Abstract
The medicinal leech, Hirudo verbana, is one of the simplest naturally occurring models for digestive-tract symbioses, where only two bacterial species, Aeromonas veronii bv. sobria (γ-Proteobacteria) and a Rikenella-like bacterium (Bacteroidetes), colonize the crop, the largest compartment of the leech digestive tract. In this study, we investigated spatial and temporal changes of the localization and microcolony structure of the native symbionts in the crop, after ingestion of a sterile blood meal, by fluorescence in situ hybridization. The population dynamics differed between the two symbiotic bacteria. A. veronii was detected mainly as individual cells inside the intraluminal fluid (ILF) during 14 days after feeding (daf) unless it was found in association with Rikenella microcolonies. The Rikenella-like bacteria were observed not only inside the ILF but also in association with the luminal surface of the crop epithelium. The sizes of Rikenella microcolonies changed dynamically through the 14-day period. From 3 daf onward, mixed microcolonies containing both species were frequently observed, with cells of both species tightly associating with each other. The sizes of the mixed microcolonies were consistently larger than the size of either single-species microcolony, suggesting a synergistic interaction of the symbionts. Lectin staining with succinylated wheat germ agglutinin revealed that the planktonic microcolonies present in the ILF were embedded in a polysaccharide matrix containing N-acetylglucosamine. The simplicity, symbiont-symbiont interaction, and mixed microcolonies of this naturally occurring, digestive-tract symbiosis lay the foundation for understanding the more complex communities residing in most animals.
Many animals, including humans, possess a diverse bacterial community in their digestive tract. For example, the human intestine contains between 500 and 1,000 species of bacteria (10, 43), and diverse microbiota have been isolated from the digestive tracts of many other animals, including mice (26), zebra fish (35), and termites (19, 44). Simpler communities, composed of 7 to 20 bacterial species, are found in some insects, such as gypsy moths (5) and mosquitoes (27). Investigators using experimentally simplified gut communities, such as germfree mice inoculated only with Bacteroides thetaiotaomicron, revealed that the resident microbes play pivotal roles throughout the host life, including the provision of essential nutrients (3, 39), energy balance (26), gut development (40), and induction of the immune system (32, 34). Despite these amazing discoveries about host-symbiont interactions, it remains relatively unclear how symbiotic bacteria interact with each other. The interaction between different species of microorganisms probably plays an important role in most digestive-tract communities yet is difficult to investigate because of the complexity of most digestive-tract communities and the lack of a natural model.
The medicinal leech, Hirudo verbana (Hirudinia, Hirudinidae), is one of the simplest naturally occurring models for digestive-tract symbioses with bacteria (17). The medicinal leech is a hematophagous parasite native to freshwater environments throughout Europe (38, 41) and is increasingly being used postoperatively in modern medicine. Recent molecular studies indicate that H. verbana is a distinct but closely related species of Hirudo medicinalis (41) and that the animals used in our previous studies, as well as animals sold for medical applications, are now considered to be H. verbana (4, 15, 20). The digestive tract of the leech consists of two major compartments, the crop and the intestinum. The crop, a large compartment, stores the large blood meals and discharges water and salts from the ingested blood, resulting in a viscous, intraluminal fluid (ILF) that is composed of densely packed erythrocytes surrounded by a little fluid that is isosmotic with the leech hemolymph. Small portions of the ILF flow gradually into the intestinum, where erythrocytes are digested and nutrients are absorbed (38).
From the leech crop, researchers repeatedly isolated a pure culture of an Aeromonas species which we identified as Aeromonas veronii bv. sobria (γ-Proteobacteria, Aeromonadaceae) and had reported that only this bacterium inhabits the leech digestive tract (11, 12, 15, 29). Recently, a PCR-based approach discovered the presence of a second symbiont, an “unculturable” Rikenella-like bacterium (clone PW3, with an 89.7% 16S rRNA gene identity to Rikenella microfusus [Bacteroidetes, Rikenellaceae]), in the crop (42). The dominance of these two symbionts in the crop was confirmed by terminal restriction fragment length polymorphism analysis. Aeromonas veronii is a facultative anaerobe and is motile (9, 21), while the members of the genus Rikenella are commonly described as being obligate anaerobes and nonmotile (23), although these features are based on a few cultured strains and have not been verified in the “unculturable” Rikenella-like symbiont. Thus, the symbiotic system of the medicinal leech is naturally simple, composed of only two bacterial species with distinct physiologies (17, 42). These symbiotic bacteria are consistently detected in the leech digestive tract and are thought to play important roles for the host, such as provision of the vitamin B complex, which is scarce in vertebrate blood, aid in the digestion of the ingested blood, prevention of other bacteria from colonizing the crop, and possibly priming by A. veronii of the digestive tract to permit the Rikenella-like bacterium to proliferate (16, 38, 42).
