Abstract
The compositions of archaeal and bacterial populations at different depths (60 m [mixolimnion-chemocline interface], 70 m [chemocline-subchemocline interface], 90 m, and 92 m [the water-sediment interface]) in the anoxic zone of the water column in Lake Pavin, a freshwater permanently stratified mountain lake in France, were determined. Phylogenetic trees were constructed from sequences to assess archaeal and bacterial diversity at the four sites.
Permanent anoxic layers in natural freshwater basins are rare and of considerable interest to microbial ecologists because of their potential undisturbed climax microbial communities and because of their relationship to an earlier biosphere. Lake Pavin in France provides such an environment. It is unusual because the water column has been stratified for a very long period and there has been a lack of mixing (meromixis) and its anoxic zone is in steady state (2). Despite its unique character, information on the distribution of microbial communities in the anoxic water column of Lake Pavin is limited to a terminal restriction fragment length polymorphism study of populations (16). Lehours et al. (16) found that the structures of both the bacterial and archaeal communities changed with depth. The results suggested that communities at interfaces played a predominant role in the water column. To obtain detailed phylogenetic information on the diverse populations in the differing anaerobic communities, 16S rRNA genes in samples collected at three interface layers in the anoxic water column of Lake Pavin were amplified, cloned, sequenced, and analyzed. Bacterial and archaeal clone libraries were also constructed from a sample collected at a depth of 90 m adjacent to the sediment to determine whether sediment fluxes influenced lake bottom community composition.
Sample collection and library construction.
Samples from depths of 60 m, 70 m, and 90 m in the water column of Lake Pavin were collected in August 2004 using an 8-liter horizontal Van Dorn bottle; samples were also collected from the sediment-water interface (Inter) at a depth of 92 m using a Jenkin-Mortimer multiple corer (21) (see reference 16 for site characteristics). Microbial samples were prepared on site from water samples (500 ml) by filtration through polycarbonate membrane filters (GTTP; Millipore) (47 mm diameter; pore size, 0.2 μm) and stored at −80°C. The sites sampled in the water column are shown in Fig. 1.
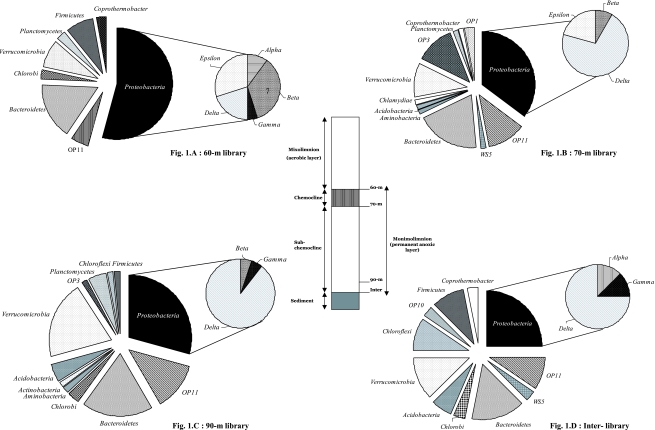
FIG. 1.
Frequencies of phylotypes affiliated with phylogenetic groups in libraries of samples collected from depths of 60 m (A), 70 m (B), and 90 m (C) and from the water-sediment interface (92 m) (D).
DNA was extracted as previously described (14). 16S rRNA genes were amplified using the archaean-specific 21f (5) and bacterium-specific 27f (8) forward primers and the universal primer 1492r (13). PCR products were cloned using a TOPO TA cloning kit according to the manufacturer's instructions (Invitrogen Corporation, San Diego, CA). Cloned inserts were PCR amplified using the M13 forward and reverse primers, and amplicons were digested with the restriction endonuclease HaeIII (Qbiogene). The banding patterns were grouped according to similarity, and plasmid DNA from a single representative of each unique restriction fragment length polymorphism pattern was isolated using a QIAprep plasmid purification kit (QIAGEN, Chatsworth, CA). Clones sequenced in one direction by MWG Biotech (Roissy CDG, France) yielded readable sequences of 800 bp on average. Because of possible sequence errors from PCR (27) or cloning (24), a conservative value of 97% sequence similarity was chosen for grouping into operational taxonomic units (OTUs). Clone libraries were screened for chimeric sequences with the Mallard program available at http://www.cardiff.ac.uk/biosi/research/biosoft/ (4), and 23 sequences identified as chimeras were excluded. The remaining 203 bacterial and 131 archaeal sequences were used in analyses.
