Abstract
The cold-water-fish pathogen Vibrio salmonicida expresses a functional bacterial luciferase but produces insufficient levels of its aliphatic-aldehyde substrate to be detectably luminous in culture. Our goals were to (i) better explain this cryptic bioluminescence phenotype through molecular characterization of the lux operon and (ii) test whether the bioluminescence gene cluster is associated with virulence. Cloning and sequencing of the V. salmonicida lux operon revealed that homologs of all of the genes required for luminescence are present: luxAB (luciferase) and luxCDE (aliphatic-aldehyde synthesis). The arrangement and sequence of these structural lux genes are conserved compared to those in related species of luminous bacteria. However, V. salmonicida strains have a novel arrangement and number of homologs of the luxR and luxI quorum-sensing regulatory genes. Reverse transcriptase PCR analysis suggests that this novel arrangement of quorum-sensing genes generates antisense transcripts that may be responsible for the reduced production of bioluminescence. In addition, infection with a strain in which the luxA gene was mutated resulted in a marked delay in mortality among Atlantic salmon relative to infection with the wild-type parent in single-strain challenge experiments. In mixed-strain competition between the luxA mutant and the wild type, the mutant was attenuated up to 50-fold. It remains unclear whether the attenuation results from a direct loss of luciferase or a polar disturbance elsewhere in the lux operon. Nevertheless, these findings document for the first time an association between a mutation in a structural lux gene and virulence, as well as provide a new molecular system to study Vibrio pathogenesis in a natural host.
Marine bioluminescent bacteria have been the subjects of considerable interest because of the biochemistry that drives light production and their ability to initiate specific, long-term cooperative symbioses with many species of squids and fishes (20, 35, 45, 51). Less is known about bioluminescence in species of bacteria that have the capacity to produce light yet are found in pathogenic associations with animal hosts (32, 33, 38). It has always been of interest to know whether luminescence plays a role in the biology of such pathogens, either to colonize the hosts or to grow in environmental niches. However, attempts to address such questions were limited because a model system in which to study the relationship between bioluminescence and pathogenesis was not available.
In the five previously characterized species of luminous bacteria (Vibrio fischeri, Vibrio harveyi, Photobacterium leiognathi, Photobacterium phosphoreum, and Photorhabdus luminescens), the six structural genes for bioluminescence are contained within a locus termed the lux operon. With the exception of a duplication of luxB (designated luxF) in one species, these genes are arranged in the order luxCDABEG (1, 9, 16, 28). luxA and luxB, respectively, encode the alpha and beta subunits of luciferase, the enzyme responsible for luminescence. luxC, luxD, and luxE each encode an enzyme required for the synthesis of an aliphatic-aldehyde substrate. luxG is not essential for luminescence but is believed to increase the capacity of the cell to synthesize flavin mononucleotide (FMN) (42). In the luminescence reaction, luciferase converts this aliphatic-aldehyde substrate, oxygen, and reduced FMN (FMNH2) into the corresponding aliphatic acid, water, and FMN, with the concomitant production of light (19, 28). In the absence of the aldehyde substrate, luciferase catalyzes a reaction that yields no light and produces oxygen radicals rather than water (15, 18).
Bacteria that carry the genes for luciferase yet do not produce a detectable level of light in culture have been referred to as cryptically bioluminescent (13), and this phenotype may be quite widespread in the environment (14, 33, 34). Cryptic bioluminescence has been best characterized with the psychrophilic fish pathogen Vibrio salmonicida (13), the only bacterium known to cause vibriosis in cold-water, farmed Atlantic salmon (Salmo salar L.), as well as rainbow trout and cod (10, 11).
Cultures of V. salmonicida become visibly luminous only upon the addition of aliphatic aldehyde (an aldehyde group attached to a linear saturated carbon chain) and induce the synthesis of luciferase 10-fold per cell as they approach stationary phase (13). Similarly, when exposed to N-3-oxohexanoyl homoserine lactone, the signal molecule that is in part responsible for quorum sensing and the induction of luminescence in V. fischeri (30, 32), V. salmonicida induces luciferase production 100-fold. Thus, the regulation of the luciferase in V. salmonicida, like that in its close congener V. fischeri (50), appears to be under the control of a quorum-sensing autoinducer mechanism. Despite these similarities, quorum-sensing regulation of the lux operon in V. fischeri strain MJ1 is responsible for a 10,000-fold increase in light production per cell (31), which is substantially greater than the 10-fold increase in V. salmonicida. Further, unlike that of V. salmonicida, the luciferase reaction of V. fischeri MJ1 is not limited by the absence of aliphatic aldehyde (31).
To examine the genetic basis for its aliphatic-aldehyde deficiency and low levels of autoinduction, the luminescence gene cluster of V. salmonicida was cloned and sequenced. The arrangement and sequence of the structural lux genes are conserved compared to those in related species of luminous bacteria; however, V. salmonicida has both a novel arrangement and a different number of homologs of the luxR and luxI quorum-sensing regulatory genes. Transcriptional analysis suggested that this novel arrangement generates antisense transcripts that may be responsible for the reduced production of bioluminescence. Further, mutagenesis of V. salmonicida luxA resulted in marked attenuation of virulence of the mutant relative to that of the wild type in both single- and mixed-strain animal challenge experiments.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The bacterial strains used in this study are listed in Table 1. Escherichia coli DH5α, grown in Luria-Bertani (LB) medium (37) at 37°C, was the host for plasmids with ColE1 or pACYC184 origins of replication. When added to LB medium for the selection of E. coli cells carrying a plasmid, ampicillin, chloramphenicol (Cam), and kanamycin (Kan) were used at concentrations of 100, 30, and 50 μg/ml, respectively. V. salmonicida strains were originally isolated from diseased Atlantic salmon (8, 39). Unless indicated otherwise, the principal strain used in this study was V. salmonicida NCMB 2262T. V. salmonicida cultures were grown at 15°C with shaking at 150 rpm for 2 to 3 days in a complex broth (SWT) that contained 10 g of tryptone, 3 g of yeast extract, and 3 ml of glycerol per liter of 70% seawater (2). SWT blood agar contained, per liter of SWT broth, 15 g of agar and 50 ml of Alsevers sheep blood (Colorado Serum Co., Denver, CO). When added to SWT for the selection of V. salmonicida cells carrying a plasmid, Cam and Kan were used at 2 and 150 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Bacterial strains | ||
| NCMB 2262 | V. salmonicida type strain isolated from diseased salmon | 8 |
| EN3 | luxA insertion mutant containing pEN124 chromosomally integrated into NCMB 2262 (Camr Kanr) | This study |
| EN4 | luxA insertion mutant derived from EN3 (Kanr) | This study |
| Plasmids | ||
| pEVS79 | Derivative of pBS SK+ (Stratagene Inc.) (mob site; ColE1 ori) | 40 |
| pEVS104 | Helper plasmid with tra and trb genes (Camr) | 40 |
| pVO8 | Vector plasmid with pACYC184 ori (Camr Ermr) | 47 |
| pKV17 | Derivative of pHV200 (16); 7.9-kbp SalI fragment encoding the V. fischeri ES114 lux region (ΔluxA) | 45 |
| pEN114 | 10.8-kbp SalI fragment with a portion of the V. salmonicida lux operon ligated into pEVS79 | This study |
| pEN115 | V. salmonicida luxAB PCR product ligated into pCR2.1 (Invitrogen Inc.) | This study |
| pEN123 | V. salmonicida luxAB PCR product ligated into pEVS79 | This study |
| pEN124 | pEN123 with a transposon (Kanr) inserted 987 bp downstream of the start of the V. salmonicida luxA ORF | This study |
| pEN133 | SalI fragment with V. salmonicida lux sequence 5′ to luxC ligated into pEVS79 | This study |
| pEN134 | BamHI fragment from pEN114 (luxDABEG) ligated into pV08 | This study |
| pEN135 | V. fischeri ES114 luxCDBEG (ΔluxA) from pHV200 in frame with the lacZ promoter on pVO8 | This study |
Camr, chloramphenicol resistance; Ermr, erythromycin resistance; Kanr, kanamycin resistance.