In this study, we investigated spatial and temporal changes of the localization and community structure of A. veronii and Rikenella-like bacteria in the crop using fluorescence in situ hybridization (FISH), which revealed detailed population dynamics of the symbionts in the leech digestive tract. Advantages of this approach are that we did not introduce bacteria during feeding and could also monitor the currently uncultured Rikenella-like symbiont.
MATERIALS AND METHODS
Leeches and feeding.
Medicinal leeches were obtained from Leeches USA (Westbury, NY) and maintained prior to feeding in leech mobile homes (Leeches USA) containing artificial pond water (38 mg/liter of Instant Ocean Salts; Aquatic Systems, Inc., Mentor, OH) at 4°C. The leeches were fed sterile, defibrinated whole sheep blood (Quad Five, Ryegate, MT) as previously described (15). After feeding, the leeches were reared at 25°C in a day-night regimen (14 h light/10 h dark).
Histology.
Leeches were collected 6 h, 12 h, 1 day, 3 days, 7 days, and 14 days after feeding (daf). Three to five replicates were prepared for each sampling time. Leeches were relaxed and stretched in 70% ethanol for 2 min and prefixed in Carnoy's solution (ethanol:chloroform:acetic acid, 6:3:1) for 2 h. A 5-mm-thick cross-section of the midbody was cut from each leech in 80% ethanol, and the tissue was postfixed overnight in Carnoy's solution. Afterwards, the tissue was bleached overnight in 80% ethanol containing 7% H2O2 to decrease autofluorescence (37; R. Koga, unpublished data). The sample was dehydrated and cleared through an ethanol-xylene series and embedded in paraffin. Three-micrometer-thick serial tissue sections were cut with a rotary microtome (Shandon Finesse microtome) and mounted on silane-coated glass slides. The sections were dewaxed through a xylene-ethanol series and air-dried prior to in situ hybridization or lectin staining.
FISH.
The leech midbody sections were subjected to FISH as described previously (24). The Aeromonas-specific probe Cy3-AER66 (5′-CTACTTTCCCGCTGCCGC-3′) (22) and the Bacteroidetes-specific probe Cy5-CF319a (5′-TGGTCCGTGTCTCAGTAC-3′) (31) targeting the 16S rRNA genes were used for detecting A. veronii and the Rikenella-like bacteria, respectively. The specificities of these probes were confirmed by a homology search performed with the DNA databases, and percent coverage for the Bacteroidetes-specific probe is listed at http://www.microbial-ecology.net/probebase/probecoverage.asp. In addition, the specificity of the hybridization was confirmed experimentally by performing the following control experiments: no-probe control, RNase digestion control, and Cy3-labeled EUB338 antisense probe (1) control, as well as cross-reactivity with cultured A. veronii bv. sobria. One hundred fifty microliters of hybridization buffer (20 mM Tris-HCl [pH 8.0], 0.9 M NaCl, 0.01% sodium dodecyl sulfate, 30% formamide) containing 50 pmol/ml of each probe and 4 nmol/ml of 4′,6-diamidino-2-phenylindole [DAPI]) (Molecular Probes, Eugene, OR) was applied to a glass slide, covered with a coverslip, and incubated in a humidified chamber at room temperature overnight. To eliminate nonspecifically bound probe, the preparation was washed in hybridization buffer for 10 min at 37°C. Slides were washed in phosphate-buffered saline (PBS) and mounted in Vectashield (Vector Labs, Burlingame, CA). The fluorescence signals were observed with an epifluorescence microscope (TE2000; Nikon) using filter sets for DAPI, Cy3, and Cy5. Images were recorded with a digital camera (Spot RT-KE; Diagnostic). For each time point for at least three animals, the number of individual cells and microcolonies (ICM) in the ILF was counted in 5 randomly selected fields, which were captured at magnification ×400, and the number of cells present in each ICM was counted in 7 to 50 randomly selected ICMs at magnification ×1,000. The counting was performed after cutting background fluorescence off by using Spot software, version 4.6 (Spot RT-KE; Diagnostic).
