Abstract
A total of 2,245 extracts, derived from 449 marine fungi cultivated in five types of media, were screened against the C4 plant enzyme pyruvate phosphate dikinase (PPDK), a potential herbicide target. Extracts from several fungal isolates selectively inhibited PPDK. Bioassay-guided fractionation of one isolate led to the isolation of the known compound unguinol, which inhibited PPDK with a 50% inhibitory concentration of 42.3 ± 0.8 μM. Further kinetic analysis revealed that unguinol was a mixed noncompetitive inhibitor of PPDK with respect to the substrates pyruvate and ATP and an uncompetitive inhibitor of PPDK with respect to phosphate. Unguinol had deleterious effects on a model C4 plant but no effect on a model C3 plant. These results indicate that unguinol inhibits PPDK via a novel mechanism of action which also translates to an herbicidal effect on whole plants.
Microorganisms have historically been a rich source of leads for pharmaceutical development, particularly for antibiotics. Over 20,000 microbial metabolites have been described, with most isolated from the terrestrial environment (21). The interface between marine and terrestrial environments is not impenetrable to microorganisms, and there is significant overlap between the microbially biodiverse populations of these two ecosystems (15). It is likely that many species, especially obligate associates of marine macroorganisms, are unique to the marine biosphere. In the increasing global research effort into marine microorganisms, those pursuing novel bioactivity endeavor to access novel biodiversity and obtain a secure and sustainable supply of bioactive metabolites. Microorganisms also enable controlled manipulation of their chemical diversity by exploiting their metabolic responses to different culture conditions (30, 34).
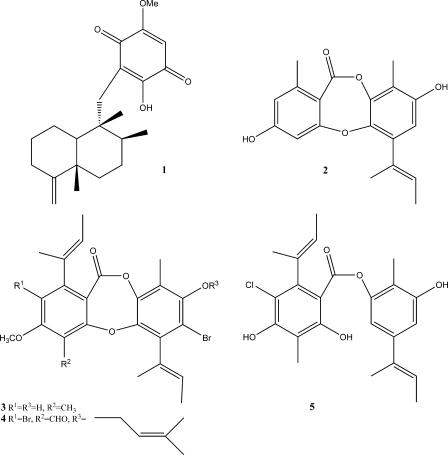
Decreasing chemical heterogeneity of herbicides targeting fewer mechanisms of action is increasing the prevalence of herbicide resistance (7, 22, 42). Inhibition of pyruvate phosphate dikinase (PPDK) significantly hinders C4 plant growth (26). PPDK has long been recognized as a potential, but as yet unused, biochemical target for herbicide development (9, 19, 20, 28), as C4 plants comprise most of the world's weeds (18). This enzyme occurs primarily in plants but has been found in protozoans (4, 27) such as Giardia (5), yet it is not detected in vertebrate or invertebrate animals, potentially minimizing the risk of PPDK inhibitors exhibiting adverse toxicological effects. Recently, we discovered marine macroorganism-derived extracts that selectively inhibited PPDK (8). From these, ilimaquinone (Fig. 1, structure 1) was isolated and found to inhibit PPDK as well as to be selectively toxic to C4 plants (16). Here we describe screening a collection of marine-derived fungi against PPDK and the reliable scale-up production of a PPDK-selective inhibitor.
FIG. 1.
Ilimaquinone (1), unguinol (2), acarogobien A (3), acarogobien B (4), and guisinol (5).
MATERIALS AND METHODS
Reagents.
Phosphoenolpyruvate carboxylase (EC 4.1.1.31) was purified from maize leaves, recombinant maize PPDK (EC 2.7.9.1) was expressed (8), and NAD-malate dehydrogenase (EC 1.1.1.37) was from Roche Diagnostics (Mannheim, Germany). Nufarm (Melbourne, Australia) supplied the herbicide formulation Uptake.
Marine fungus growth and preparation for primary screening.
The Australian Institute of Marine Science (AIMS) houses a collection of marine-derived fungi (2). Fungal isolates (n = 449) were streaked onto solid malt extract agar and incubated at 25°C until confluent. Each isolate was cultivated in 10 ml of five different types of medium. Media were as follows (all concentrations are in g liter−1 in artificial seawater unless otherwise stated): high-nutrient medium consisted of dextrose 10, malt extract 10, yeast extract 4, unbuffered; low-nutrient medium consisted of dextrose 2, malt extract 0.2, yeast extract 0.1, unbuffered; high-pH (pH 9.5) medium consisted of dextrose 2, malt extract 2, yeast extract 1; low-pH (pH 3.5) medium consisted of dextrose 2, malt extract 2, yeast extract 1; and no-salt medium consisted of dextrose 2, malt extract 2, yeast extract 1, unbuffered in deionized and sterile water. Isolates were incubated for 8 days at 27°C in a shaking incubator at 100 rpm. Microbial cells were lysed by three consecutive freeze-thaw cycles, and the entire broth was lyophilized. The broth was extracted overnight with 10 ml ethanol (EtOH), clarified by centrifugation and decanting prior to solvent evaporation, and reconstituted in 1 ml dimethyl sulfoxide (DMSO) for bioassay (original screening extract).
