig. 2). As natural environments are rarely as rich in nutrients as laboratory culture conditions, the cellular rRNA content of most cells in environmental samples is low, and the ability to detect these cells is important.
Water was collected from a nontidal region of the river Exe (Ordnance Survey grid reference SX914923) in July 2004. The river flow was normal for the time of year. To maximize target cell numbers, the contents of 25 liters of river water were concentrated using tangential flow filtration and centrifugation to a 10-ml volume. Culturable counts were made before fixation (as above).
For both river water and activated sludge samples, molecular beacons detected more target cells than linear probes in every sample analyzed (Table 1). However, the increases in cell numbers detected with Ps440MB were not significant, probably due to the large variability within the samples and due to target cells being very rare (<0.01%).
TABLE 1.
Detection of indigenous pseudomonads in environmental samples by FISH and culturable counts
| Samplea | Confirmed countb | Cells detected/ml byc:
|
Pd | % of confirmed count
|
||
|---|---|---|---|---|---|---|
| Ps440MB | Ps440LP | Ps440MB | Ps440LP | |||
| AS1 | 1.2 × 104 | 1.8 × 104 (4.4 × 103) | 9.2 × 103 (6.6 × 103) | 0.118 | 151 | 76 |
| AS2 | 1.1 × 104 | 1.2 × 104 (7.1 × 103) | 4.0 × 103 (1.1 × 103) | 0.121 | 111 | 37 |
| AS3 | 3.3 × 103 | 3.9 × 103 (2.9 × 103) | 2.2 × 103 (8.5 × 102) | 0.380 | 119 | 67 |
| RW1 | 1.4 | 0.62 (0.22) | 0.36 (0.23) | 0.226 | 43 | 25 |
| RW2 | 1.7 | 1.73 (0.62) | 1.05 (0.42) | 0.193 | 103 | 62 |
AS, activated sludge; RW, river water.
Confirmed counts are 67% of the average of duplicate fluorescent CFU counts (12 out of 18 fluorescent isolates were confirmed to be pseudomonads by using restriction digest of PCR products).
FISH-flow cytometry counts have been converted to specific cells detected per ml of original sample. One standard deviation is shown in parentheses.
P values are from independent-sample t tests between cells detected by FISH with Ps440MB and Ps440LP.
One thousand target events (and nontarget events) were sorted by flow cytometry from activated sludge samples hybridized with both Ps440LP and Ps440MB. Target events were confirmed as pseudomonads by PCR and restriction digests (18). The PCR products from samples hybridized with Ps440MB gave pseudomonad-specific products with higher relative intensities (RI; calculated by Jasc Paint Shop Pro) than those hybridized with Ps440LP (Ps440MB RI of 32.5 compared to Ps440LP RI of 9.8). For negative controls, nonspecific events were also sorted and were used as a PCR template (Ps440MB nonspecific RI of 6.2 and Ps440LP nonspecific RI of 7.2). These results again confirm the higher discriminatory power of molecular beacons over standard linear probes.
Alternative protocols are available that allow better detection of target cells using flow cytometry. One such protocol uses dual-laser flow cytometry, where only events stained with a DNA dye are analyzed for the presence of the specific FISH probe (17). Dual-laser flow cytometry requires both a substantially more expensive instrument and the use of more complex staining procedures. FISH using a probe labeled with an enzyme to amplify the signal intensity has also been used (13). Again, this technique introduces more steps to the staining protocol and, due to the size of the enzyme complex, is limited to easily permeabilized bacteria. The technique presented in this work uses a simple protocol which has a greater potential for automation. Thus, a much-higher sample throughput can be achieved.
PNA beacons have also been used to achieve better discriminatory power (using signal-laser flow cytometry), but low signal-to-noise ratios and long hybridization times reportedly hindered DNA beacons (20). DNA beacons cost substantially less than PNA beacons, and thus are more accessible to a higher number of microbial ecology labs. This study demonstrates that DNA beacons can be used for FISH in laboratory and environmental samples to obtain better discriminatory power than that of standard monolabeled fluorescent linear probes. The use of DNA beacons together with single-laser flow cytometry presents a method that does not have the associated expense of PNA beacons or dual-laser flow cytometry. The protocol is also simpler than those using washing steps and/or enzymatically amplified signals.
Acknowledgments
J.L. was supported by the Biotechnological and Biological Sciences Research Council (Swindon, United Kingdom).
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Amann, R. I. 1995. Fluorescently labeled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-553. [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braunhowland, E. B., P. A. Vescio, and S. A. Nierzwickibauer. 1993. Use of a simplified cell blot technique and 16S rRNA-directed probes for identification of common environmental isolates. Appl. Environ. Microbiol. 59:3219-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broude, N. E. 2002. Stem-loop oligonucleotides: a robust tool for molecular biology and biotechnology. Trends Biotechnol. 20:249-256. [DOI] [PubMed] [Google Scholar]
- 6.DeLong, E. F., L. T. Taylor, T. L. Marsh, and C. M. Preston. 1999. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Environ. Microbiol. 65:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: rRNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 8.Forster, S., J. R. Snape, H. M. Lappin-Scott, and J. Porter. 2002. Simultaneous fluorescent Gram staining and activity assessment of activated sludge bacteria. Appl. Environ. Microbiol. 68:4772-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonald, R., and V. S. Brozel. 2000. Community analysis of bacterial biofilms in a simulated recirculating cooling-water system by fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes. Water Res. 34:2439-2446. [Google Scholar]
- 11.Monroe, W. T., and F. R. Haselton. 2003. Molecular beacon sequence design algorithm. BioTechniques 34:68-73. [DOI] [PubMed] [Google Scholar]
- 12.Schonhuber, W., B. Fuchs, S. Juretschko, and R. Amann. 1997. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63:3268-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 15.Valeur, N., P. Engel, N. Carbajal, E. Connolly, and K. Ladefoged. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 70:1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 17.Wallner, G., R. Erhart, and R. Amann. 1995. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl. Environ. Microbiol. 61:1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Widmer, F., R. J. Seidler, P. M. Gillevet, L. S. Watrud, and G. D. Di Giovanni. 1998. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl. Environ. Microbiol. 64:2545-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winstanley, C., J. P. Carter, M. Seasman, J. A. W. Morgan, R. W. Pickup, and J. R. Saunders. 1993. A comparison of the survival of stable and unstable chromosomally located xylE marker cassettes as an indicator of cell division within populations of Pseudomonas putida released into lake water and soil. Microb. Releases 2:97-107. [Google Scholar]
- 20.Xi, C. W., M. Balberg, S. A. Boppart, and L. Raskin. 2003. Use of DNA and peptide nucleic acid molecular beacons for detection and quantification of rRNA in solution and in whole cells. Appl. Environ. Microbiol. 69:5673-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]