Using the ICM number and the numbers of cells present in sections of these ICMs, we estimated the A. veronii and the Rikenella-like bacterial densities in the ILF. The amount of bacterial cells per 1 ml of the ingested blood was estimated by the following calculation: density of A. veronii (bacterial cells/ml) = [(Aeromonas-ICM number × mean number of cells in Aeromonas-ICM sections) + (Mixed-ICM number × mean number of Aeromonas cells in mixed-ICM)] × 863,400. The corresponding calculation was done for the Rikenella-like bacterium.
The constant, 863,400, is a conversion factor to convert the value obtained for a section to the number of cells per ml. It was calculated as follows: 1 ml/1.16 nl, the observed volume (= 320 μm × 241 μm × 3 μm thickness × 5 fields). Because we used 3-μm sections, our counts would overestimate the number of cells if only a portion of a cell was contained in a section. We also assume that cells located in the center of the section were detectable and their signal not masked by other cells. The theoretical detection limit of the FISH observation was 863 symbiont cells per 1 μl of the ILF.
The number of the symbionts associated with the crop epithelial tissue was counted in five randomly selected fields at magnification ×1,000. In the captured images, the length of the sectioned epithelial surface was measured by using the Spot software, and then the number of bacterial cells/mm of the leech epithelium was calculated.
Lectin staining.
Lectin staining was performed for tissue sections and cultured A. veronii. For A. veronii samples, A. veronii strain HM21 (15) cultured in LB broth was spread on slide glasses, air dried, and then dehydrated through an ethanol series. Dewaxed tissue sections and A. veronii samples were blocked in blocking solution (1% bovine serum albumin-0.3% Triton X-100 in PBS) for 1 h. Sections were then incubated overnight with 10 μg/ml of either concanavalin A, Dolichos biflorus agglutinin, Griffonia simplicifolia I-isolectin B4, Lens culinaris agglutinin, Phaseolus vulgaris erythroagglutinin, Phaseolus vulgaris leucoagglutinin, peanut agglutinin, Pisum sativum agglutinin, Ricinus communis agglutinin I, soybean agglutinin, Sophora japonica agglutinin, Ulex europeaus agglutinin, wheat germ agglutinin (WGA), and succinylated WGA (WGA-S), all of which were labeled with rhodamine (Vector Labs), in a blocking solution containing 4 nmol/ml of DAPI. Slides were washed in PBS and mounted in Vectashield (Vector Labs) for imaging. A no-lectin probe control was performed for the negative control. The fluorescence signals were observed and counted at magnification ×400 in the manner described for FISH.
Statistical analysis.
The statistical analyses used in this study, median test, two-way analysis of variance (ANOVA), and Kendall's correlation analysis, were performed by using the software program R, version 1.14 (36).
RESULTS
The symbionts form microcolonies in the crop ILF.
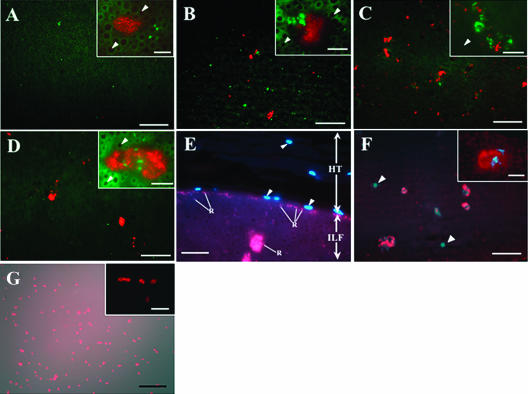
The colonization dynamics of native A. veronii and Rikenella-like bacteria residing in the ILF of the crop was observed for 14 daf by using FISH. Although no signals were detected 6 and 12 h after feeding, both species were continuously detected from 1 daf onward (Fig. 1A to D), suggesting that they were present initially below the limit of detection (863 cells/μl). The Rikenella-like bacteria were detected in clusters, suggesting that they formed microcolonies in the ILF. Comparison of the images captured on DAPI-stained sections to FISH images suggested that A. veronii is stained comparatively little with DAPI (data not shown). From 3 daf onward, A. veronii and the Rikenella-like bacteria were often detected together, suggesting that they formed mixed microcolonies (Fig. 1B to D, insets). We classified the microcolonies as Aeromonas-, Rikenella- or mixed microcolonies. Because Aeromonas occurred often as single cells, we used the number of individual cells and microcolonies (ICM) to simplify the description of these data. No signals were detected when cultured A. veronii strain HM21 (15) was observed with CF319a, the probe for the Rikenella-like bacteria (the inability to culture the Rikenella-like bacteria prevented us from performing this control with the Cy3-AER66 probe), nor in any of the negative controls (data not shown), indicating that the FISH specifically detected the two symbionts.