Analyses of diversity and comparisons of libraries.
Calculations were performed on a personal computer with the freeware program aRarefactWin (S. Holland, University of Georgia, Athens). Shannon-Weiner index (H′) (11), Sorensen similarity index (Cs) (20), and SChao1 (the nonparametric Chao species richness estimator) (12) values were computed using EstimateS software version 7.5 (K. Colwell; http://purl.oclc.org/estimates). Coverage (C) and Margalef index (DMg) values were calculated as previously described (11, 22).
Rarefaction analyses showed that there was a broad diversity of bacteria in samples but less diversity of archaea (see Fig. S.1.a in the supplemental material). Coverage values for bacteria in all four libraries were found to be low (≤57%) (Table 1). This, and results from rarefaction curve analyses (see Fig. S.1.b in the supplemental material), indicated that a greater number of clones would have provided more robust information on bacterial diversity within sites. Our data showed that bacterial diversity increased with depth (Table 1) in agreement with findings from a previous terminal restriction fragment length polymorphism study of Lake Pavin populations (16). Coverage values for archaea were high (≥92%, Table 1) at all four sites, and rarefaction curves showed that asymptotes were almost reached (see Fig. S.1.b in the supplemental material). The diversity indices for archaea ranged from 0.8 to 1.4 for H′ and 0.8 to 1.6 for DMg, indicating low diversities at all sites (Table 1). The results with respect to composition of archaea did not differ greatly between sites (≥67%), whereas Sorensen similarity indices of the bacterial phylotypes were low, ranging from 11.1% to 27.3% (Table 2).
TABLE 1.
Properties of the distribution of phylotypes in clone libraries from the anoxic zone of Lake Pavina
| Sample source depth (m) | Results for archaea
|
Results for eubacteria
|
||||||
|---|---|---|---|---|---|---|---|---|
| % Coverage | H′ | Dmg | SChao1 | % Coverage | H′ | Dmg | SChao1 | |
| 60 | 92 | 1.2 | 1.6 | 6-12 | 43 | 3.1 | 6.9 | 48-153 |
| 70 | 94 | 1.4 | 1.4 | 6-20 | 57 | 3.6 | 10.0 | 67-152 |
| 90 | 95 | 1.4 | 1.4 | 6-14 | 45 | 3.6 | 10.3 | 100-269 |
| 92b | 97 | 0.8 | 0.8 | 4-4 | 25 | 3.3 | 7.8 | 52-155 |
H′, Shannon-Weiner index; Dmg, Margalef index; SChao1, nonparametric Chao species richness estimator.
The depth of 92 m represents the water-sediment interface.
TABLE 2.
Similarity matrix for the compositions of bacterial and archaeal sequences in samples collected at different depths in Lake Pavin
| Sample source depth (m) | Sorensen similarity index (%) for samples froma:
|
|||
|---|---|---|---|---|
| 60 m | 70 m | 90 m | 92 mb | |
| 60 | 14.4 (73) | 14.2 (91) | 14.5 (89) | |
| 70 | 18.3 (67) | 11.1 (80) | ||
| 90 | 27.3 (80) | |||
Values in parentheses are Sorensen index values for archaeal libraries; values without parentheses are those for bacterial libraries.
The depth of 92 m represents the water-sediment interface.
Phylogenetic analyses.
By the use of the phylogenetic software package ARB (19), neighbor-joining trees were constructed and partial sequences from clone libraries inserted while overall tree topology was retained. The robustness of inferred topologies was tested by bootstrap analysis using PHYLIP (PHYLIP [Phylogeny Inference Package] version 3.5c, 1993; J. Felsenstein, Department of Genetics, University of Washington, Seattle, WA) and 1,000 resamplings of trees. The sequences of the cloned inserts were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/). For accession numbers, see Fig. 2 and Fig. S.2.A to S.2.E in the supplemental material.