Triparental mating for V. salmonicida.
We developed a version of the triparental mating procedure described by Valla et al. (44), adjusted for the differences in the optimal growth temperatures of E. coli and V. salmonicida (8). The transfer of plasmids to V. salmonicida was performed using pEVS104 as a helper plasmid (40) contained in E. coli DH5α. E. coli and V. salmonicida strains were grown to an optical density (OD) at 600 nm of between 0.5 and 0.8 in LB (37°C) and SWT (15°C) broths, respectively. The cells in 1 ml of each culture were pelleted, washed three times in chilled (4°C) SWT, and resuspended in 5 μl of chilled SWT. The resuspended cells were combined, spotted onto a chilled SWT plate, and placed in a 23°C incubator for 6 h. The plate was subsequently incubated at 15°C for another 12 h. The resulting confluent growth of cells was scraped off the plate, resuspended in 1 ml of chilled SWT broth, and incubated with shaking at 150 rpm for 12 h at 15°C. Following the incubation, the suspension was plated onto antibiotic-containing SWT blood agar plates. After 10 days of growth, colonies of V. salmonicida transconjugates were streaked for purification.
Molecular manipulation of the lux region. (i) Cloning and sequencing of the lux gene cluster.
Using standard PCR methods with consensus primers for the luxAB region (forward, O-LUXDFP2 [5′-CATGTCATTCGCTA-3′], and reverse, [O-LUXDRP1 5′-AGATAAGATCATCA-3′]), we generated a PCR product that was cloned into the TA cloning vector (Invitrogen, Carlsbad, CA) to make the luxAB plasmid pEN115 (Table 1). A Southern probe analysis based on the internal luxAB sequence in pEN115 was used to screen a library of SalI genomic fragments of V. salmonicida cloned into the vector pEVS79 for luxAB-positive clones. One such clone, pEN114, was isolated and sequenced. Because pEN114 lacked the region upstream of luxC, we subcloned the luxAB fragment from pEN114 into pEVS79 to make pEN123 and marked luxAB by using the in vitro EZ::TN <KAN-2> insertion kit (Epicenter Technologies Inc., Madison, WI) to generate pEN124. pEN124 contained a transposon (Kanr) insertion near the middle of the luxAB fragment (886 bp into the luxA open reading frame [ORF]); this transposon has transcriptional terminators at each end. The marked copy of luxAB was introduced into the genome of V. salmonicida by triparental mating, and the single recombinant, EN3, was selected by sequentially patching colonies onto SWT-Kan, SWT-Cam, and nonselective SWT blood plates. Genomic DNA from EN3 was purified; digested with SacI, which cuts upstream of luxC; and ligated into the SacI site in pEVS79. A Kan-resistant clone harboring a pEVS79 derivative containing the marked copy of luxAB was found to contain a small portion of the genomic sequence upstream of luxC. Primers were designed from this upstream sequence and used to rescreen the V. salmonicida SalI library by PCR, resulting in the identification of the plasmid pEN133. The plasmid pEN133 was found to contain a large region immediately upstream of luxC, adjacent to the SalI fragment cloned into pEN114. pEN133 was sequenced by standard methods.
(ii) Screening for a double-recombinant luxA mutant.
Strain EN3 (Cam and Kan resistant) was grown without Cam selection in SWT. Approximately 10,000 colonies from this culture were screened (Cam sensitive, Kan resistant) for the loss of the integrated pEN124 plasmid by double recombination. Subsequent clones were screened by PCR for the integration of the Kanr marker. The resulting clone, EN4, was examined by sequencing and Southern blot analysis to confirm that a single integration of the Kan resistance marker had occurred in luxA.
(iii) Complementing the luxA mutation.
pEN114 was digested with BamHI, and the luxDABEG fragment was cloned into the BamHI site of pV08. The resulting plasmid (pEN134) and the parent vector were separately moved into the luxA mutant EN4 and the wild-type parent by triparental mating.
DNA sequence analysis of lux regions.
Stem-loop structures in lux gene clusters of several bacterial species were identified using DNA Strider V1.2. The Gibbs free-energy (ΔG) value for each stem-loop was calculated in units of kilocalories per mole by using the program Mfold (GCG, Madison, WI). ΔG values were calculated for the optimal growth temperatures for the following bacterial species: V. salmonicida (15°C), V. fischeri MJ1 (30°C), V. harveyi (35°C), and P. leiognathi (35°C).
PCR analysis of the lux region arrangement in five V. salmonicida strains.
PCR was used to putatively identify and map the luxR1, luxR2, and luxI regions (Fig. 1) of V. salmonicida strains isolated from independent sources (Table 1). PCRs were performed with genomic DNA by using the primer pairs O-EN11 (5′-GCCAGATCAAATGTTTGCTG-3′) and O-EN20 (5′-GTCACTTGGCTACCGCTCG-3′); O-EN26 (5′-TAAATGAGTTGAGCCACG-3′) and O-EN23 (5′-CTCCATCGCTGTCCAACCG-3′); and O-PEN115M13R1F1 (5′-GTAAATACATGAATGAGC-3′) and O-EN14 (5′-CCAAAATACTCCATTGCGAG-3′). These three primer pairs spanned the regions between luxR1 and luxC, luxE and luxR2, and luxR2 and ribG, respectively.