Fungi whose extracts, when retested, reproduced the primary assay results were recultivated and extracted as described above to determine the reproducibility of bioactivity.
Fungal growth scale-up.
Fungal isolates with identified reproducible activity were cultivated in 3 × 250 ml of the medium that elicited the initial bioactivity. After incubation (8 days shaking at 100 rpm at 27°C) cells were lysed as described previously. The entire broth was lyophilized and extracted overnight with 250 ml EtOH, filtered using glass wool, and the solvent evaporated under vacuum. The dried EtOH extract was reconstituted in 10 ml DMSO for bioassay. The amount used for the bioassay was proportional to that in the original screening extract. For nuclear magnetic resonance (NMR), EtOH extracts were dissolved in 10 ml EtOH and partitioned twice with H2O (100 ml) and dichloromethane (90 ml). Both the dichloromethane and H2O extracts were tested for PPDK inhibition at the same relative concentration as the original screening extract.
One bioactive isolate (F3000054) was grown in 7 × 1 liter of no-salt medium to provide sufficient biomass. Cultures were shaken (100 rpm) at 27°C for 8 days prior to the addition of equal volumes of ethyl acetate (EtOAc), stirring for 24 h, and decanting of the EtOAc layers. This EtOAc extraction was repeated twice, and the combined EtOAc extracts were evaporated under vacuum. Aqueous extracts were combined, frozen, and lyophilized. Both extracts were tested for PPDK inhibition at the same concentration as the original screening extract.
Isolation and identification of the bioactive metabolite.
A 1.0-g portion of the large-scale EtOAc extract was chromatographed on Sephadex LH20 (35 × 3 cm) with methanol (MeOH) to give 18 fractions. Fine white needles (81.7 mg, 8.1%) were crystallized from the active fraction. NMR spectra were acquired on a Bruker AC 300 spectrometer operating at 300 MHz for 1H and 75 MHz for 13C in methanol-d4 (δH 3.31 and δC 49.0, respectively) and acetone-d6 (δH 2.00 and δC 30.0 and 205.0, respectively). Electrospray ionization mass spectrometry (ESI MS) was done using an Esquire3000plus quadrupole ion trap with an Apollo source at 40 eV.
Genomic extraction and molecular taxonomy.
DNA was extracted from a 10-ml culture of the bioactive fungal isolate F3000054 by use of the hexadecyltrimethyl ammonium bromide procedure (36). Phylogenetic identification was performed by PCR amplification (GeneAmp 9700 temperature cycler; Perkin Elmer Cetus, Shelton, CT) and sequencing of near-complete 18S rRNA gene and internal transcribed spacer 1 (ITS1) regions of the fungal isolate. Eukaryote primers Ns1F/Ns8R were used for PCR amplification of the 18S rRNA gene and primers ITS1/ITS4 used for amplification of the ITS1 region (41). Sequencing was performed with a Dynamic ET dye sequencing kit (Amersham Pharmacia Biotech, Piscataway, NJ), and reactions were analyzed on a MegaBACE DNA analysis system (Amersham Pharmacia Biotech, Piscataway, NJ) following standard methods. A complete 18S ribosomal DNA (rDNA) sequence of F3000054 was obtained using conserved eukaryotic primers NS3, NS4, NS5, and NS7 (41).
Sequences were checked and compared to available sequences within the NCBI GenBank database using the BLAST algorithm (1). 18S rRNA sequence data were analyzed with the ARB software package (25). Framework trees were constructed with fastDNAml with only near-complete sequences. Missing sequence data and uncertainties in all near-complete sequences were omitted with a generated filter. The stability of branching patterns was tested using alternative treeing methods including evolutionary distance (Jukes and Cantor model) and maximum parsimony (DNAPARS) (25). Selected ITS sequence data were aligned using ClustalX (v1.83) (38) followed by manual checking. Evolutionary distance was determined using the Jukes and Cantor model to construct unrooted trees by the neighbor-joining method using PHYLIP v. 3.6 (13, 14).
Host sponge extract.
Ianthella reticulate (family Ianthellidae, order Verongida, class Demospongiae) was collected (June 1996) from Joseph Bonaparte Gulf, Australia (30-m depth, 128°39.53′E, 14°11.68′S). Approximately 2 g (wet weight) was freeze-dried; extracted with 10 ml EtOH, which was decanted and dried under vacuum; and then dissolved in 10 ml DMSO for assay.
PPDK assay.
