Abstract
We describe a new degenerate real-time PCR approach to simultaneously quantify phylogenetically different butyrate-producing bacteria based on the detection of butyryl-coenzyme A (CoA) CoA transferase genes. This pathway is present in numerically important groups of butyrate producers within the human colon, and thus this assay estimates the butyrate-producing ability of the microbiota.
Butyrate-producing bacteria within the human large intestine are of increasing interest because of their potential involvement in maintaining colonic health (12, 15). The ability of the gut microbiota to produce butyrate can vary considerably in response to environmental factors, as has been shown in vitro in response to pH (16) and in vivo in response to diet (9, 13, 14). Two alternative pathways for butyrate formation have been described in rumen bacteria (8). A range of human butyrate-producing strains belonging mainly to clostridial clusters XIVa and IV (7) were screened for the two pathways (11). Only 4 of 38 strains carried the butyrate kinase/phosphotransbutyrylase pathway for butyrate formation, indicating that the majority of human colonic butyrate producers use butyryl-coenzyme A (CoA) CoA transferase for the last step of butyrate formation. The corresponding gene has now been identified from several human gut bacteria (6). Crucially, all of the strains tested that are related to the Roseburia-Eubacterium rectale cluster and to Faecalibacterium prausnitzii, which are believed to be the numerically important butyrate producers (2, 10), carried the butyryl-CoA CoA transferase gene (6; unpublished data). This indicates that the butyryl-CoA CoA transferase gene is a good marker gene to target the main butyrate-producing bacteria in the human colon. It has been difficult to estimate the number of butyrate-producing bacteria in complex fecal samples by targeting the 16S rRNA gene, as these bacteria do not form a homogeneous phylogenetic group, and often butyrate producers and non-butyrate producers are found within the same phylogenetic clusters (12). Therefore, numerous different groups of bacteria have to be quantified. The concentration of butyrate in fecal samples may reflect the relative abundance of butyrate-producing bacteria (9) but will be strongly influenced by butyrate uptake by the host. The aim of the present study was to develop a semiquantitative assay for the detection of the butyryl-CoA CoA transferase gene in complex samples in order to estimate the butyrate-producing ability of fecal microbiota with a single assay that can be applied in high throughput.
Design of degenerate primers and optimization of real-time PCR conditions for detection of the butyryl-CoA CoA transferase gene.
Deduced amino acid sequences of butyryl-CoA CoA transferase genes from Roseburia hominis A2-183 (accession no. AAX19660), Anaerostipes caccae L1-92 (accession no. ABA39273), Eubacterium hallii L2-7 (accession no. AAZ23219), and Faecalibacterium prausnitzii A2-165 (accession no. AAZ23220); 4-hydroxybutyrate CoA transferase genes from A. caccae L1-92 (accession no. ABA39275), Clostridium kluyveri (accession no. P38942), C. tetani (accession no. NP_781174), and C. aminobutyricum (accession no. CAB60036); and a putative acetyl-CoA hydrolase sequence of Desulfitobacterium hafniense (accession no. BAE85155) were aligned and inspected for regions conserved in butyryl-CoA CoA transferases only. Several degenerate primers were designed and tested in real-time PCR experiments with the CoA transferase genes of R. hominis A2-183, A. caccae L1-92, E. hallii L2-7, and F. prausnitzii A2-165. Gene templates were amplified with gene-specific primers and purified with a QIAquick kit (QIAGEN). The DNA concentration was determined with a Nanodrop ND 1000 spectrophotometer (NanoDrop Technologies), and amplicons were diluted in 5 μg ml−1 herring sperm DNA (Promega) to 107, 106, 105, and 104 gene copies assuming an average molecular weight of 660 per nucleotide pair. Real-time PCR experiments were performed with an iCycler (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad) in a total volume of 25 μl in optical-grade 96-well plates sealed with optical sealing tape. Initial experiments revealed that the primer concentration had to be high in order to lead to good amplification, probably because of the degeneracy of the primers. Therefore, the primer concentration was routinely set to 2.5 μM per primer.