FIG. 1.
Microscopic fluorescence images of A. veronii, Rikenella-like bacteria, and polysaccharide matrix in the leech crop. (A to D) FISH visualizations of A. veronii (green) and Rikenella-like bacteria (red) colonizing in the ILF, using the specific probes Cy3-AER66 and Cy5-CF319a, respectively. One daf (A), 3 daf (B), 7 daf (C), or 14 daf (D). After 3 days, A. veronii and Rikenella-like bacteria often formed mixed microcolonies. Bars, 50 μm. Insets are enlarged images (bar, 10 μm). The arrowheads in the insets indicate erythrocytes which have a dark center surrounded by green autofluorescence. (E) Crop epithelial region of leech 7 daf. Rikenella-like bacterium stained with probe Cy5-CF319a and host nuclei (blue, indicated by arrowheads) were counterstained with DAPI. Bar, 20 μm. Abbreviations: HT, host tissue; ILF, intraluminal fluid; R, Rikenella-like bacterium. (F and G) Lectin staining with WGA-S (red). (F) ILF at 3 daf. Bacterial microcolonies (blue) were counterstained with DAPI. Arrowheads indicate nucleated cells in the ILF. Bars, 50 μm. Inset is an enlarged image (bar, 10 μm). (G) Cultured A. veronii. The lectin staining image was merged with a differential interference contrast image. Bar, 20 μm. The inset is an enlarged fluorescence image (bar, 10 μm).
(i) Dynamics of ICM number.
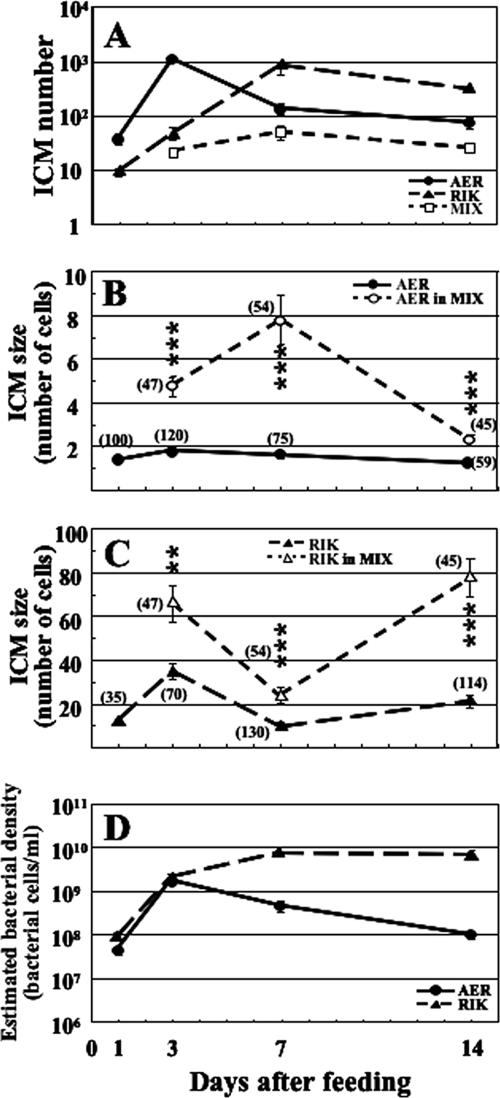
The number of Aeromonas and Rikenella ICMs revealed different temporal patterns (Fig. 2A). The number of Aeromonas ICMs increased quickly between 1 and 3 daf, subsequently decreased by 7 daf, and leveled off. On the other hand, the Rikenella ICM number increased until 7 daf and then decreased by half a log. At 3 daf, mixed microcolonies were detected, and the number of these mixed microcolonies remained relatively constant during the 14 daf (median test, P = 0.2). The number of the mixed microcolonies represents a lower limit, because if the microcolonies are not uniformly mixed with both species, a particular cross section of a mixed microcolony could, by chance, not contain one of the two species and be counted as a single-species microcolony. In contrast, the number of Aeromonas and Rikenella ICM changed significantly (median test, Aeromonas, P < 0.005; Rikenella, P < 0.0001).
FIG. 2.