Extract screening, enzyme selectivity testing, and all mechanistic experiments to determine inhibition parameters for substrates were conducted as detailed elsewhere (8, 16). Kinetic experiments were conducted at 5 and 10 μg ml−1 of the bioactive metabolite in the presence of six substrate concentrations for the following substrates: pyruvate (40, 80, 160, 320, 480, and 720 μM), ATP (20, 40, 60, 120, 240, and 480 μM), and phosphate (0.3, 0.6, 1.2, 1.8, 2.7, and 4.32 mM). The noncompetitive inhibitor (I) binds to either the free enzyme (E) or the enzyme-substrate complex (ES), and the dissociation constants are Kic = [E][I]/[EI] and Kiu = [ES][I]/[ESI].
Whole-plant phytotoxicity.
Whole-plant phytotoxicity assessment was performed as described previously (16) with minor modifications. Seedlings of Digitaria ciliaris and barley (Hordeum vulgare) were used as models of the C4 and C3 plants, respectively. The bioactive metabolite was prepared in 0.5% DMSO and 0.08% Uptake in MeOH to give 10 mg ml−1 and serially diluted down to 0.1 mg ml−1 in the same formulation. Dilutions were applied to the middle section of individual flag leaves on duplicate plants by use of 5 μl per C4 leaf and 20 μl per C3 leaf, corresponding to absolute amounts applied ranging from 50 μg to 0.5 μg for C4 leaves and 200 μg to 2 μg for C3 leaves. Control plants were treated with formulation only. Plants were observed within 6 h of application of the compound and then at 24-h intervals for 5 days. Each sample was tested in duplicate with at least four plants per treatment per replicate.
Antimicrobial assays.
The antimicrobial activity of the metabolite was tested against Staphylococcus aureus (ATCC 25923), Escherichia coli (ATCC 25922), and Vibrio harveyi (strain C071 [3]). Assays were conducted in a similar fashion with all manipulations of S. aureus performed in peptone yeast extract medium (peptone 10 g liter−1, yeast extract 5 g liter−1, NaCl 5 g liter−1) and for manipulations of E. coli and V. harveyi performed in Luria-Bertani medium (DIFCO, Becton Dickinson; Australia). Inoculated medium (20 ml) was grown overnight at 37°C with shaking (150 rpm) and diluted 1,000-fold into fresh medium. Culture (198 μl) was then transferred into wells of a 96-well microtiter plate. Serial twofold dilutions of the bioactive metabolite ranging from 100 to 0.2 μg ml−1 in DMSO (final concentration, 1% [vol/vol]) were added in duplicate to the 96-well microtiter plate. Solvent only was added to column 11 and antibiotic (500 μg ml−1 ampicillin for S. aureus and E. coli and 500 μg ml−1 streptomycin for V. harveyi) was added to column 12 to control for no growth. An initial absorbance reading was taken at 595 nm, and the plates were incubated overnight at 37°C. A final absorbance reading was taken after 24 h, and levels of growth inhibition were determined by comparing results to those for solvent controls after subtraction of background.
Anti-tumor cell assays.
MCF-7 (breast; pleural effusion adenocarcinoma, ATCC HTB-22), SF-268 (central nervous system; glioblastoma), and H460 (lung; large-cell carcinoma, ATCC HTB-177) cells were grown in RPMI 1640 medium with l-glutamine supplemented with 5% fetal bovine serum and maintained in a humidified incubator at 37°C with 5% CO2.
Cells were seeded in 96-well microtiter plates at a density of 4,000 (H460) or 5,000 (MCF-7 and SF-268) cells per well in 100 μl medium and allowed to attach for 24 h. Unguinol in DMSO was serially diluted in medium, added to the cells in triplicate so that the final doses ranged from 3 μg ml−1 to 250 μg ml−1, and returned to the incubator. Cell number was measured at 0 h and 48 h after sample addition via total cellular protein concentration with the sulforhodamine B (SRB) assay. Cells were fixed with 30 μl of 50% trichloroacetic acid for 30 min at 4°C, rinsed in running water (×5), air dried, and then stained with 50 μl 0.4% SRB in 1% acetic acid for 30 min at room temperature. Plates were washed with 1% acetic acid (×5) and then air dried. SRB dye was solubilized in 10 mM Tris (100 μl), and A490 was read on a Wallac Victor plate reader. Inhibition of growth by 50% (GI50) was determined by comparing the sample-treated values to those of the vehicle-only control and to time zero readings.
Theoretical chemical property calculations.
The theoretical logarithm of the n-octanol/water partition coefficient (log P) was calculated using ClogP (Daylight Chemical Information Systems), which implements the method of Leo (23), and the LOGKOW program (29) of the Environmental Science Centre of the Syracuse Research Corporation (Syracuse, NY). The numbers of hydrogen bond donors and acceptors were calculated by the Lipinski approach (24), and the number of rotatable bonds was counted using the interactive software provided by Molinspiration Cheminformatics (Bratislava, Slovak Republic). This same software was used to calculate molecular polar surface area (PSA) based on the method of Ertl et al. (11). This calculation was independently confirmed using Marvin (version 3.4.2; Chemaxon, Budapest, Hungary).
