Abstract
In the present work we develop a multiplex PCR assay for the detection and identification of the fish pathogen Vibrio vulnificus biotype 2 with discriminating potential for zoonotic strains (serovar E). The PCR assay allowed the identification of two new biotype 2 serovar E human isolates from culture collections. Finally, the multiplex was successfully applied to both diagnosis and carrier detection in field samples.
Vibrio vulnificus is an aquatic bacterium from warm and tropical ecosystems, with pathogenic potential for humans and fish (mainly eels). Human vibriosis occurs after eating contaminated seafood or exposing open wounds to seawater (7, 16, 19), and fish vibriosis occurs after immersion in contaminated water or contact with diseased animals or carriers (2). The species is subdivided into three biotypes on the basis of differences in biochemical, genetic, and serological tests, as well as on the basis of the host range (4, 20). At present, biotyping involves tedious and time-consuming tests (3, 4). For this reason, the clinical strains from fish are directly classified as biotype 2 (BT2), and those from humans as biotype 1, with the exception of the cellobiose-negative isolates from Israel, considered to be biotype 3.
The first link between diseased fish manipulation and human diseases was established by Veenstra et al. in The Netherlands in 1992 (23). These authors hypothesized that diseased eels could constitute a risk for public health because the fish pathogen V. vulnificus BT2 could sporadically infect humans. The hypothesis was confirmed after the identification of one human isolate from the ATCC as belonging to BT2 and serovar E (BT2-SerE) (1). Although new human cases of vibriosis, acquired after fish manipulation, have been reported in northern Europe (5, 6, 9, 14, 22), none of these isolates has been identified at a subspecies level. These cases have been related to an increase in seawater temperature surrounding Baltic countries (water with salinity adequate for V. vulnificus survival) due to atypically warm years.
The main objective of the present work was to develop a biotyping procedure based on a PCR assay that simplifies the identification of the V. vulnificus BT2 fish pathogen and, at the same time, allows for the discrimination of those isolates with human pathogenic potential (SerE). A secondary objective was to adapt the PCR protocol to vibriosis-diagnostic and -sensitive detection of the pathogen in subclinical carrier fish.
For the design of the PCR primer sets, we selected the cytolisin gene vvhA, which is present in all V. vulnificus strains regardless of the biotype (13, 24, 25), and two DNA sequences specific for BT2 and SerE. These sequences were obtained from a study in which genomic DNA of eel-virulent and of eel-avirulent strains of different biotypes was compared by suppression subtractive hybridization (15). The BT2-specific sequences could be considered virulence markers since they are present only in the fish-virulent strains (15). Table 1 shows the sequences of the three primer pairs used in this study. PCRs were performed in 50-μl reaction volumes that contained 1× PCR buffer (Roche Diagnostics), 1.5 mM MgCl2, 200 μmol of each deoxynucleoside triphosphate (Roche), 1.5 U of Taq polymerase (Roche Diagnostics), primer concentrations specified in Table 1, and 2 to 5 μl of sample (culture supernatant, cell lysates, extracted or purified DNA). The multiplex PCR was performed on a TC-312 thermal cycler (Techne, Duxford, Cambridge, United Kingdom), and the selected parameters were as follows: an initial denaturation step at 94°C for 4 min, followed by 35 serial cycles of 1 min at 94°C for denaturing, 45 s at 64°C for annealing, and 1 min at 72°C for extension, and a final extension step at 72°C for 10 min. A negative control (no template DNA) and a positive control (2 ng of purified DNA from the BT2-SerE strain CECT 4602) were included in each batch of PCR. The amplified products were separated by electrophoresis on 1.8% agarose gels and visualized by staining with ethidium bromide.
TABLE 1.