Primers BCoATscrF (GCIGAICATTTCACITGGAAYWSITGGCAYATG) and BCoATscrR (CCTGCCTTTGCAATRTCIACRAANGC) led to the best amplification of all four butyryl-CoA CoA transferase genes while not amplifying the A. caccae 4-hydroxybutyrate CoA transferase. An alignment of the primer binding sites with the gene sequences used for primer design is shown in Fig. 1. An annealing temperature of 53°C was found to be optimal, and an extension step at 72°C was included, as this primer pair amplified a region of approximately 530 bp. While long amplicons are generally regarded as suboptimal for real-time PCR experiments, targeting ideally suited sequences is more important for a degenerate PCR approach and we obtained good results with this primer pair despite the amplicon length (see below). The amplification cycle used was 1 cycle of 95°C for 3 min; 40 cycles of 95°C, 53°C, and 72°C for 30 s each with data acquisition at 72°C; 1 cycle each of 95°C and 55°C for 1 min; and a stepwise increase of the temperature from 55 to 95°C (at 10 s/0.5°C) to obtain melting curve data. Data were analyzed with the iCycler IQ software version 3.1. Real-time PCR amplifications for each of the four different butyryl-CoA CoA transferase amplicons with primers BCoATscrF and BcoATscrR resulted in PCR efficiencies of between 88.0 and 94.9%. The butyryl-CoA CoA transferase template from R. hominis A2-183 was used to generate standard curves in subsequent experiments, as this bacterium represents a dominant group of butyrate producers within the human gut microbiota (2).
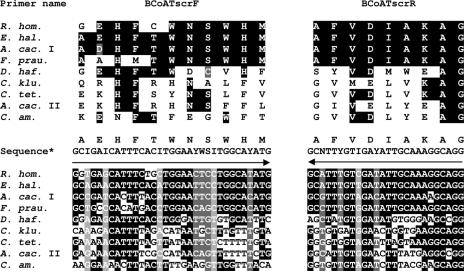
FIG. 1.
Nucleic acid and amino acid sequence alignments of primer binding sites of degenerate primers BCoATscrF and BCoATscrR. Butyryl-CoA CoA transferases: R. hom., R. hominis A2-183; E. hal., E. hallii L2-7; A. cac. I, A. caccae L1-92; F. prau., F. prausnitzii A2-165. Putative acetyl-CoA hydrolase: D. haf., D. hafniense. 4-Hydroxybutyrate CoA transferases: C. klu., C. kluyveri; C. tet., C. tetani; A. cac. II, A. caccae L1-92; C. am., C. aminobutyricum (for accession numbers, see the text). Primer binding sites correspond to nucleotide positions 217 to 249 for primer BCoATscrF and 748 to 773 for primer BCoATscrR of the butyryl-CoA CoA transferase gene from R. hominis A2-183. (*) The sequence for primer BCoATscrR is given in reverse to enable comparison to the alignment. Amino acids encoded by the primer sequence are indicated. Arrows indicate primer direction. Black shading indicates positions identical to the butyryl-CoA CoA transferase consensus, and gray shading indicates matches due to degeneracy of the primers. I, inosine; N, A, C, G or T; S, C or G; W, A or T; Y, T or C.
Amplification of butyryl-CoA CoA transferase from pure bacterial genomic DNA.
Genomic DNA isolated from a range of human gut bacteria with a DNeasy kit (QIAGEN) was amplified with the degenerate butyryl-CoA CoA transferase primers. All of the bacteria used that are not available from strain collections have been previously described (3, 4, 11). The copy number detected for 12 cluster XIVa bacteria carrying the butyryl-CoA CoA transferase gene (R. hominis A2-183, R. intestinalis L1-82, R. faecis M72/1, R. inulinivorans A2-194, E. rectale A1-86, Butyrivibrio fibrisolvens 16.4, A. caccae L1-92, E. hallii L2-7, E. hallii SM6/1, isolate SSC/2, isolate GM2/1, and isolate M62/1) was between 1.3 × 104 and 3.4 × 105 copies ng DNA−1, except for isolate GM2/1 (7.9 × 102 copies ng DNA−1). Cluster XIVa strains not carrying this gene (Coprococcus sp. strain L2-50, isolate A2-162, and isolate A2-232) were 2 to 4 logarithmic units lower (3.3 × 101 to 2 × 102 copies ng DNA−1). Of the three F. prausnitzii-related strains tested, A2-165 showed the best amplification with 4.5 × 104 copies ng DNA−1, while strains M21/2 and L2-6 were amplified at 5.3 × 102 and 2.2 × 103 copies ng DNA−1, respectively.