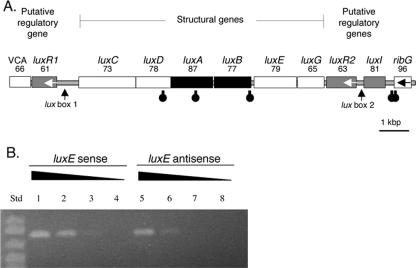
FIG. 1.
Organization of bacterial lux genes in V. salmonicida (Vs) and V. fischeri (Vf). VCA represents homologs of the genes for the V. cholerae VCA0181 protein (21) and the V. fischeri VFA0926 protein (36). ribG is a riboflavin synthesis gene (24). Unless indicated otherwise by an arrow, ORFs are predicted to be transcribed from left to right. Black and gray highlighting denotes ORFs corresponding to luciferase genes and lux regulatory genes, respectively (27, 29).
Genetic complementation of the aldehyde deficiency.
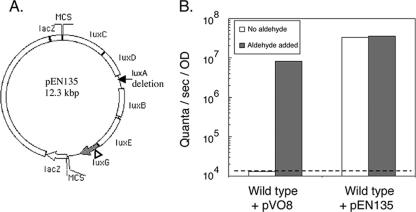
A derivative of V. salmonicida NCMB 2262T that harbored pEN135, which constitutively expressed the V. fischeri ES114 aliphatic-aldehyde synthesis (AAS) genes, luxCDE, was constructed. To construct the plasmid pEN135, the V. fischeri lux gene region containing luxRICDBEG (luxA was previously deleted) was excised from pKV17 (45) by using SalI and ligated into the V. fischeri cloning vector pVO8 (47). The luxR and luxI genes were removed from the resulting plasmid by KpnI and BglII double digestion, followed by ligation of the digest with a double-stranded oligonucleotide linker that contained KpnI- and BglII-complementing ends; a SalI site was in the middle of the linker. E. coli DH5α was transformed with the resulting product, and transformants were selected on LB-Cam medium. A plasmid that carried the V. fischeri luxCDBEG genes under the control of the lacZ promoter was isolated (see Fig. 2A). The in vitro EZ::TN <KAN-2> insertion kit (Epicenter Technologies Inc., Madison, WI) was used to create a null insertional mutation located 302 bp into the luxG ORF, resulting in pEN135. The pEN135 plasmid was mated into V. salmonicida in a triparental mating as described above.
FIG. 2.
Bioinformatic and RT-PCR transcriptional analysis of the V. salmonicida lux region motifs. (A) The putative direction of transcription is from left to right unless indicated otherwise by an arrow. Black and gray highlighting denotes ORFs corresponding to luciferase genes and lux regulatory genes, respectively. The number listed below each gene name is the percentage of identity between the predicted amino acid sequence for the ORF and the sequence of the equivalent lux protein in V. fischeri strain MJ1. Stem-loop symbols below the ORFs represent the approximate locations of putative rho-independent transcriptional terminators. VCA is a homolog of the gene for the VCA0181 protein, a V. cholerae hypothetical protein with homologs found in both V. salmonicida and V. fischeri. (B) RT-PCR products of sense (lanes 1 to 4) and antisense (lanes 5 to 8) transcripts of luxE amplified from V. salmonicida total cellular RNA. The RNA extract was either undiluted (lanes 1 and 5), diluted 1:10 (lanes 2 and 6), diluted 1:100 (lanes 3 and 7), or diluted 1:1,000 (lanes 4 and 8). RT-PCR performed with no added RNA gave no product (data not shown). In lanes 1, 2, 5, and 6, a band matching the predicted 803-bp luxE product can be seen. Std, standard size markers; from top: 1,000, 900, 700, and 500 bp.
Measurement of bacterial culture luminescence.
Luminescence was measured with a TD-20/20 luminometer (Turner Designs Inc., Sunnyvale, CA). The luminescence of late-log-phase cultures (OD, 0.7 to 0.9) at the time of the assay both with and without the addition of decyl aldehyde (4.5-μg/ml final concentration) was measured.
Total RNA isolation.
V. salmonicida was streaked onto SWT agar and incubated for 4 days at 16°C. An isolated colony was inoculated into 10 ml of SWT broth and grown with shaking at 16°C to an OD of 0.5. Two milliliters of broth (containing about 2 × 108 cells) was pelleted, and total RNA was extracted from the pelleted cells by using the RNeasy kit (QIAGEN, Valencia, CA). The resulting total RNA fraction (about 1 μg) was mixed with 2 μg of DNase I in a 75-μl reaction mixture containing 30 mM Tris-HCl (pH 7.8), 50 mM NaCl, and 10 mM MgCl2 in nuclease-free water. The reaction mixture was incubated at 37°C for 1 h to digest any contaminating DNA. Following this incubation, total RNA was reisolated using the RNeasy protocol and quantified using a Biophotometer (Eppendorf, Hamburg, Germany). PCR was used to confirm that DNase I had removed DNA from the sample.
cDNA synthesis and reverse transcriptase PCR (RT-PCR) analysis.
The expression of lux genes was determined using the SuperScript II RNase H− reverse transcriptase kit (Invitrogen, Carlsbad, CA). The following components were combined in a first-strand cDNA synthesis reaction mixture: 2 pmol of sense or antisense gene-specific primers, 1 to 1,000 ng of total RNA, and each deoxynucleoside triphosphate to a concentration of 10 mM in a final volume of 12 μl. The cDNA was synthesized according to the manufacturer's protocol. For second-strand PCR, 1 μl of the resulting cDNA mixture was combined with 10× PCR buffer (200 mM Tris-HCl [pH 8.4] and 500 mM KCl), 2 mM MgCl2, deoxynucleoside triphosphates (250 μM [each]), gene-specific primers (10 μM [each]), and Taq polymerase to a final volume of 50 μl. The mixture was subjected to the following amplification conditions: 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 30 cycles, followed by a one-time final extension step at 72°C for 7 min. Gene-specific primers were as follows: luxE sense primer (CPO7), 5′-ATTTATGAGTACGCCACAAG-3′; luxE antisense primer (CPO8), 5′-GGTACTCGCTTTCTTTGAAA-3′; luxR2 sense primer (CPO11), 5′-ATATAACGGGTTCATTGCTC-3′; luxR2 antisense primer (CPO12), 5′-TGCCTACAAGAACTAACCAA-3′.