Nucleotide sequence accession numbers.
Nucleotide sequence data were submitted to the NCBI GenBank database under the accession numbers EF067336 and EF067337.
RESULTS
The PPDK primary screening assay proved robust for the 2,245 fungal extracts having a Z′ factor of 0.59, putting it into the “excellent” category (43). Extracts that caused statistically very significant inhibition of PPDK (P < 0.001) were those with inhibitions of <71.4% of the control. To minimize the probability of obtaining false positives, the cutoff point for an extract being declared active was reduced further to 50%. By use of this criterion, 297 extracts from 197 isolates were classified as active, with 258 extracts returning a similar result upon confirmatory retesting. This assay can be manipulated to determine selective PPDK inhibition (8); thus, the 258 confirmed bioactive extracts were subjected to selectivity profiling against the three enzymes involved in the coupled enzyme reaction. This approach identified 22 extracts from 19 different fungal isolates that selectively inhibited PPDK.
All 19 isolates were regrown in the same five media, and only 11 extracts, representing eight isolates, reproduced PPDK inhibition. Activity was predominant in the high-nutrient and no-salt media. The eight fungal isolates were cultivated on a larger scale, in 250 ml, with only one isolate (F3000054) producing bioactivity under these scaled-up conditions in all media except low-pH (pH 3.5) medium. 1H NMR spectra of the active extracts of F3000054 showed similar chemical fingerprints, indicating similar chemistries.
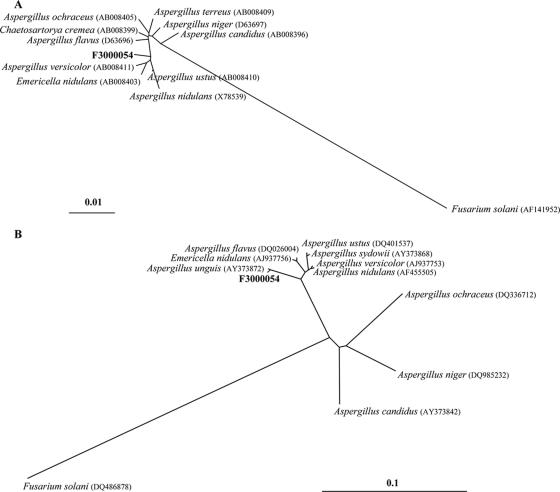
Phylogenetic affiliation (Fig. 2A) of fungal isolate F3000054 by sequencing of the near-complete amplified fragment of the 18S rRNA indicated that it belongs to the Trichocomaceae family, with Emericella nidulans (Aspergillus nidulans) demonstrating the highest identity (99% over 1,716 bp). Comparison of ITS regions demonstrated the highest identity to Aspergillus unguis (99% over 532 bp), with the relationship to other species shown in Fig. 2B.
FIG. 2.
Phylogenetic analyses of selected aligned 18S rDNA (A) and ITS1 (B) sequences. Unrooted trees were generated by means of the neighbor-joining method and the Jukes and Cantor algorithm model. Fusarium solani (DQ486878) was used as the out-group for both analyses. The scale bars indicate the expected numbers of base substitutions per site, with bar lengths corresponding to 1% and 10% differences for 18S rDNA and ITS1 regions, respectively.
Culturing in the no-salt medium produced the strongest inhibitory activity and was chosen for subsequent scale-up cultures. Large-scale (7 × 1 liter) growth in the no-salt medium produced 3.57 g of an EtOAc extract, which inhibited PPDK. EtOAc was used in preference to EtOH to increase the efficiency of the extraction, specifically to enable partitioning of the aqueous/organic layers to separate the medium components from those produced by the fungus.
Bioassay-guided fractionation of 1.0 g of the EtOAc extract yielded 81.7 mg (8.2% by weight) of a single bioactive natural product whose spectroscopic data (ESI MS and one- and two-dimensional NMR) were consistent with those reported for unguinol (Fig. 1, structure 2), previously isolated from mycelia of A. unguis (37) and A. nidulans (35).