Population dynamics of A. veronii and Rikenella-like bacteria in the ILF. (A) Dynamics of ICM number. (B and C) Dynamics of ICM size. A. veronii (B) or Rikenella-like bacteria (C). Asterisks indicate a significant difference between the numbers of symbionts in single-species and mixed microcolonies (two-way ANOVA: **, P < 0.0005; ***, P < 0.0001). (D) Changing patterns of estimated bacterial densities (bacterial cells per 1 ml of the ingested blood). Abbreviations: AER, Aeromonas ICM; AER in MIX, A. veronii in mixed microcolony; MIX, mixed microcolony; RIK, Rikenella ICM; RIK in MIX, Rikenella-like bacteria in mixed microcolony. Means ± standard errors are plotted. The number of animals used is 5, 5, 3, or 4 at 1 daf, 3 daf, 7 daf, or 14 daf, respectively. In panels B and C, the combined number of observed microcolonies is shown in parentheses.
(ii) Dynamics of ICM size.
The number of bacteria present in cross-sections of the ICMs was determined by counting the bacterial cells in random ICMs (Fig. 2B and C). Throughout the first 14 days, Aeromonas was often observed as individual cells (Fig. 2B). In contrast, mixed microcolonies always contained multiple A. veronii cells, and the total number of these cells was significantly greater than the total number of Aeromonas cells present in ICM at each time point (two-way ANOVA: 3 daf, F1,162 = 73.2 and P < 0.0001; 7 daf, F1,124 = 42.4 and P < 0.0001; 14 daf, F1,100 = 36.7 and P < 0.0001). The number of A. veronii cells in mixed microcolony cross sections increased between 3 daf and 7 daf and dropped to around two cells afterwards. The number of A. veronii cells in single-species ICMs and mixed microcolonies changed significantly (median test. single-species, P < 0.0001; mixed, P < 0.0001). The pattern of the Rikenella-like bacteria changed more dramatically (Fig. 2C). The Rikenella ICM size increased by 3 daf, followed by a decrease 7 daf, and then recovered. The number of the Rikenella-like bacteria in mixed microcolonies showed a pattern similar to that of Rikenella ICMs, except that the number of Rikenella-like cells in mixed microcolonies was always greater than in Rikenella ICMs (two-way ANOVA, 3 daf, F1,112 = 14.7 and P < 0.0005; 7 daf, F1,199 = 26.9 and P < 0.0001; 14 daf, F1,155 = 66.9 and P < 0.0001). The number of Rikenella-like bacteria in single-species ICM and mixed microcolonies changed significantly (median test, single-species, P < 0.0001; mixed, P < 0.0001). These microcolony sizes were not significantly different between leech replicates except for those of Rikenella at 14 daf, where one animal had low Rikenella-like bacteria numbers (two-way ANOVA, F2,155 = 4.7 and P = 0.011).
(iii) Estimated density of the symbionts.
Using the ICM number and the number of cells present in sections of these ICMs, we estimated the A. veronii and Rikenella-like bacterium densities in the ILF (bacterial cells per 1 ml of the ingested blood). Figure 2D shows the patterns of the estimated A. veronii and Rikenella-like bacteria densities. Although both species increased similarly between 1 and 3 daf, the subsequent pattern was quite different. While A. veronii slowly decreased below 108 cells/ml, the Rikenella-like bacteria leveled off around 1010 cells/ml. The estimated densities changed significantly for A. veronii (median test, P < 0.0001) and Rikenella-like bacteria (P < 0.0001).
Symbionts associated with the crop epithelium.
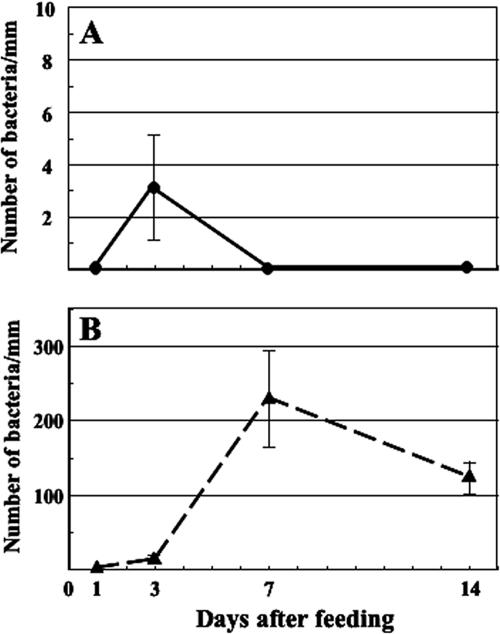
Three daf, symbionts associated with the luminal side of the crop epithelium were detected (Fig. 1E). To follow the colonization dynamics of the association, we counted the number of A. veronii and Rikenella-like bacteria associated with the host epithelial tissue by FISH. While few Aeromonas associated with the epithelium (Fig. 3A), many Rikenella-like bacteria associated with the crop tissues (Fig. 3B). Epithelium-associated Rikenella-like bacteria reached more than 200 cells/mm between 3 daf and 7 daf.