Preexperiment passage of V. salmonicida strains in fish and dosage titration.
An animal model was developed that controlled for variability due to water conditions (temperature and osmolarity), equitable feeding, and fish stock. The protocol was approved by the institutional review board for the ethical treatment of animals at the Norwegian School of Veterinary Science. Atlantic salmon fry used for the challenge averaged 50 g (wet weight) and were presmolts. Salmon were kept in 200-liter freshwater tanks at 6 to 7°C with standard oxygenation and a flow rate of 50 liters/h. Both the luxA mutant EN4 and the wild type NCMB 2262 were separately passaged for 2 days in Atlantic salmon fry to confirm that the strains had not lost pathogenicity during in vitro culture (49). Mutant and wild-type clones were recovered from their hosts by inserting a sterile probe into the head kidneys and streaking for single colonies. These passaged strains were tested in dosage titration experiments to find the optimum infective doses. A series of twofold dilutions of these fish-passaged strains (1 × 108 to 5 × 106 CFU per fish) was injected into the prepelvic abdominal regions of the fry. Mortality was monitored each day over a 25-day time course.
Single-strain challenge infection of Atlantic salmon.
Fish were injected in the abdominal cavity with 1 × 108 CFU of the wild type or the luxA mutant grown overnight in SWT broth. Fish were fin tagged according to the inoculum received and placed in a single tank. The tank was observed twice daily for dead fish. V. salmonicida does not survive in freshwater and does not transfer among individuals kept in the same tank (8, 39; unpublished data). Mortality was tracked over 1 month, and V. salmonicida cells were recovered from the head kidney of each dead fish by plating homogenates onto SWT agar. The identity of the infecting strain (wild type or mutant) was confirmed by sequential plating on SWT-Kan and then SWT agar plates.
Competition between luxA mutant and wild-type V. salmonicida strains infecting Atlantic salmon.
The fish-passaged luxA mutant EN4 and its wild-type parent strain were grown overnight in SWT. The two strains were combined in an approximately 1:1 ratio, and a total of 1.1 × 108 CFU per fish was injected into the abdominal cavities of 50 fish. Fish were fin tagged accordingly and placed in a single tank. The tank was observed twice daily for diseased and dead fish. Tissue from the head kidney of each dead fish was first streaked onto nonselective SWT plates for single colonies. Subsequently, colonies were patched onto SWT-Kan and SWT, and their differential growth was used to calculate the ratio of the mutant cells to the wild-type cells in the infected fish. A similar procedure was used in an in vitro competition experiment performed with SWT broth. Cultures were inoculated to an OD of 0.01 and grown to stationary phase.
Nucleotide sequence accession number.
The nucleotide sequence of the V. salmonicida lux gene cluster has been submitted to GenBank (accession no. AF452135).
RESULTS
The structural lux genes are conserved in V. salmonicida.
The order of the structural genes of the V. salmonicida lux operon is luxCDABEG, which agrees with the gene orientation in lux operons from other luminous bacterial species (Fig. 1). The amino acid sequences corresponding to these six genes from V. salmonicida and V. fischeri MJ1 are also highly conserved, with levels of identity ranging between 65 and 87% (Fig. 2A). In addition, there are no detectable deletions or insertions in this six-gene locus.
The V. salmonicida luxR and luxI homologs are arranged differently from those in the V. fischeri lux region.
In contrast to the conserved arrangement of the lux structural genes, homologs of the quorum-sensing regulatory genes luxR and luxI have a novel arrangement in V. salmonicida (Fig. 1) compared to V. fischeri (1, 17). There are two copies of the luxR homolog, one located upstream and one downstream of the structural gene cluster. These homologs were designated luxR1 and luxR2, and their predicted protein sequences showed 61 and 63% amino acid identity to the V. fischeri MJ1 luxR gene product, respectively (Fig. 2A). The luxR1 and luxR2 predicted protein sequences showed only 61% amino acid identity to each other, suggesting that there has been considerable divergence since the apparent gene duplication event. Like the luxR gene of V. fischeri, both luxR1 and luxR2 of V. salmonicida appear to be transcribed in a direction opposite to that of the structural lux genes. In addition, there is a luxI homolog adjacent to, but apparently divergent from, the luxR2 homolog (Fig. 2A). The product of this V. salmonicida luxI homolog showed 81% amino acid identity with the product of V. fischeri MJ1 luxI. Between the luxR1 and luxC V. salmonicida ORFs, the position where luxI is located in V. fischeri, are 559 bp of sequence that do not contain any apparent ORFs (Fig. 2A).
The arrangement of the V. salmonicida lux cluster is conserved among different strains.
PCR was used to determine whether the lux gene arrangement present in V. salmonicida strain NCMB 2262T was found in four other strains of this species. The three sets of PCRs (see Materials and Methods) covering the luxR1-luxC, luxE-luxR2, and luxR2-ribG regions all produced products of the predicted 1.6-kbp, 1.9-kbp, and 1.9-kbp lengths, respectively, for each of the five V. salmonicida strains (data not shown). These results suggest that the arrangement of the lux gene cluster in the regions amplified is conserved within V. salmonicida.
The aliphatic-aldehyde deficiency can be genetically complemented.
One explanation for the aliphatic-aldehyde deficiency and the reduced luminescence of V. salmonicida is that the metabolism of this species is unable to provide the substrate(s) required for the synthesis of aliphatic aldehyde. To examine this hypothesis, V. fischeri AAS genes were expressed in trans in V. salmonicida and the resulting luminescence per cell was determined. V. salmonicida, expressing in trans the AAS gene-carrying plasmid pEN135 (Fig. 3A), was detectably luminous and produced at least 1,200-fold more luminescence than the wild-type strain (Fig. 3B). The addition of exogenous aliphatic aldehyde did not result in a significant increase in luminescence in V. salmonicida cells harboring pEN135 (Fig. 3B). These data suggest that V. salmonicida is not physiologically limited in its ability to produce the substrates required for aliphatic-aldehyde synthesis. In addition, because V. salmonicida cells harboring pEN135 and those harboring the vector plasmid pV08 have the same growth rate (3.3 h per generation), the production of additional aliphatic aldehyde does not appear to be toxic to V. salmonicida. It is unlikely that the collateral presence of a copy of the V. fischeri luxB gene in the construct (Fig. 3A) is responsible for the enhanced luminescence expression observed because aldehyde addition alone increased the luminescence of wild-type cells by several orders of magnitude (Fig. 3B).
FIG. 3.