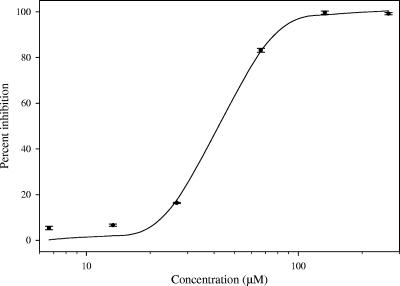
Dose-response analysis of unguinol on PPDK revealed a 50% inhibitory concentration of 42.3 ± 0.8 μM with a Hill slope of 3.4 ± 0.5 (Fig. 3). Kinetic evaluation indicated unguinol to be a mixed noncompetitive inhibitor with respect to pyruvate (Kiu = 67.3 ± 14.4 μM, Kic = 52.0 ± 33.6 μM), a mixed noncompetitive inhibitor with respect to ATP (Kiu = 122.3 ± 52.0 μM, Kic = 140.6 ± 143.7 μM), and an uncompetitive inhibitor with respect to phosphate (Kiu = 1,070 ± 14.4 μM).
FIG. 3.
Dose response of unguinol on PPDK. Data points are the means ± standard errors of two measurements. The data were fitted with a sigmoidal (Hill) dose-response curve according to the following equation: % inhibition = xn/(IC50n + xn), where n is the Hill slope and x is the concentration of unguinol. IC50 = 42.3 ± 0.8 μM; Hill slope = 3.4 ± 0.5.
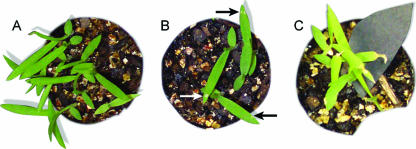
Unguinol caused a photosynthetic effect, evident as small dead patches at the site of application, on D. ciliaris down to an absolute applied amount of 0.5 μg leaf−1 after 24 h (Fig. 4), but it had no effect on barley at the highest test amount of 200 μg leaf−1, even after 5 days.
FIG. 4.
Phytotoxic effect of unguinol on C4 plants. (A) Five microliters of formulation (0.5% DMSO and 0.08% Uptake in MeOH); (B) 0.5 μg unguinol leaf−1 (lowest dose tested); (C) 50 μg unguinol leaf−1 (highest dose tested). Plants were photographed 48 h after application.
Unguinol inhibited growth of S. aureus and V. harveyi, with GI50s of 8.7 ± 0.2 and 69.5 ± 15.2 μM, respectively. No inhibition against E. coli was observed at the highest tested concentration. All three cancer cell lines were affected by unguinol, with GI50s of 28.2 ± 1.3 μM (H460), 50.8 ± 11.3 μM (MCF-7), and 44.3 ± 7.0 μM (SF-268).
The sponge Ianthella reticulate, from which F3000054 was originally isolated, was also tested. The EtOH extract did not inhibit PPDK. Unguinol was not detectable by 1H NMR or ESI MS analysis in the EtOH extract.
Theoretical chemical property calculations on unguinol gave a ClogP value of 5.7, a log KOW of 4.92, and a PSA of 83.8 Å2. Unguinol also possesses two H-bond donors, five H-bond acceptors, and one rotatable bond (Table 1).
TABLE 1.
Comparison of the calculated theoretical properties and PPDK-inhibitory activities of unguinol and ilimaquinone
| Parameter | Desired range | Value for:
|
|
|---|---|---|---|
| Ilimaquinone | Unguinol | ||
| IC50 (PPDK) | 292 ± 23 μM | 42.3 ± 0. 8 μM | |
| Mol wt | ≥150-≤500 | 358.2 | 326.35 |
| ClogP value | ≤3.5 | 6.0 | 5.7 |
| No. of hydrogen bond donors | ≤3 | 1 | 2 |
| No. of hydrogen bond acceptors | ≥2-≤12 | 4 | 5 |
| No. of rotatable bonds | ≤12 | 3 | 1 |
| PSA (Å2) | 50-60 | 63.6 | 83.8 |
DISCUSSION
Fungally produced depsidones, including unguinol, are confined to the morphologically indistinguishable fungi Aspergillus nidulans (teleomorph of Emericella nidulans) and Aspergillus unguis (teleomorph of Emericella unguis) (10, 37). The fungus from which unguinol was isolated, F3000054, possesses 99% sequence identities to A. nidulans and A. unguis in the 18S rDNA and ITS sequenced regions, respectively. At present, no 18S sequence data exist for A. unguis in the NCBI GenBank database to enable any sequence comparison. This sequence data and the fact that the isolate produces unguinol indicate that the fungal species isolated is an Aspergillus species closely related to both A. unguis and A. nidulans.
Unguinol selectively inhibited PPDK and showed mixed noncompetitive inhibition of PPDK with respect to the substrates pyruvate and ATP and was uncompetitive with respect to phosphate. The value of PPDK as an herbicide target is that it is potentially specific to C4 plants, enabling application of a weed-selective herbicide inactive against C3 crops without having to produce genetically modified crops. The spot application of unguinol caused localized leaf bleaching of D. ciliaris, a model C4 plant, indicating that the formulated unguinol could penetrate the plant cuticle and cell wall to inhibit photosynthesis, but had no effect on barley, a model C3 plant, even at much higher concentrations. The lack of toxicity against barley and the selective inhibition of PPDK, which translates into phytotoxicity on C4 plants, make unguinol worthy of further investigation as an herbicide.