Sequences and concentrations of the primers used in the multiplex PCR assay
To test the specificity of the multiplex PCR, a total of 108 V. vulnificus strains (Table 2), most of which have been previously biotyped (1, 3, 15), together with 37 strains of other Vibrio species and related genera (Table 3) were used. No amplicon was obtained from the strains belonging to the species listed in Table 3, whereas all V. vulnificus strains gave the expected amplification products (Table 2 and Fig. 1). Specificity of the method, calculated as a percentage of coincident results with those produced by the traditional methods, was 100%. Interestingly, two new isolates of human origin, not previously biotyped, presented three bands after PCR amplification (Table 2). The isolates were subjected to the conventional subtyping procedure that involves biochemical and serological identification and virulence tests in eels (1). The results confirmed that isolates CIP 81.90 from human blood (France, 1980) and CCUG 38521 from wound infection (Sweden, 1997) belonged to BT2-SerE, since they were virulent for eels (50% of the lethal dose < 104 CFU/fish), agglutinated with anti-SerE-specific serum, and gave an API20E profile (Biomerieux, France) similar to those previously published for BT2-SerE isolates (5106005 and 5306005, respectively). No information about the source of infection has been reported for strain CIP 81.90 (Institute Pasteur Collection), but in the case of strain CCUG 38521 (Swedish Type Culture Collection), the patient was wounded with a fishhook while fishing. The rest of the human strains identified as BT2-SerE in previous works (1, 3, 15) came from Australia, Denmark, and the United States, and only those from Denmark were clearly related to eel manipulation. Our results suggest that seawater or other fish species apart from eels should be considered as putative reservoirs for this pathogen.
TABLE 2.
Strains of Vibrio vulnificus, original biotyping, and results of multiplex PCR
| Origin | Source | Original biotypinga | No. of strains | PCR resultc
|
||
|---|---|---|---|---|---|---|
| SE | VV | BT2 | ||||
| Humans | Blood/wound | Biotype 3 | 6 | − | + | − |
| Wound | Biotype 1 | 3 | − | + | − | |
| BT2-SerE | 2 | + | + | + | ||
| ND | 1b | + | + | + | ||
| Lung | Biotype 1 | 1 | − | + | − | |
| Blood | Biotype 1 | 13 | − | + | − | |
| BT2-SerE | 2 | + | + | + | ||
| ND | 1b | + | + | + | ||
| Aquatic | Diseased eels | Biotype 1 | 5 | − | + | − |
| animals | BT2-SerE | 18 | + | + | + | |
| BT2-no-SerE | 12 | − | + | + | ||
| Healthy eels | Biotype 1 | 1 | − | + | − | |
| BT2-SerE | 2 | + | + | + | ||
| Diseased shrimps | BT2-SerE | 1 | + | + | + | |
| Healthy fish | Biotype 1 | 1 | − | + | − | |
| Oysters | Biotype 1 | 9 | − | + | − | |
| Water | Seawater/brackish | Biotype 1 | 8 | − | + | − |
| water | BT2-SerE | 2 | + | + | + | |
| Tank water from | Biotype 1 | 14 | − | + | − | |
| a fish farm | BT2-SerE | 6 | + | + | + | |
ND, not done.
New human clinical cases due to BT2-SerE.
SE, serovar E; VV, Vibrio vulnificus.
TABLE 3.