Genetic complementation of the aliphatic-aldehyde deficiency in wild-type V. salmonicida. (A) The plasmid used for complementation studies, pEN135, contains the V. fischeri ES114 AAS genes, luxCDE, as well as the luciferase subunit gene luxB. The other luciferase subunit gene, luxA, is deleted, and luxG (gray) is inactivated by a transposon insertion (triangle). MCS, multiple cloning site. (B) Comparison of luminescence produced by wild-type V. salmonicida harboring the vector plasmid pVO8 and that produced by wild-type V. salmonicida harboring the AAS gene-carrying plasmid pEN135. Aliquots of cultures (late exponential phase of growth; OD at 600 nm, 0.7 to 0.9) were either immediately measured photometrically or supplemented with aliphatic aldehyde before measurement. The dashed line represents the limit of photometric detection. Data shown are representative of the results of three independent experiments.
The intergenic regions of V. salmonicida lux genes share conserved elements with similar regions in V. fischeri.
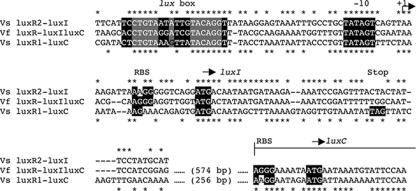
The intergenic regions between luxR1 and luxC and between luxR2 and luxI were analyzed for possible transcriptional promoter elements. The putative transcriptional initiation loci upstream of both luxC and luxI in V. salmonicida contain substantial similarities to the region upstream of luxI in V. fischeri MJ1 (Fig. 4). There are two putative regulatory elements, lux box 1 (between luxR1 and luxC) and lux box 2 (between luxR2 and luxI), in V. salmonicida. Both of these elements precede a −10 region that is identical to that found in V. fischeri MJ1 (Fig. 4). In addition, V. salmonicida lux box 1, V. salmonicida lux box 2, and the V. fischeri lux box are centered at −42.5, −43.5, and −42.5 bp, respectively, upstream of their predicted transcriptional start sites (6) (Fig. 4). Putative ribosomal binding sites and start codon loci, determined by sequence similarities, are conserved between V. fischeri MJ1 and V. salmonicida for luxI, luxC, luxA, luxB, and luxG (Fig. 4 and data not shown). However, unlike V. fischeri MJ1, V. salmonicida has no apparent start codon at the 5′ end of the luxD ORF.
FIG. 4.
Sequence comparison of the intergenic regions between luxR2 and luxI of V. salmonicida (Vs luxR2-luxI), luxR and luxIC of V. fischeri MJ1 (Vf luxR-luxIluxC), and luxR1 and luxC of V. salmonicida (Vs luxR1-luxC). The V. fischeri luxR-luxIC and V. salmonicida luxR1-luxC sequences have been extended into their luxC ORFs. Identical nucleotides in the V. salmonicida luxR2-luxI and V. fischeri luxR-luxIC sequences are indicated with asterisks above the alignment. Identical nucleotides in the V. fischeri luxR-luxIC and V. salmonicida luxR1-luxC sequences are indicated with asterisks below the alignment. The highlighted sequences are based on motifs conserved around V. fischeri lux genes (17). These regions include a lux box, a −10 promoter region, and known or putative ribosome binding sites (RBS). Start (ATG) and stop (TAG) codons are also highlighted. Gray boxes highlight sequences required for a functional lux box in V. fischeri MJ1 (6).
Stem-loop structures in the V. salmonicida lux region are conserved.
To assess how transcription of the V. salmonicida lux operon may be terminated, the sequence data were screened for stem-loop structures that may function as rho-independent transcriptional-termination factors. We identified three putative stem-loop structures in the lux gene cluster of V. salmonicida. These structures are located in the middle of the luxD and luxA coding regions and at the 3′ end of luxB (Fig. 2A). The sequence of the stem-loop at the luxB terminus in V. salmonicida (AAAAGAATGACAGAATTAACTCTGCCATTCTTTT) is similar to those in other luminous bacteria but was predicted to have greater thermostability (ΔG = −15 kcal/mol) than the equivalent structures in P. leiognathi (−12 kcal/mol), V. harveyi (−10 kcal/mol), and V. fischeri MJ1 (−5 kcal/mol). In V. salmonicida, there are two additional predicted stem-loops, between luxI and ribG (Fig. 2A). It was previously shown by reporter gene analysis that an equivalent stem-loop in V. fischeri, also located between the lux operon and ribG, is a bidirectional transcriptional terminator (42).
Sense and antisense transcripts of luxE and luxR2 are produced by V. salmonicida.
The organization of the V. salmonicida lux operon, coupled with its aldehyde-deficient luminescence physiology (13), suggested that a long transcript from the rightward luxR2 promoter (Fig. 2A) might exert antisense control over the expression of luxE, one of the genes required for aldehyde synthesis. In support of this hypothesis, when total RNA isolated from growing cells of V. salmonicida was mixed with primers to amplify either sense or antisense luxE or luxR2 transcripts by reverse transcriptase PCR, we detected sense and antisense transcripts of both genes (Fig. 2B; luxR2 data not shown). The relative amounts of the sense and antisense luxE products resulting from the semiquantitative RT-PCR suggested that the level of the sense transcripts was higher than that of the antisense transcripts.
Complementation of the luxA mutation.
Complementation of the luxA mutation in vitro was observed in the luxA mutant EN4 harboring the plasmid pEN134 compared to EN4 harboring the vector parent plasmid pV08. Specifically, in the presence of decyl aldehyde (10 μl/ml), EN4 harboring the plasmid pEN134 or pV08 produced 109 or <104 quanta/s/OD unit, respectively. In the absence of added decanal, an aliphatic aldehyde, EN4 harboring either pEN134 or pV08 produced 5 × 105 or <104 quanta/s/OD unit, respectively.
Atlantic salmon infected with a luxA mutant show delayed mortality.