The modified Lipinski's “rule of five” proposed for postemergent herbicides (39) requires a compound to meet the following physical criteria: a molecular weight of between 150 and 500; a log P value of less than 3.5; a number of hydrogen bond donors equal to or lower than three; and a number of hydrogen bond acceptors between 2 and 12, inclusive. Another important descriptor used to predict herbicide potential is the PSA, which should lie between 50 and 60 Å2 (40). Unguinol satisfies three of the five criteria, but its log P is above 3.5 and it has a PSA of greater than 60 Å2 (Table 1). Comparison of the theoretical and biological properties of unguinol with those of ilimaquinone, a PPDK inhibitor isolated from a marine sponge, indicates that although unguinol has a much greater PSA, it is a more potent inhibitor of PPDK.
The original screening extract of F3000054 inhibited the growth of S. aureus (gram-positive bacteria) and V. harveyi (gram-negative bacteria) but not that of E. coli (gram-negative bacteria). Unguinol has a similar activity profile. Interestingly, unguinol analogues acarogobien A (Fig. 1, structure 3), acarogobien B (Fig. 1, structure 4), and guisinol (Fig. 1, structure 5) also inhibit the growth of S. aureus (31, 32). Given the antimicrobial activity of unguinol, one could postulate it may provide a defense for the fungus against other microorganisms. Unguinol exhibited in vitro cytotoxicity against the three human tumor cell lines MCF-7, SF-268, and H460. Unguinol has previously been shown to act as an animal growth permittant (12) and to inhibit bile salt hydrolase (12). A few derivatives of unguinol have exhibited phospholipase A2 inhibition (17), or cytotoxicity (HOP-18, lung non-small cell line) (Pubchem [http://pubchem.ncbi.nlm.nih.gov]); however, most have no reported bioactivity (Pubchem [http://pubchem.ncbi.nlm.nih.gov]). Unlike ilimaquinone, unguinol and its derivatives have a narrow range of bioactivity and so are more attractive PPDK-selective herbicide candidates.
Preliminary 1H NMR analysis of all the extracts from the 10-ml cultures of F3000054 indicated that those that were inactive had spectral fingerprints different from those that inhibited PPDK activity. Comparison of the 1H NMR spectra of the EtOH extracts from the 250-ml cultures of F3000054 and of the combined EtOAc extracts of the 1-liter cultures of F3000054 with that of the isolated bioactive metabolite unguinol correlated to the reproducibility of selective PPDK inhibition and therefore to the presence of unguinol.
NMR and ESI MS analyses could not detect unguinol in the EtOH extract of the host sponge I. reticulate. In the laboratory, however, we found that the fungus produced unguinol in high yield when cultured in the high-nutrient or the no-salt media. From this, we conclude that a saline environment is not essential for unguinol production. It is well documented that aspergilli are prolific sources of metabolites as well as being salt tolerant, fast growing, and easily obtained from most substrates (6). No evidence was found to indicate this fungus produces unguinol in the fungus/sponge matrix, and to date there are no other reports of unguinol being produced by any other marine-derived organism. Guisinol (Fig. 1, structure 5), a closely related precursor depside, however, was isolated from a marine-derived E. unguis (teleomorph of A. unguis) (31).
Although this study focused on isolating PPDK-selective compounds, the issue of supply, whether by synthetic, semisynthetic, microbial, cell culture, or whole-organism production, must be addressed when commercial potential is considered. The fact that the production of unguinol is easily scaled up using a number of different media lends the process to further optimization. The commercial yield of thiocoraline, an anticancer agent isolated from the bacterium Micromonospora marina, is approximately 9 mg liter−1 (33). With minimal manipulation, a yield of approximately 40 mg liter−1 was achieved in this study, indicating that with further optimization of the growth conditions, such as light, temperature, time, periodicity, and medium type, commercial-scale supply of unguinol and its derivatives may be readily achievable.
During the present study, we identified a number of microorganism-derived extracts that inhibited PPDK activity. By using an integrated in vitro/in vivo screening strategy and taking into consideration the need for a commercially sustainable supply, unguinol was identified as a new lead herbicide. Unguinol also has the potential to be used as a biochemical probe for the structure and function of PPDK as well as to provide a template quite different from that of ilimaquinone for structure-activity relationship studies to improve its herbicidal properties.
Acknowledgments
This research was supported by the James Cook University Collaborative Research Grants Scheme and Nufarm Pty. Ltd.