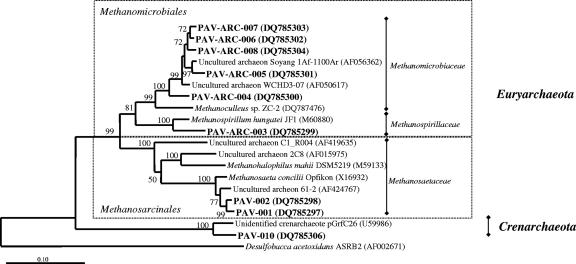
FIG. 2.
Neighbor-joining tree showing inferred phylogenetic relationships of 16S rRNA archaeal gene sequences cloned from the anoxic zone of Lake Pavin. Scale bars indicate Jukes-Cantor distances. Bootstrap values of >50 (for 1,000 iterations) are shown. GenBank accession numbers are given in parentheses.
Unusual archaeal phylotypes were not detected except for one OTU at the 70-m depth (Pav-Arc-010) affiliated with the phylum Crenarchaeota (see Table S.1 in the supplemental material). The archaeal communities were dominated by sequences related to methanogens belonging to the Methanosarcinales and the Methanomicrobiales (see Fig. 2 and Table S.1 in the supplemental material). This finding is consistent with the results of in situ hybridization analyses performed in a previous study of Lake Pavin water samples (16). Lehours et al. (16) also noted that methane concentrations in the anoxic zone of Lake Pavin correlated with detection of methanogens belonging to Methanosarcinales. In the present study, 71% of methanogen sequences were related (>95%) to Methanosaeta concilii (Fig. 2). We postulate that acetoclastic methanogenesis is an important process in the anoxic zone of the water column in Lake Pavin.
Bacterial sequences were found to align with 16 of the 52 phylogenetic divisions (see Table S.1 in the supplemental material). The majority of the 113 OTUs were most closely related to bacteria or clone-library sequences associated with Proteobacteria, Bacteroidetes, Verrucomicrobia, and candidate division OP11 (Fig. 1). Because of the wide diversity, the inferred relatedness of bacterial OTUs was examined in five separate phylogenetic trees (see Fig. S.2.A to S.2.E in the supplemental material). In the present study, many sequences were found to be only distantly related to previously cultivated organisms (see Table S.1 in the supplemental material). Nevertheless, some sequences were sufficiently related to known bacteria to enable reasonable hypotheses of function to be formulated. At the 60-m depth, three sequences were related (≥98.9%) to sequences from known methylotrophs, suggesting that some bacteria are involved in methane oxidation in the upper part of the chemocline. Three sequences (Pav-008) closely related (97.5%) to that of the microaerophilic iron-oxidizing bacterium Gallionella ferruginea (10) were retrieved from the 60-m library. Because high concentrations of ferric and ferrous iron have been detected in the monimolimnion of Lake Pavin (16, 25), it is likely that microaerophilic iron-oxidizing bacteria are involved in recycling ferrous iron in the chemocline in Lake Pavin.
In the present study, bacterial sequences were dominated by sequences from δ-proteobacteria; a number of bacteria in this subdivision are known to reduce Fe (III) (7). Dissimilatory Fe (III) reduction appears to be a significant biological process in systems containing high concentrations of ferrous iron (18). Three sequences (Pav-087) were found to be related (94.4% to 94.6%) to those of Geothrix fermentans, an Fe (III)-reducing bacterium isolated from a petroleum-contaminated aquifer (17). A feature of our study was the finding that many sequences (at least 40%) had their best matches with clone sequences obtained from contaminated sites and sediments (1, 3, 6, 9, 15, 23, 26; see Table S.1 in the supplemental material). This included archaeal sequences closely related (97%) to strains of Methanosaeta concilii recovered from hydrocarbon and chlorinated-solvent contaminated systems (Fig. 2). This apparent relationship was unexpected, because Lake Pavin is a mountain lake in a protected environment. We suggest that the link is ferric-iron reduction, a process often involved in detoxification processes (18).
Concluding remarks.
Because of prolonged meromixis, the anoxic zone of Lake Pavin represents an unusual microbial ecosystem. The anoxic communities have evolved since the formation of Lake Pavin 6,000 years ago, and there has been little influence from the external environment except for the sedimentation fluxes. Only 30% of the sequences of the bacterial OTUs retrieved in this study were more similar to sequences from isolated bacteria or published clone sequences than to those represented by the 93% genus proxy cutoff. It appears that the microbial communities which inhabit the anoxic zone of Lake Pavin are rich in new types of bacteria. Culture-dependent studies will be necessary to reveal their ecological roles.