Sources and origins of the non-Vibrio vulnificus strains used in this study
| Straina | Source | Country |
|---|---|---|
| Aeromonas allosaccharophila CECT 4199T | Diseased elver | Spain |
| Aeromonas encheleia CECT 4342T | Healthy elver | Spain |
| Aeromonas hydrophila CECT 839T | Tin of milk with fishy odor | |
| Aeromonas jandaei CECT 4338 | Diseased elver | Spain |
| Aeromonas sobria CECT 4245T | Diseased carp | |
| Aeromonas jandaei M6 | Eel mucus | Spain |
| Edwarsiella tarda CECT 886 | Diseased eel | U.S. |
| Photobacterium damselae subsp. damselae RG191 | Internal organ of turbot | Spain |
| Plesiomonas shigelloides CECT 4354 | Liver from healthy eel | Spain |
| Pseudomonas spp. | Diseased tilapia | Spain |
| Shewanella spp. | Diseased tilapia | Spain |
| Vibrio aestuarianus CECT 625T | Oyster | |
| Vibrio alginolyticus CECT 521T | Horse mackerel | |
| Listonella anguillarum 775 | Diseased fish | |
| Vibrio campbellii CECT 523T | U.S. | |
| Vibrio carchariae CECT 4215T | Kidney of brown shark | U.S. |
| Vibrio cholerae CECT 653 | Water | India |
| Vibrio cincinnatiensis CECT 4216T | Human blood | U.S. |
| Vibrio diazotrophicus CECT 627T | Gastrointestinal tract of sea urchin | Canada |
| Vibrio fischeri CECT 524T | ||
| Vibrio fluvialis CECT 4217T | Seawater | Spain |
| Vibrio furnissii CECT 4203T | Human feces | Japan |
| Vibrio furnissii CECT 4349 | Brackish water of an eel farm | Spain |
| Vibrio harveyi CECT 605 | Marine plankton | U.S. |
| Vibrio mediterranei CECT 621T | Coastal marine sediment | Spain |
| Vibrio mimicus CECT 4218T | Human ear | U.S. |
| Vibrio mytili CECT 632T | Mussels | Spain |
| Vibrio natriegens CECT 526T | Salt marsh mud | U.S. |
| Vibrio nereis CECT 595T | Seawater | U.S. |
| Vibrio nigripulchritudo CECT 628T | Seawater | U.S. |
| Vibrio ordalii CECT 582T | Kidney of coho salmon | U.S. |
| Vibrio orientalis CECT 629T | Seawater | China |
| Vibrio parahaemolyticus CECT 611 | Spain | |
| Vibrio proteolyticus CECT 630T | Wood-boring isopod | U.S. |
| Vibrio salmonicida CECT 4195T | Atlantic salmon | Norway |
| Vibrio splendidus CECT 528T | Marine fish | |
| Vibrio scophthalmi CECT 4638T | Turbot | U.S. |
CECT, Spanish Type Culture Collection.
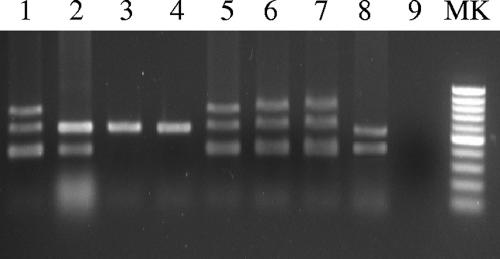
FIG. 1.
Agarose gel electrophoresis of the multiplex PCR products obtained for different samples. Lane MK, molecular weight DNA ladder, low range (Fermentas); lanes 1 to 4, V. vulnificus BT2 Ser E (CECT 4602), BT2 Ser A (CECT 5198), biotype 1 (ATCC 27562T), and biotype 3 (VV12) control strains; lane 5, DNA extracted from the liver of an eel infected with strain CECT4602; lane 6, DNA extracted from the kidney of an eel infected with strain CECT 4602; lane 7, DNA extracted from enrichment broth from gill mucus of an asymptomatic carrier (farm B); 8, DNA from the kidney of a naturally infected eel from fish farm A; lane 9, negative control (no DNA).
To adapt the multiplex PCR to fish vibriosis diagnosis, we artificially infected eels by intraperitoneal injection and processed liver and kidney from moribund animals recovered before 48 h postinfection for both microbiological and PCR analysis. For DNA extractions, samples of 100 μl of homogenated fish tissue (approximately 1.5 to 2.5 mg in phosphate-buffered saline [PBS]-1) plus 300 μl of lysis solution (10 mM Tris-HCl, 5 mM EDTA, 200 mM NaCl, 0.4% sodium dodecyl sulfate, 100 μg/ml proteinase K, pH 8) were incubated at 55°C for 2 h in a water bath. Cell debris was discarded by centrifugation at 10,000 × g for 5 min, and two volumes of ice-cold absolute ethanol was added, drop by drop, to the supernatant. Samples were placed at −20°C for 20 min, DNA was precipitated by centrifugation at 1,000 × g for 15 min, washed in 70% ethanol, and resuspended in 30 to 40 μl of 10 mM Tris-HCl, pH 8 (extracted DNA samples). All tissue samples were positive for microbiological and PCR identification (Fig. 1). In order to test the sensitivity of the assay, we contaminated eel tissue with 10-fold dilutions in PBS-1 of an overnight culture in Trypticasein soy broth plus 0.5% NaCl of the BT2-SerE strain CECT 4602 and processed the sample as described above. The minimal number of cells that gave a clear, positive reaction was 15 CFU/mg tissue, which is similar to the detection limits described by other authors (10, 17).