All other strains tested as negative controls (cluster IV isolate L2-63, Bacteroides thetaiotaomicron DSM 2079, and Bifidobacterium adolescentis DSM 20083), as well as an isolate related to E. cylindroides, T2-87 (cluster XVI), were amplified at less than 102 copies ng DNA−1. This was in the same range as the negative control and is mostly due to primer dimer formation, as could be determined from melting curves. It was shown previously that isolate T2-87 does not seem to carry the butyrate kinase pathway (11), and it also did not reveal a PCR product with degenerate CoA transferase primers described previously (6; data for T2-87 unpublished). Its mode of butyrate formation therefore remains to be resolved, and the existence of an unrelated CoA transferase in this strain cannot be ruled out. Otherwise, quantitative differences in amplification between different bacteria may be partly due to differences in genome size. Furthermore, the primers were designed on the basis of the gene sequences from four bacteria, so PCR efficiency might not be optimal for some of the bacteria tested because of sequence differences between strains. Overall, the butyryl-CoA CoA transferase real-time PCR primers worked well with most cluster XIVa bacteria, while they somewhat underamplified some F. prausnitzii-related strains. Crucially, little cross-reaction with genomes of bacteria believed not to carry the CoA transferase gene was observed.
Application of the degenerate real-time PCR assay to human fecal samples.
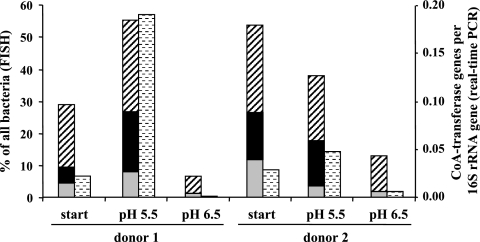
Complex samples from a previous fermentor-based study into the effect of pH on the human fecal microbiota, for which fluorescence in situ hybridization (FISH) quantification for several microbial groups was available (16), were used here to test the new degenerate PCR primers. DNA was isolated from the fermentor samples with a Fast DNA spin kit for soil (Qbiogene) and amplified under the PCR conditions described above. The number of 16S rRNA genes of all bacteria was determined with universal primers as described previously (5), in parallel with the butyryl-CoA CoA transferase gene quantification, and data are expressed as the number of CoA transferase genes detected per 16S rRNA gene. As different bacteria can carry different numbers of 16S rRNA genes (1), this ratio might reflect somewhat different bacterial numbers in different samples with their unique microbiota, and an exact comparison to the FISH data (expressed as a percentage of all bacteria) is not possible. However, the quantification of butyrate-producing bacterial groups by FISH mostly agreed well with the data obtained for the quantification of the butyryl-CoA CoA transferase gene (Fig. 2). Both methods revealed high numbers of butyrate producers at the low pH and very low numbers at the higher pH. Furthermore, the change observed between samples was much more pronounced in donor 1 than in donor 2 with both detection methods. It has to be kept in mind that the FISH probes used by Walker et al. (16), while detecting the two major groups of butyrate-producing bacteria commonly found in human volunteers (Roseburia spp. and E. rectale from clostridial cluster XIVa and F. prausnitzii from cluster IV), did not include butyrate-producing bacteria belonging to other phylogenetic groups (12) that might also use the butyryl-CoA CoA transferase route. While the cluster XIVa group can be enumerated by FISH, this group consists of both butyrate producers and non-butyrate producers. Therefore, the available FISH probes only estimate the abundance of certain groups of butyrate-producing bacteria, and a semiquantitative assay targeting a functional gene present in different phylogenetic groups of butyrate producers provides a potentially more comprehensive approach for detecting this functional group. In agreement with the results from the degenerate butyryl-CoA CoA transferase screen, butyrate concentrations in the pH 5.5 fermentor samples were high (26.3 mM for donor 1 and 23.5 mM for donor 2) (16) compared to the pH 6.5 samples (6.3 mM for donor 1 and 4.9 mM for donor 2) (16).
FIG. 2.