Supplementary Material
Acknowledgments
We gratefully acknowledge the French Embassy in New Zealand, the New Zealand Ministry for Research Science and Technology, and the French Ministry for Foreign Affairs for financial support for scientist exchanges (G.F., K.J., and A.-C.L.).
We thank J. C. Romagoux and G. Demeure for their skilled technical assistance.
Footnotes
Published ahead of print on 19 January 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aeschbag-Hertig, W., M. Hofer, R. Kipfer, D. M. Imboden, and R. Wieler. 1999. Accumulation of mantle gas in a permanently stratified volcanic lake (Lac Pavin, France). Geochim. Cosmochim. Acta 63:3357-3372. [Google Scholar]
- 3.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Béjà, O., E. V. Koonin, L. Aravind, L. T. Taylor, H. Seitz, J. L. Stein, D. C. Bensen, R. A. Feldman, R. V. Swanson, and E. F. DeLong. 2002. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brofft, J. E., J. V. McArthur, and L. J. Shimkets. 2002. Recovery of novel bacterial diversity from a forested wetland impacted by reject coal. Environ. Microbiol. 4:764-769. [DOI] [PubMed] [Google Scholar]
- 7.Coleman, M. L., D. B. Hedrick, D. R. Lovley, D. C. White, and K. Pye. 1993. Reduction of Fe(III) in sediments by sulphate-reducing bacteria. Nature 361:436-438. [Google Scholar]
- 8.Cottrell, M. T., and D. L. Kirchman. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66:5116-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanert, H., H. 1991. The genus Gallionella, p. 4082-4088. In A. Ballows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. 4. Springer-Verlag, New York, NY. [Google Scholar]
- 11.Hill, A. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, J., B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jardillier, L., M. Basset, I. Domaizon, A. Belan, C. Amblard, M. Richardot, and D. Debroas. 2004. Bottom-up and top-down control of bacterial community composition in the euphotic zone of a reservoir. Aquat. Microb. Ecol. 35:259-273. [Google Scholar]
- 15.Joynt, J., M. Bischoff, R. Turco, and A. Konopka. 2006. Microbial community analysis of soils contaminated with lead, chromium and petroleum hydrocarbons. Microb. Ecol. 51:209-219. [DOI] [PubMed] [Google Scholar]
- 16.Lehours, A.-C., C. Bardot, A. Thénot, D. Debroas, and G. Fonty. 2005. Anaerobic microbial communities in Lake Pavin, a unique meromictic lake in France. Appl. Environ. Microbiol. 71:7389-7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. P. Phillips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe (III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovley, D. R. 2006. Dissimilatory Fe(III) and Mn(IV) reducing prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 2. Ecophysiology and biochemistry. Springer, New York, NY.
- 19.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, et al. 2004. ARB: a software environment for sequence data. Nucleic Acid Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 21.Mallet, C., and D. Debroas. 2000. Regulation of aminopeptidase activity in the sediment of a eutrophic lake. Arch. Hydrobiol. 149:327-335. [Google Scholar]
- 22.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 23.Schlötelburg, C., F. Von Wintzingerode, R. Hauck, W. Hegemann, and U. B. Göbel. 2000. Bacteria of an anaerobic 1,2-dichloropropane-dechlorinating mixed culture are phylogenetically related to those of other anaerobic dechlorinating consortia. Int. J. Syst. Evol. Microbiol. 50:1505-1511. [DOI] [PubMed] [Google Scholar]
- 24.Speksnijder, A. G. C. L., G. A. Kowalchuck, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viollier, E., G. Michard, D. Jézéquel, M. Pèpe, and G. Sarazin. 1997. Geochemical study of a crater lake: Lake Pavin, Puy de Dôme, France. Constraints afforded by the particulate matter distribution in the element cycling within the lake. Chem. Geol. 142:225-241. [Google Scholar]
- 26.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Wintzingerode, F., U. B. Göbel, and E. Staeckebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.