The PCR protocol was then tested with eels naturally affected with vibriosis. These eels came from an intensive fish farm that cultures eels in freshwater (farm A). Four moribund eels were processed for both microbiological and PCR analyses. In all cases, a BT2-no-SerE amplification profile was obtained from the DNA of the infected tissues (Fig. 1). The bacterial isolates were serologically identified with specific antisera as serovar A (11, 12). This serovar was described for the first time in 2000 in Spain (11) and has spread to Northern Europe, producing vibriosis in eels cultured in freshwater (12). Thus, the PCR protocol would allow the diagnosis of natural vibriosis from eel tissues in less than 5 h from the point of DNA extraction to observation in an agarose gel.
To adapt the multiplex PCR to the detection of healthy carriers, we sampled apparently healthy eels from another local farm that had registered vibriosis 1 year before (farm B). Gills were selected for sampling since the fish pathogen preferentially colonizes this organ (21). Gills were extracted and processed for DNA extraction and PCR analysis as described before. In parallel, gill mucus was sampled with swabs soaked in eel serum broth, a selective enrichment broth designed for the isolation of BT2 strains from environmental samples, or in alkaline peptone water (18). Swabs were incubated in the same medium for 6 h, and DNA was extracted from the cultures (500 μl). The isolation of V. vulnificus from enrichment broths and its identification and biotyping were performed according to conventional procedures (8, 18). V. vulnificus Bt2-SerE was detected by multiplex PCR from 2 of 10 samples but only after enrichment in the appropriate enrichment broth (Fig. 1). The same PCR-positive samples were also positive for the isolation of the pathogen, confirming the feasibility of the protocol. This result is in accordance with previous studies that suggested that survivors of vibriosis can act as carriers (21). Thus, the PCR protocol would allow the detection of carriers without killing the animals. In this case, PCR would be recommended after preenrichment in alkaline peptone water (eel serum broth is more difficult to use since it contains eel serum that contains more PCR inhibitors) for 6 h.
In conclusion, the multiplex PCR developed in the present work is a useful tool to detect and identify the fish pathogen V. vulnificus BT2 from multiple types of sample. The assay would allow discrimination of those pathological cases that constitute a risk to public health. In addition, the multiplex assay could be used in epidemiological studies to correctly biotype clinical isolates and clarify the status of BT2 as a human pathogen.
Acknowledgments
This work was supported by grants AGL 2002-01291 and AGL200504688 from the Spanish Ministry of Science and Technology and grant MTKD-CT-2004-0145019 from the EU.
The experiments with eels were performed in the Experimental Aquaculture Plant of the University of Valencia (Spain).
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Amaro, C., and E. G. Biosca. 1996. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl. Environ. Microbiol. 62:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaro, C., E. G. Biosca, B. Fouz, E. Alcaide, and C. Esteve. 1995. Evidence that water transmits Vibrio vulnificus biotype 2 infections to eels. Appl. Environ. Microbiol. 61:1133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biosca, E. G., C. Amaro, J. L. Larsen, and K. Pedersen. 1997. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl. Environ. Microbiol. 63:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. N. Cameron, D. L. Wykstra, D. L. Swerdlow, and J. J. Farmer III for the Israel Vibrio Study Group. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 5.Bock, T., N. Christensen, N. H. Eriksen, S. Winter, H. Rygaard, and F. Jorgensen. 1994. The first fatal case of Vibrio vulnificus infection in Denmark. APMIS 102:874-876. [PubMed] [Google Scholar]
- 6.Bruun, B. G., N. Frimodt-Moller, A. Dalsgaard, H. E. Busk, H. Friis, H. J. Kolmos, E. Laursen, J. Prag, N. Rosdahl, P. Schouenborg, and P. Sogaard. 1996. Vibrio vulnificus infections in Denmark during the summer of 1994. Ugeskr Laeg. 158:4291-4294. (In Danish.) [PubMed] [Google Scholar]
- 7.CDC Division of Bacterial and Mycotic Diseases. 2005. Vibrio vulnificus: general information, frequently asked questions. www.cdc.gov/ncidod/dbmd/diseaseinfo/vibriovulnificus_g.htm.