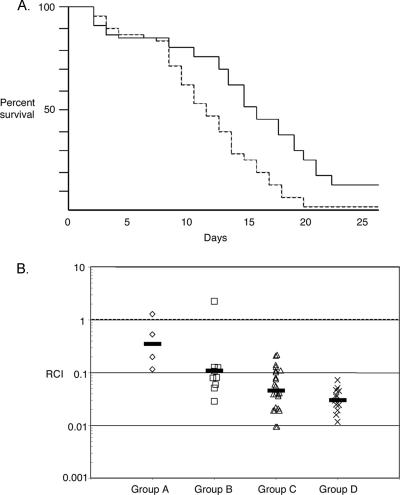
Dose titration data showed that inoculation with approximately 1 × 108 CFU of V. salmonicida produced a consistent level of mortality in salmon such that the criterion for a 50% lethal dose would be observed by approximately day 10 (data not shown). This dose is at the upper range of the 50% lethal doses determined previously (5 × 106 to 1 × 108 CFU) by Wiik et al. (49), perhaps because the fry we used were particularly robust. In the large-scale studies, each fish was injected with 1 × 108 CFU of either the luxA mutant (EN4) or its wild-type parent. Mortality was observed starting at day 2 and continued until day 25 (Fig. 5A). There was no evidence of cross contamination between infected animals: the postmortem examination of fish injected with wild-type cells did not reveal the presence of the mutant strain; the converse was also true. Dead fish did not display pathology on their external surfaces; however, their livers were atypically fatty, and points of hemorrhaging were observed in both the intestines and the kidneys. Levels of mortality in the two branches of the experiment were identical until day 9 (Fig. 5A), at which time the mutant showed a dramatic delay in its ability to kill fish compared to the wild type. The difference between the survival plots for the mutant and the wild type was statistically significant (log rank test, P = 0.0002; Wilcoxon test, P = 0.0015).
FIG. 5.
Data from fish virulence experiments comparing the luxA mutant EN4 and its wild-type parent. (A) Survival plot for Atlantic salmon fry infected with either the luxA mutant EN4 (solid line) or the wild type (dashed line). Mortality among the fish infected with the mutant was significantly delayed compared to that among the fish infected with the wild type (log rank test, P = 0.0002; Wilcoxon test, P = 0.0015). Inputs were 9.7 × 107 and 1 × 108 CFU per fish for the mutant and the wild type, respectively. (B) Competition experiment with the luxA mutant EN4 and the wild type. Fish were grouped according to the duration of infection before death (A, 1 to 5 days; B, 6 to 10 days; C, 11 to 15 days; and D, 16 to 20 days). The relative competitive index (RCI) of the luxA mutant for each fish was determined by dividing the output ratio for the two strains (mutant cells to wild-type cells) in the head kidney of the dead fish by the input ratio in the mixed inoculum. An RCI of less than 1.0 indicates that the wild type outcompetes the mutant. Each symbol on the graph represents the RCI calculated for one fish, and the geometric mean for each group is indicated by a bar.
The luxA mutant is outcompeted by the wild type during infection.
Cells of the luxA mutant EN4 and its wild-type parent were combined in approximately equal numbers (1.07:1), and a portion of this mixed inoculum was injected into the abdominal cavities of 50 fish. The final ratio of mutant cells to wild-type cells in the infection was assessed for each fish that died over the 25-day experiment. Mortality groupings were made according to the duration of infection as a method of identifying stages of the disease (group A, 1 to 5 days; group B, 6 to 10 days; group C, 11 to 15 days; and group D, 16 to 20 days). The mutant was attenuated approximately threefold during the first day postinoculation, and the competitive disadvantage continued throughout the experiment, with the competitive index decreasing from 0.35 to 0.11 to 0.05 to 0.03 for groups A to D, respectively (Fig. 5B). To determine whether there was a significant growth rate difference between the strains during growth in culture, mixed 1:1 inocula of mutant and wild-type cells (OD, 0.01) were added to SWT broth and replicate cultures were grown overnight. The ratios of mutant to wild-type cells at stationary phase ranged from 0.85 to 0.96.
DISCUSSION
We have characterized the luminescence gene cluster of V. salmonicida with the goal of understanding the mechanism and role of this organism's cryptic bioluminescence. To this end, we discovered a unique gene structure that includes an unusual arrangement of quorum-sensing genes. We also showed that a mutation in luxA could attenuate V. salmonicida pathogenesis. Although the nature of the association between lux gene expression and virulence remains unknown, the implications of uncovering a new class of virulence factors are significant. The introduction and development of V. salmonicida genetics now permits future investigators to explore virulence in what perhaps is the only truly natural vertebrate model system for studying pathogenesis in the genus Vibrio.
The prevalence of luciferase genes in Vibrionaceae species that do not produce detectable light has led to the question of what role, if any, there is for the activities encoded by these genes in nonluminescent bacteria. In at least some of these bacteria, the luciferase genes are expressed but the reaction catalyzed by their corresponding proteins is limited by the availability of the aliphatic-aldehyde substrate. The experimental addition of an aliphatic aldehyde results in detectable light emission from these cells (12, 13; unpublished results). Such cryptic luminescence in V. salmonicida has been described previously and may occur as a result of several possible explanations: (i) one (or more) of the genes encoding AAS enzymes is absent or nonfunctional; (ii) all the enzymes are synthesized, but the cell produces insufficient substrates for the AAS reaction; and/or (iii) the relative level of expression of the AAS genes is reduced.
The V. salmonicida lux operon contains all of the structural lux genes that are required for bioluminescence (Fig. 1). These genes, luxCDABE, are organized in the typical arrangement observed in other known lux operons. The V. salmonicida lux nucleotide sequences are most similar to those of its close relative, V. fischeri, and there are no detectable deletions or insertions in the structural lux genes. These data suggest that the aliphatic-aldehyde deficiency is not caused by the absence of the AAS genes.
We also determined that the synthesis of aliphatic aldehyde is not limited by the ability to provide substrates for AAS (Fig. 3B). The precursors for aliphatic aldehyde, namely, saturated long-chain fatty acids and reducing equivalents (5, 43, 48), are apparently readily available for AAS in V. salmonicida. Therefore, we explored the hypothesis that reduced or aberrant expression of the AAS genes may explain cryptic bioluminescence in V. salmonicida.
The transcription of bioluminescence genes is complex but has been well studied with other models. For example, the 218-bp intergenic region between the start codons for luxR and luxI in V. fischeri is the site where the transcription factors LitR (12) and LuxR (41) bind to divergently promote the transcription of luxR and luxI, respectively. Although the exact binding site for LitR is not known, LuxR, in complex with an acyl-homoserine lactone autoinducer, binds at a specific region called the lux box (7). The intergenic region between luxR1 and luxC in V. salmonicida also contains a conserved lux box and a −10 region that is identical to that of V. fischeri. In addition, there is a similar translational initiation region for a luxI gene (Fig. 4); however, there is no luxI homolog following this locus in V. salmonicida. Instead, the region between lux box 1 and luxC in V. salmonicida consists of only half the number of nucleotides found between the lux box and luxC in V. fischeri. We also detected evidence for a 2-bp frameshift that may result in a premature stop codon (TAG) (Fig. 4). Following this intergenic region in V. salmonicida, elements of the translational promoter for luxC are conserved between V. salmonicida and V. fischeri. Thus, the presence of the conserved transcriptional initiation elements suggests that the transcription of the V. salmonicida lux operon may be initiated in a fashion similar to that of the V. fischeri operon but that the first gene transcribed in V. salmonicida is the luxC homolog and not a luxI homolog. Strong stem-loop structures are predicted in the V. salmonicida lux gene cluster that are shared across bioluminescent taxa. The role that these stem-loops play in transcriptional modification remains unknown. Additional findings that V. salmonicida produces an as yet undescribed autoinducer (13; data not shown) and encodes a homolog of the V. fischeri luminescence regulatory gene litR (12; data not shown) suggest that V. fischeri and V. salmonicida share other genetic control mechanisms for bioluminescence expression. However, the natures of the expression differ.