FIG. 3.
Association of A. veronii (A) or Rikenella-like bacteria (B) with host epithelial tissue. The number of Rikenella-like bacteria attaching to the host epithelia significantly changed during the period (median test, P < 0.0001). Means ± standard errors are plotted (n = 5, 5, 3, or 4 at 1 daf, 3 daf, 7 daf, or 14 daf, respectively).
Detection of polysaccharide matrix associated with microcolonies by lectin staining.
The spatial separation of the bacterial cells within the microcolonies and surrounding the microcolonies in the ILF (Fig. 1A to D, insets) suggested the presence of an associated matrix. We investigated the presence of a polysaccharide matrix in the leech digestive tract by performing lectin staining on the ILF of a leech 3 daf. Out of the 14 lectins tried in this study, WGA and WGA-S showed positive signals associated with bacterial microcolonies in 100% of the examined microcolonies (Fig. 1F; Table 1). The results for WGA-S were verified with multiple leeches during a time course. We detected some signals with peanut agglutinin, P. vulgaris leucoagglutinin, and R. communis agglutinin I, but there were no signals associated with bacterial microcolonies (Table 1). L. culinaris agglutinin, P. sativum agglutinin, and P. vulgaris erythroagglutinin produced a high background signal and were not further considered in this study. When cultured A. veronii were subjected to lectin staining, positive signals were observed only in WGA and WGA-S (Fig. 1G). The signals were detected only closely associated with the bacterial cell, indicating the lipopolysaccharide (LPS) or capsule structure of the symbiont was stained. The size and the distribution of the WGA and WGA-S signals was not consistent with the matrix observed in the ILF, but these cells were cultured in broth or on agar plates. No fluorescence signal was detected in the no-lectin control performed on leech sections and cultured A. veronii (data not shown).
TABLE 1.
Results of lectin staining in a medicinal leech 3 days after feeding
| Lectin | Glycan specificitya | Resultb |
|---|---|---|
| Concanavalin A | α-Mannose residues | −c |
| Dolichos biflorus agglutinin | α-N-Acetylgalactosamine | −c |
| Griffonia simplicifolia I—isolectin B4 | α-Galactose residues | −c |
| Phaseolus vulgaris leucoagglutinin | Not defined | − (0/3, 0) |
| Peanut agglutinin | Galactosyl (β-1,3)N-acetylgalactosamine | − (0/28, 0) |
| Ricinus communis agglutinin I | Galactose/N-acetylgalactosamine | − (0/25, 0) |
| Soybean agglutinin | Galactose/α- or β-N-acetylgalactosamine | −c |
| Sophora japonica agglutinin | Galactose/N-acetylgalactosamine | −c |
| Ulex europaeus agglutinin | α-Fucose | −c |
| Wheat germ agglutinin | N-Acetylglucosamine and sialic acid | + (40/47, 100) |
| Wheat germ agglutinin, succinylated | N-Acetylglucosamine | + (47/54, 100) |
Referenced from Vector Labs homepage.
Values in parentheses are as follows: number of lectin signals associated with microcolonies/total number of lectin signals, percentage of microcolonies associated with lectin signals among detected microcolonies.
No lectin signal.
Dynamics of WGA-S lectin signals in the ILF.
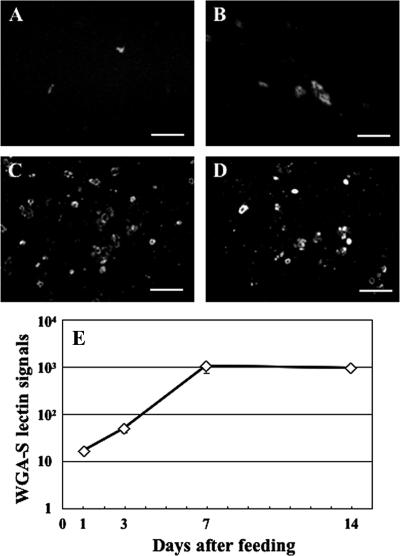
The lectin signals of WGA-S were observed over time in the ILF (Fig. 4). The number of the lectin signals increased to around 1,000 by 7 daf and then slightly decreased, following the dynamics of the Rikenella-like bacteria density (Fig. 2D). Correlation analysis showed that the number of the lectin signals significantly correlated with the number of Rikenella-like bacteria (Kendall's correlation analysis, τ = 0.717 and P < 0.0001) but not with the number of A. veronii (τ = 0.25; P = 0.195).