Authority to collect the sponge was provided by the Commonwealth of Australia legislation Australian Institute of Marine Science Act 1972. Sponge taxonomy was undertaken by P. Bergquist. We thank C. Hooi, R. Anderson, and C. Cullinane of the Peter MacCallum Cancer Centre for a gift of the H460 and MCF-7 cells. We also thank those AIMS staff, both past and present, involved in the acquisition of samples for the production of marine natural product extracts.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourne, D. G., E. Evans-Illidge, and L. E. Llewellyn. 2004. Marine microbes for biodiscovery: just the tip of an iceberg, p. 185-205. In I. Kurtboke and J. Swings (ed.), Microbial genetic resources and biodiscovery. World Federation for Culture Collections, Surrey, United Kingdom.
- 3.Bourne, D. G., L. Høj, N. S. Webster, S. Swan, and M. R. Hall. 2006. Biofilm development within a larval rearing tank of the tropical rock lobster, Panulirus ornatus. Aquaculture 260:27-38.
- 4.Bringaud, F., D. Baltz, and T. Baltz. 1998. Functional and molecular characterization of a glycosomal PPi-dependent enzyme in trypanosomatids: pyruvate, phosphate dikinase. Proc. Natl. Acad. Sci. USA 95:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruderer, T., C. Wehrli, and P. Kohler. 1996. Cloning and characterization of the gene encoding pyruvate phosphate dikinase from Giardia duodenalis. Mol. Biochem. Parasitol. 77:225-233. [DOI] [PubMed] [Google Scholar]
- 6.Bugni, T. S., and C. M. Ireland. 2004. Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 21:143-163. [DOI] [PubMed] [Google Scholar]
- 7.Donaldson, D., T. Kiely, and A. Grube. 2002. Pesticides industry sales and usage: 1998 and 1999 market estimates. U.S. Environmental Protection Agency, Washington, DC.
- 8.Doyle, J. R., J. N. Burnell, D. S. Haines, L. E. Llewellyn, C. A. Motti, and D. M. Tapiolas. 2005. A rapid screening method to detect specific inhibitors of pyruvate orthophosphate dikinase as leads for C4 plant-selective herbicides. J. Biomol. Screen. 10:67-75. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, G. E., H. Nakamoto, J. N. Burnell, and M. D. Hatch. 1985. Pyruvate Pi dikinase and NADP-malate dehydrogenase in C4 photosynthesis: properties and mechanism of light/dark regulation. Annu. Rev. Plant Physiol. 36:255-286. [Google Scholar]
- 10.Eltem, R., T. Askun, S. Nermin, E. Ozkale Taskin, and H. Efendiler. 2004. Colonial and morphological characteristics of some Aspergillus Fr.:Fr. species isolated from vineyards in Manisa and Izmir provinces (Turkey). Turk. J. Bot. 28:287-298. [Google Scholar]
- 11.Ertl, P., B. Rohde, and P. Selzer. 2000. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 43:3714-3717. [DOI] [PubMed] [Google Scholar]
- 12.Feighner, S. D., G. M. Salituro, J. L. Smith, and N. N. Tsou. September. 1994. Unguinol and analogs are animal growth permittants. U.S. patent 5,350,763.
- 13.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6 ed. Distributed by the author, Department of Genome Sciences, University of Washington, Seattle.
- 15.Fennical, W., and P. R. Jensen. 1993. Marine microorganisms: a new biomedical resource, p. 419-45. In D. H. Attaway and O. R. Zaborsky (ed.), Marine biotechnology, pharmaceutical and bioactive natural products, vol. 1. Plenum Press, New York, NY. [Google Scholar]
- 16.Haines, D. S., J. N. Burnell, J. R. Doyle, L. E. Llewellyn, C. A. Motti, and D. M. Tapiolas. 2005. Translation of in vitro inhibition by marine natural products of the C4 acid cycle enzyme pyruvate P(i) dikinase to in vivo C4 plant tissue death. J. Agric. Food Chem. 53:3856-3862. [DOI] [PubMed] [Google Scholar]
- 17.Hamano, K., M. Kinoshita-Okami, A. Hemmi, A. Sato, M. Hisamota, K. Matsuda, K. Yoda, H. Haruyama, T. Hosoya, and K. Tanzawa. 1992. Folipastatin, a new depsidone compound from Aspergillus unguis as an inhibitor of phospholipase A2. Taxonomy, fermentation, isolation, structure determination and biological properties. J. Antibiot. 45:1195-1201. [DOI] [PubMed] [Google Scholar]
- 18.Holm, L. G., D. L. Plucknett, J. V. Pancho, and J. P. Herberger. 1977. The world's worst weeds: distribution and biology. University of Hawaii Press, Honolulu.