Homologs of the luxR and luxI regulatory genes in V. salmonicida have a novel arrangement. First, there are luxR homologs located both upstream and downstream of the V. salmonicida structural lux genes (Fig. 1). Second, there is a luxI homolog downstream of luxR2. While this arrangement of luxR::luxI is found in V. fischeri as well, the location of the regulatory gene pair downstream rather than upstream of the structure genes is unique to V. salmonicida. In addition, the bidirectional transcriptional terminator at the end of luxG in other bioluminescent bacteria is decoupled from luxG in V. salmonicida but remains upstream of ribG. This novel genetic structure suggests a transcriptional model in which rightward-sense transcription from luxC to luxE may continue, generating antisense luxR2. Conversely, leftward-sense transcription from luxR2 may produce antisense luxE transcripts. RT-PCR detected both sense and antisense transcripts for luxE and luxR2. These data support the model that antisense gene regulation drives cryptic bioluminescence by reducing the expression of luxE and, therefore, AAS. Preliminary data suggest that a mutation in luxR2 delays the onset of peak bioluminescence but eventually produces a higher peak level than that for the wild type (data not shown). Further mutational and quantitative PCR analyses of each lux gene will be needed to test this model in which antisense RNA contributes to the cryptic bioluminescence phenotype of V. salmonicida.
Given the novelty of the arrangement of the luminescence gene cluster in V. salmonicida, is cryptic bioluminescence merely a remnant of an ancestral phenotype, or does it serve a current biological function? All five V. salmonicida strains tested appear to share the same lux gene cluster organization. The conservation of the arrangement within the species suggests that a function of the lux gene cluster may exist. To begin answering questions of functionality, we asked whether an insertional mutation in luxA would affect the virulence of V. salmonicida.
In a single-strain challenge experiment, the mutagenesis of luxA resulted in a marked delay in mortality among V. salmonicida-infected Atlantic salmon compared to that induced by the wild type. Similarly, in a mixed-strain competition experiment with the mutant and the wild type, the luxA mutant was attenuated 3- to 50-fold depending on the duration of infection. These data demonstrate that the disruption of luxA attenuates V. salmonicida colonization. However, the mechanism underlying this attenuation is not known. We hypothesize that the attenuation may be directly due to the loss of luciferase, which results in the elimination of the dark luciferase reaction. The dark luciferase reaction produces toxic oxygen radicals that may serve as a direct virulence agent or a stimulant for bacterial DNA repair (15, 22, 23, 25). Alternatively, the luciferase reaction may function as an alternative pathway to provide oxidized flavin at low oxygen tensions which may aid colonization if oxygen becomes limited (4, 26).
In the symbiosis of V. fischeri with the Hawaiian squid Euprymna scolopes (3), bioluminescence has been shown to be a colonization factor for the bacterium (46). At the cellular level, the luciferase reaction of V. fischeri is associated with symbiosis-induced host development including epithelial-cell swelling, as well as with the maintenance of persistent colonization of the host squid tissue (46). Thus, a similar, but nonbeneficial, role in pathophysiology may underlie the attenuation observed in the luxA mutant of V. salmonicida. In summary, the development of molecular tools for V. salmonicida, together with the creation of a natural salmon infection model, have led to a new system with which to evaluate the genetic structure, function, and evolution of bacterial bioluminescence in pathogenesis.
Acknowledgments
We thank S. Chang, M. McFall-Ngai, P. Patek, and C. Wimpee for critical evaluation and direction throughout this project. We thank K. Visick for providing pKV13 and E. Stabb for providing pEVS79 and pEVS104. We thank Stig Larsen for guidance on the statistical analysis.
This work was supported by grant number RR12294 from the National Center for Research Resources of the National Institutes of Health to E.G.R. and M. McFall-Ngai and by a National Science Foundation grant (IBN 9904601) to M. McFall-Ngai and E.G.R. A grant from the Norwegian Science Foundation (NFR 158882/I10) supported the virulence studies.