FIG. 4.
Number of lectin signals in the ILF after feeding. (A to D) Microscopy images of lectin (WGA-S) staining of the ILF. One daf (A), 3 daf (B), 7 daf (C), or 14 daf (D). Bars, 50 μm. (E) The number of lectin signals changed significantly during the observed period (median test, P < 0.0001). Means ± standard errors are plotted (n = 5, 5, 3, or 4 at 1 daf, 3 daf, 7 daf, or 14 daf, respectively).
DISCUSSION
Symbiont populations change dynamically in the crop.
Medicinal leeches feed infrequently and store the large blood meal in the crop, from where it is gradually transported into the intestinum. Our results confirm the presence of A. veronii and the Rikenella-like bacterium in the leech using a PCR-independent approach and demonstrate that the localization and microcolony structure of the symbionts change dynamically in the leech crop after a blood meal. Interestingly, the majority of the symbiotic bacteria were detected in the ILF (Fig. 1A to D and 2), although Rikenella-like bacteria associated with the crop epithelium (Fig. 1E and 3B). This suggests that the symbiotic bacteria mainly colonize the ILF rather than the epithelial tissue. Considering the slow-flowing ILF that is moved gradually over weeks into the intestinum, adhesion to epithelial tissue may not be as important for maintaining a symbiotic community in the leech as in other gut communities.
In the ILF, A. veronii and the Rikenella-like bacteria showed different population dynamics (Fig. 2). While both organisms proliferated rapidly during the first 3 daf, subsequently, A. veronii decreased in abundance while the Rikenella-like symbionts remained at a high population level. This pattern is consistent with observations in previous culture-based studies where A. veronii was introduced in a blood meal and the number of culturable bacteria monitored over time after feeding (15, 20). Differences in the physiology of A. veronii and R. microfusus, the culturable type species of the genus Rikenella, provide the basis for testable hypotheses. For example, A. veronii is beta-hemolytic and motile (9, 21), while R. microfusus is an obligate anaerobe and nonmotile (23).
A. veronii was detected either as individual cells or in association with Rikenella inside the ILF (Fig. 1A to D and 2B). Considering that A. veronii is motile (9, 21), this result suggests that this symbiont could actively swim in the ILF until it encounters Rikenella microcolonies. In contrast to Aeromonas, the Rikenella symbionts were consistently detected in clusters (Fig. 1A to D and 2C), as one may expect for a nonmotile bacterium. For A. veronii, this hypothesis can be tested by generating nonmotile mutants and introducing them into the leech.
From 3 daf onward, the population dynamics were different between the two symbiotic bacteria: A. veronii decreased in abundance, while the Rikenella-like bacteria were maintained at a stable level (Fig. 2D). It is unclear what led to the decrease of A. veronii cells, but it indicates that there is a loss of A. veronii cells compared to Rikenella-like cells. The transport into the intestinum would affect both symbionts similarly, but perhaps aspects of the innate immune response, such as antimicrobial peptides or macrophage-like cells, hemocytes, target A. veronii more efficiently. In addition, the growth rate of A. veronii could decrease after the removal of a key nutrient or the depletion of oxygen. An important effect of oxygen concentrations on the localization of the microorganisms was shown in the termite gut, where microaerophilic protozoans lined the epithelium and anaerobes were found in the deeper lumen of the hindgut (6-8).
The symbionts form mixed microcolonies in the ILF.
One of the exciting findings is that A. veronii and the Rikenella-like bacterium formed mixed microcolonies in the ILF, where the two symbionts tightly associated with each other (Fig. 1B to D, insets). The mixed microcolonies were detected from 3 daf onward (Fig. 2C). The presence of individual A. veronii cells suggests an invasion by A. veronii into semimatured Rikenella microcolonies, resulting in the mixed microcolonies.