- 19.Jenkins, C. L. D. 1989. Effects of the phosphoenolpyruvate carboxylate inhibitor 3,3-dichloro-2-(dihydroxyphosphinoylmethyl)propenoate on photosynthesis. Plant Physiol. 89:1231-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins, C. L. D., R. L. N. Harris, and H. G. McFadden. 1987. 3,3-Dichloro-2-dihydroxyphospinoylmethyl-2-propenoate, a new, specific inhibitor of phosphoenolpyruvate carboxylase. Biochem. Int. 14:219-226. [Google Scholar]
- 21.Knight, V., J. J. Sanglier, D. DiTullio, S. Braccili, P. Bonner, J. Waters, D. Hughes, and L. Zhang. 2003. Diversifying microbial natural products for drug discovery. Appl. Microbiol. Biotechnol. 62:446-458. [DOI] [PubMed] [Google Scholar]
- 22.Lein, W., F. Bornke, A. Reindl, T. Ehrhardt, M. Stitt, and U. Sonnewald. 2004. Target-based discovery of novel herbicides. Curr. Opin. Plant Biol. 7:219-225. [DOI] [PubMed] [Google Scholar]
- 23.Leo, A. J. 1993. Calculating log POCT from structures. Chem. Rev. 93:1281-1304. [Google Scholar]
- 24.Lipinski, C. A., F. Lombardo, B. W. Dominy, and P. J. Feeney. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 23:3-25. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Struckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K-H. Schleifer. 2004. ARB: a software package environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maroco, J. P., M. S. B. Ku, P. J. Lea, L. V. Dever, R. C. Leegood, R. T. Furbank, and G. E. Edwards. 1998. Oxygen requirement and inhibition of C4 photosynthesis—an analysis of C4 plants deficient in the C3 and C4 cycles. Plant Physiol. 116:823-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, J. S., A. R. Ashton, F. Govers, and A. R. Hardham. 2001. Isolation and characterization of four genes encoding pyruvate, phosphate dikinase in the oomycete plant pathogen Phytophthora cinnamomi. Curr. Genet. 40:73-81. [DOI] [PubMed] [Google Scholar]
- 28.McFadden, H. G., R. L. N. Harris, and C. L. D. Jenkins. 1989. Potential inhibitors of phosphoenolpyruvate carboxylase. II. Phosphonic acid substrate analogues derived from reaction of trialkyl phosphites with halomethacrylates. Aust. J. Chem. 42:301-314. [Google Scholar]
- 29.Meylan, W. M., and P. H. Howard. 1995. Atom/fragment contribution method for estimating octanol-water partition coefficients. J. Pharm. Sci. 84:83-92. [DOI] [PubMed] [Google Scholar]
- 30.Moore, B. S. 2005. Biosynthesis of marine natural products: microorganisms (part A). Nat. Prod. Rep. 22:580-593. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen J. L., P. H. Nielsen, and J. C. Frisvad. 1999. Fungal depside, guisinol, from a marine derived strain of Emericella unguis. Phytochemistry 50:263-265. [Google Scholar]
- 32.Rezanka, T. 1999. Brominated depsidones from Acarospora gobiensis, a lichen of Central Asia. J. Nat. Prod. 62:1675-1677. [Google Scholar]
- 33.Romero, F., F. Espliego, J. Perez Baz, T. Garcia De Quesada, D. Gravalos, F. de la Calle, and J. L. Fernandez-Puentes. 1997. Thiocoraline, a new depsipeptide with antitumor activity produced by a marine Micromonospora. I. Taxonomy, fermentation, isolation, and biological activities. J. Antibiot. 50:734-737. [DOI] [PubMed] [Google Scholar]
- 34.Sherman, D. H., and J. L. Fortman. 2005. Utilizing the power of microbial genetics to bridge the gap between the promise and the application of marine natural products. Chembiochem 6:1-19. [DOI] [PubMed] [Google Scholar]
- 35.Sierankiewicz, J., and S. Gatenbeck. 1972. A new depsidone from Aspergillus nidulans. Acta Chem. Scand. 26:455-458. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, C. N. J., and L. E. Via. 1993. A rapid CTAB isolation technique useful for RAPD fingerprinting and other PCR applications. BioTechniques 14:748-758. [PubMed] [Google Scholar]
- 37.Stodola, F. H., R. F. Vesonder, D. I. Fennell, and D. Weisleder. 1972. Fungi Moniliales. A new depsidone from Aspergillus unguis. Phytochem. Rep. 11:2107-2108. [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tice, C. M. 2001. Selecting the right compounds for screening: does Lipinski's rule of 5 for pharmaceuticals apply to agrochemicals. Pest Manag. Sci. 57:3-16. [DOI] [PubMed] [Google Scholar]
- 40.Tice, C. M. 2002. Selecting the right compounds for screening: use of surface-area parameters. Pest Manag. Sci. 58:219-233. [DOI] [PubMed] [Google Scholar]
- 41.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA.
- 42.Woodburn, A. T. 2000. Glyphosate: production, pricing and use worldwide. Pest Manag. Sci. 56:309-312. [Google Scholar]
- 43.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]