This work does not necessarily represent the official views of the funding agencies.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Baldwin, T. O., J. H. Devine, R. C. Heckel, J. W. Lin, and G. S. Shadel. 1989. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence. J. Biolumin. Chemilumin. 4:326-341. [DOI] [PubMed] [Google Scholar]
- 2.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boettcher, K. J., and E. G. Ruby. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgois, J. J., F. E. Sluse, F. Baguet, and J. Mallefet. 2001. Kinetics of light emission and oxygen consumption by bioluminescent bacteria. J. Bioenerg. Biomembr. 33:353-363. [DOI] [PubMed] [Google Scholar]
- 5.Byers, D., and E. Meighen. 1984. Vibrio harveyi aldehyde dehydrogenase. Partial reversal of aldehyde oxidation and its possible role in the reduction of fatty acids for the bioluminescence reaction. J. Biol. Chem. 259:7109-7114. [PubMed] [Google Scholar]
- 6.Callahan, S. M., and P. V. Dunlap. 2000. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 182:2811-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devine, J. H., G. S. Shadel, and T. O. Baldwin. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. USA 86:5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egidius, E., R. Wiik, K. Andersen, K. A. Hoff, and B. Hjeltnes. 1986. Vibrio salmonicida sp. nov., a new fish pathogen. Int. J. Syst. Bacteriol. 36:518-520. [Google Scholar]
- 9.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enger, O., B. Husevag, and J. Goksoyr. 1989. Presence of the fish pathogen Vibrio salmonicida in fish farm sediments. Appl. Environ. Microbiol. 55:2815-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enger, O., B. Husevag, and J. Goksoyr. 1991. Seasonal variation in presence of Vibrio salmonicida and total bacterial counts in Norwegian fish-farm water. Can. J. Microbiol. 37:618-623. [DOI] [PubMed] [Google Scholar]
- 12.Fidopiastis, P. M., C. M. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 13.Fidopiastis, P. M., H. Sørum, and E. G. Ruby. 1999. Cryptic luminescence in the cold-water fish pathogen Vibrio salmonicida. Arch. Microbiol. 171:205-209. [DOI] [PubMed] [Google Scholar]
- 14.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez-Flecha, B., and B. Demple. 1994. Intracellular generation of superoxide as a by-product of Vibrio harveyi luciferase expressed in Escherichia coli. J. Bacteriol. 176:2293-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray, K. M., and E. P. Greenberg. 1992. Sequencing and analysis of luxR and luxI, the luminescence regulatory genes from the squid light organ symbiont Vibrio fischeri ES114. Mol. Mar. Biol. Biotechnol. 1:414-419. [Google Scholar]
- 18.Hastings, J. W., and C. Balny. 1975. The oxygenated bacterial luciferase-flavin intermediate. Reaction products via the light and dark pathways. J. Biol. Chem. 250:7288-7293. [PubMed] [Google Scholar]
- 19.Hastings, J. W., C. J. Potrikus, S. C. Gupta, M. Kurfurst, and J. C. Makemson. 1985. Biochemistry and physiology of bioluminescent bacteria. Adv. Microb. Physiol. 26:235-291. [DOI] [PubMed] [Google Scholar]
- 20.Haygood, M. G. 1993. Light organ symbioses in fishes. Crit. Rev. Microbiol. 19:191-216. [DOI] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsev, A. M., G. Wegrzyn, and H. Szpilewska. 2004. Effects of hydrogen peroxide on light emission by various strains of marine luminescent bacteria. J. Basic Microbiol. 44:178-184. [DOI] [PubMed] [Google Scholar]
- 23.Koga, K., T. Harada, H. Shimizu, and K. Tanaka. 2005. Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol. Genet. Genomics 274:180-188. [DOI] [PubMed] [Google Scholar]
- 24.Lee, C. Y., R. B. Szittner, C. M. Miyamoto, and E. A. Meighen. 1993. The gene convergent to luxG in Vibrio fischeri codes for a protein related in sequence to RibG and deoxycytidylate deaminase. Biochim. Biophys. Acta 1143:337-339. [DOI] [PubMed] [Google Scholar]
- 25.Lyzen, R., and G. Wegrzyn. 2005. Sensitivity of dark mutants of various strains of luminescent bacteria to reactive oxygen species. Arch. Microbiol. 183:203-208. [DOI] [PubMed] [Google Scholar]
- 26.Makemson, J. C. 1986. Luciferase-dependent oxygen consumption by bioluminescent vibrios. J. Bacteriol. 165:461-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meighen, E. A. 1993. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 7:1016-1022. [DOI] [PubMed] [Google Scholar]
- 28.Meighen, E. A. 1991. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 55:123-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meighen, E. A., and P. V. Dunlap. 1993. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv. Microb. Physiol. 34:1-67. [DOI] [PubMed] [Google Scholar]
- 30.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 31.Nealson, K. H. 1977. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch. Microbiol. 112:73-79. [DOI] [PubMed] [Google Scholar]
- 32.Oliver, J. D., D. M. Roberts, V. K. White, M. A. Dry, and L. M. Simpson. 1986. Bioluminescence in a strain of the human pathogenic bacterium Vibrio vulnificus. Appl. Environ. Microbiol. 52:1209-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, L. M., and R. R. Colwell. 1991. Detection of luciferase gene sequence in nonluminescent Vibrio cholerae by colony hybridization and polymerase chain reaction. Appl. Environ. Microbiol. 57:1286-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramaiah, N., J. Chun, J. Ravel, W. L. Straube, R. T. Hill, and R. R. Colwell. 2000. Detection of luciferase gene sequences in nonluminescent bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 33:27-34. [DOI] [PubMed] [Google Scholar]
- 35.Ruby, E. G. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 50:591-624. [DOI] [PubMed] [Google Scholar]
- 36.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lostroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schmidt, T. M., K. Kopecky, and K. H. Nealson. 1989. Bioluminescence of the insect pathogen Xenorhabdus luminescens. Appl. Environ. Microbiol. 55:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sørum, H., T. T. Poppe, and O. Olsvik. 1988. Plasmids in Vibrio salmonicida isolated from salmonids with hemorrhagic syndrome (Hitra disease). J. Clin. Microbiol. 26:1679-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 41.Stevens, A. M., K. M. Dolan, and E. P. Greenberg. 1994. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA 91:12619-12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swartzman, A., S. Kapoor, A. F. Graham, and E. A. Meighen. 1990. A new Vibrio fischeri lux gene precedes a bidirectional termination site for the lux operon. J. Bacteriol. 172:6797-6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulitzur, S., and J. W. Hastings. 1978. Myristic acid stimulation of bacterial bioluminescence in “aldehyde” mutants. Proc. Natl. Acad. Sci. USA 75:266-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valla, S., K. Frydenlund, D. H. Coucheron, K. Haugan, B. Johansen, T. Jorgensen, G. Knudsen, and A. Strøm. 1992. Development of a gene transfer system for curing of plasmids in the marine fish pathogen Vibrio salmonicida. Appl. Environ. Microbiol. 58:1980-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visick, K. G., and E. G. Ruby. 1996. Construction and symbiotic competence of a luxA-deletion mutant of Vibrio fischeri. Gene 175:89-94. [DOI] [PubMed] [Google Scholar]
- 46.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Visick, K. L., and E. G. Ruby. 1997. New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri, p. 119-122. In J. W. Hastings, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence: molecular reporting with photons. John Wiley & Sons, Inc., Chichester, United Kingdom.
- 48.Wall, L. A., D. M. Byers, and E. A. Meighen. 1984. In vivo and in vitro acylation of polypeptides in Vibrio harveyi: identification of proteins involved in aldehyde production for bioluminescence. J. Bacteriol. 159:720-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiik, R., K. Andersen, F. L. Daae, and K. A. Hoff. 1989. Virulence studies based on plasmid profiles of the fish pathogen Vibrio salmonicida. Appl. Environ. Microbiol. 55:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiik, R., E. Stackebrandt, O. Valle, F. L. Daae, O. M. Rodseth, and K. Andersen. 1995. Classification of fish-pathogenic vibrios based on comparative 16S rRNA analysis. Int. J. Syst. Bacteriol. 45:421-428. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, T., and J. W. Hastings. 1998. Bioluminescence. Annu. Rev. Cell Dev. Biol. 14:197-230. [DOI] [PubMed] [Google Scholar]