- 8.Cerda-Cuellar, M., J. Jofre, and A. R. Blanch. 2000. A selective medium and a specific probe for detection of Vibrio vulnificus. Appl. Environ. Microbiol. 66:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalsgaard, A., N. Frimodt-Moller, B. Bruun, L. Hoi, and J. L. Larsen. 1996. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227-232. [DOI] [PubMed] [Google Scholar]
- 10.del Cerro, A., I. Marquez, and J. A. Guijarro. 2002. Simultaneous detection of Aeromonas salmonicida, Flavobacterium psychrophilum, and Yersinia ruckeri, three major fish pathogens, by multiplex PCR. Appl. Environ. Microbiol. 68:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouz, B., and C. Amaro. 2003. Isolation of a new serovar of Vibrio vulnificus pathogenic for eels cultured in freshwater farms. Aquaculture 217:677-682. [Google Scholar]
- 12.Fouz, B., J. L. Larsen, and C. Amaro. 2006. Vibrio vulnificus serovar A: an emerging pathogen in European anguilliculture. J. Fish Dis. 29:285-291. [DOI] [PubMed] [Google Scholar]
- 13.Hor, L. I., C. T. Gao, and L. Wan. 1995. Isolation and characterization of Vibrio vulnificus inhabiting the marine environment of the southwestern area of Taiwan. J. Biomed. Sci. 2:384-389. [DOI] [PubMed] [Google Scholar]
- 14.Hoyer, J., E. Engelmann, R. M. Liehr, A. Distler, H. Hahn, and T. Shimada. 1995. Septic shock due to Vibrio vulnificus serogroup 04 wound infection acquired from the Baltic Sea. Eur. J. Clin. Microbiol. Infect. Dis. 14:1016-1018. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C.-T., C. Amaro, E. Sanjuán, and L.-I. Hor. 2005. Identification of DNA sequences specific for Vibrio vulnificus biotype 2 strains by suppression subtractive hybridization. Appl. Environ. Microbiol. 71:5593-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 17.Mata, A. I., A. Gibello, A. Casamayor, M. M. Blanco, L. Dominguez, and J. F. Fernandez-Garayzabal. 2004. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water streptococcosis in fish. Appl. Environ. Microbiol. 70:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanjuán, E., and C. Amaro. 2004. Protocol for specific isolation of virulent strains of Vibrio vulnificus serovar E (biotype 2) from environmental samples. Appl. Environ. Microbiol. 70:7024-7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 20.Tison, D. L., M. Nishibuchi, J. D. Greenwood, and R. J. Seidler. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valiente, E., and C. Amaro. 2006. A method to diagnose the carrier state of Vibrio vulnificus serovar E in eels: development and field studies. Aquaculture 258:173-179.
- 22.Veenstra, J., P. J. Rietra, J. Goudswaard, J. A. Kaan, P. H. van Keulen, and C. P. Stoutenbeek. 1993. Extra-intestinal infections caused by Vibrio spp. in The Netherlands. Ned. Tijdschr. Geneeskd. 137:654-657. (In Dutch.) [PubMed] [Google Scholar]
- 23.Veenstra, J., P. J. Rietra, C. P. Stoutenbeek, J. M. Coster, H. H. de Gier, and S. Dirks-Go. 1992. Infection by an indole-negative variant of Vibrio vulnificus transmitted by eels. J. Infect. Dis. 166:209-210. [DOI] [PubMed] [Google Scholar]
- 24.Wright, A. C., J. G. Morris, Jr., D. R. Maneval, Jr., K. Richardson, and J. B. Kaper. 1985. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect. Immun. 50:922-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto, K., A. C. Wright, J. B. Kaper, and J. G. Morris, Jr. 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae E1 Tor hemolysin gene. Infect. Immun. 58:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]