Quantification of butyryl-CoA CoA transferase genes within samples from fermentors inoculated with human fecal slurries (start) from two donors grown at two pH values and in the presence of 0.6% peptide as described by Walker et al. (16). Data for quantification of bacterial groups based on FISH are taken from reference 16. Gray bars, F. prausnitzii group; black bars, Roseburia spp. and E. rectale group; hatched bars, other clostridial cluster XIVa bacteria; stippled bars, results of quantification by real-time PCR, expressed as the number of CoA transferase genes detected per 16S rRNA gene.
In conclusion, the assay developed here will facilitate the estimation of the butyrate-producing ability of the microbiota present in human gut samples. This semiquantitative method can be used in conjunction with other techniques but will be particularly useful for screening large numbers of samples from dietary studies to obtain information on which samples are particularly interesting for a more thorough analysis by more laborious techniques such as FISH. For example, individuals could be identified with particularly high or low numbers of butyrate producers, or major changes in the microbiota could be monitored over time within the same individual. Samples can be stored frozen until processing, which is a major benefit compared to other approaches, such as direct measurement of butyryl-CoA CoA transferase activity. The primers described here might also be useful for other applications, such as RNA-based detection to facilitate analysis of the activity of butyrate-producing bacteria and phylogenetic analysis of butyryl-CoA CoA transferase-carrying strains from different environments. More generally, this work demonstrates the feasibility of using functionally relevant genes as targets for enumerating important groups of bacteria in complex microbial communities.
Acknowledgments
This work was supported by the Scottish Executive Environment and Rural Affairs Department.
We thank Kathleen Slezak, Sheila McCrae, and Carlett Ramirez for technical help and provision of genomic DNA.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Acinas, S. G., L. A. Marcelino, V. Klepac-Ceraj, and M. F. Polz. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186:2629-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aminov, R. I., A. W. Walker, S. H. Duncan, H. J. Harmsen, G. W. Welling, and H. J. Flint. 2006. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl. Environ. Microbiol. 72:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcenilla, A. 1999. Ph.D. thesis. Robert Gordon University, Aberdeen, United Kingdom.
- 4.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belenguer, A., S. H. Duncan, A. G. Calder, G. Holtrop, P. Louis, G. E. Lobley, and H. J. Flint. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72:3593-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charrier, C., G. J. Duncan, M. D. Reid, G. J. Rucklidge, D. Henderson, P. Young, V. J. Russell, R. I. Aminov, H. J. Flint, and P. Louis. 2006. A novel class of CoA-transferase involved in short-chain fatty acid metabolism in butyrate-producing human colonic bacteria. Microbiology 152:179-185. [DOI] [PubMed] [Google Scholar]
- 7.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. E. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 8.Diez-Gonzalez, F., D. R. Bond, E. Jennings, and J. B. Russell. 1999. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch. Microbiol. 171:324-330. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, S. H., A. Belenguer, G. Holtrop, A. M. Johnstone, H. J. Flint, and G. E. Lobley. 2007. Reduced dietary intake of carbohydrate by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 73:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lay, C., L. Rigottier-Gois, K. Holmstrøm, M. Rajilic, E. E. Vaughan, W. M. de Vos, M. D. Collins, R. Thiel, P. Namsolleck, M. Blaut, and J. Doré. 2005. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71:4153-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis, P., S. H. Duncan, S. I. McCrae, J. Millar, M. S. Jackson, and H. J. Flint. 2004. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J. Bacteriol. 186:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryde, S. E., S. H. Duncan, G. L. Hold, C. S. Stewart, and H. J. Flint. 2002. The microbiology of butyrate formation in the human colon. FEMS Microbiol. Lett. 217:133-139. [DOI] [PubMed] [Google Scholar]
- 13.Schwiertz, A., U. Lehmann, G. Jacobasch, and M. Blaut. 2002. Influence of resistant starch on the SCFA production and cell counts of butyrate-producing Eubacterium spp. in the human intestine. J. Appl. Microbiol. 93:157-162. [DOI] [PubMed] [Google Scholar]
- 14.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 15.Wächtershäuser, A., and J. Stein. 2000. Rationale for the luminal provision of butyrate in intestinal diseases. Eur. J. Nutr. 39:164-171. [DOI] [PubMed] [Google Scholar]
- 16.Walker, A. W., S. H. Duncan, E. C. McWilliam Leitch, M. W. Child, and H. J. Flint. 2005. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl. Environ. Microbiol. 71:3692-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]