Notably, the number of Rikenella-like bacteria in mixed microcolonies was consistently greater than that in single-species microcolonies (Fig. 2C), suggesting that the Rikenella-like bacteria might benefit from the invading A. veronii, possibilities due to syntrophy or the removal of oxygen by A. veronii. Although a similar situation was observed for A. veronii (Fig. 2B), the higher A. veronii cell number could be caused by multiple invasions, increased proliferation, or reduced loss after the invasion.
The symbionts associated with the crop epithelial tissues.
Many Rikenella-like bacteria and fewer A. veronii cells were detected on the crop epithelial tissues from 3 daf onwards (Fig. 1E and 3). These results indicate that the Rikenella-like bacteria can adhere to the crop epithelium. In a previous culture-based study, we reported that prior to 12 h after feeding, A. veronii cells were associated with the dissected epithelium in numbers that were unlikely to be due to carryover from the ILF (15). It is possible that the limit of detection of the FISH analysis prevented us from detecting these cells, they adhere to a specific location that was not included in our analysis, or in the culture-based study the viscous ILF contaminated the tissue sample.
The symbionts form biofilms in the leech digestive tract.
In this study, we detected signals of the lectins WGA and WGA-S that were associated and correlated with the bacterial microcolonies (Fig. 1F; Table 1), revealing that the symbiotic bacteria were embedded in a thick polysaccharide matrix containing N-acetylglucosamine. One possibility is that the signal is due to the LPS and/or capsule of the symbiotic bacteria. While cultured A. veronii cells were stained with WGA-S (Fig. 1G), indicating at least that this symbiont possesses an LPS/capsule structure composed of N-acetylglucosamine, the signal was always closely associated with the bacterial cells and was not similar to the thick, irregular matrix that we detected in vivo (Fig. 1F, inset). This observation is suggestive of biofilms.
A common definition of “biofilms” is that they are dense cohesive communities of microbes that embed themselves within surface-associated matrices and resist hydrodynamic shear forces, which are commonly found in natural aquatic environments and inside animal bodies (18, 33). The crop environment has very low flow rates and thus lower shear forces than other gut environments. The microcolonies inside the crop could be either attached to the erythrocytes or floating freely in the ILF. The polysaccharide-embedded microcolonies are closely associated with the surrounding erythrocytes, supporting the notion of the microcolonies being a biofilm. Alternatively, the symbiotic bacteria form self-immobilized biofilms, called “granular biofilms,” in the ILF, as has been observed in engineered bioreactors that are a model system for the digestive tract, where bacteria and archaea frequently form granular biofilms which develop without carrier materials (2, 25, 30). In either case, the observed microcolony structures are embedded in polysaccharides and resemble biofilms in a slow-flowing environment.
The lectin signals correlated with the Rikenella-like bacterial density (Fig. 2D and 4) but not A. veronii bacterial density, even though A. veronii produces an N-acetylglucosamine-containing surface molecule, which may be the LPS or capsule but not an extracellular matrix. Assuming that the Rikenella-like bacteria form the biofilms in the ILF, the following results obtained in this study are explained consistently. (i) Both microcolony number and size increased early on after feeding. During this period, the biofilms appeared to mature. (ii) Between 3 daf and 7 daf, the number of microcolonies increased but their size decreased, suggesting that the microcolonies broke apart, analogous to biofilms dispersing. (iii) During the next 7 days, the microcolony number decreased while their size increased, suggesting that some dispersers actively proliferated and others disappeared, possibly by aggregation. Such a biofilm development process, especially the maturation-dispersion cycle shown in results i and ii, is similar to the typical pattern of biofilm development found in many natural environments and under in vitro conditions (18, 33). We cannot rule out the possibility that A. veronii contributes to the biofilm formation inside the leech, just as Aeromonas spp. are reportedly able to form biofilms under certain in vitro conditions, but the composition of the matrix has not been determined (13, 14, 28).
This study for the first time directly followed the symbiont dynamics inside leeches without introducing bacteria in the blood meal. Our results suggest a model for spatial-temporal dynamics of A. veronii and the Rikenella-like bacterium in the leech crop (Fig. 5). Based on the feeding and starvation cycle of leeches in nature, where leeches feed approximately every 2 to 8 weeks (38) and both symbionts remain in the leech crop 90 days after a blood meal (42), it seems likely that this cycle repeats itself with each blood meal. We are currently pursuing the role of bacterial motility and biofilm formation using genetically modified A. veronii, which may provide us with novel insights into the symbiont-symbiont interactions in digestive tract systems.
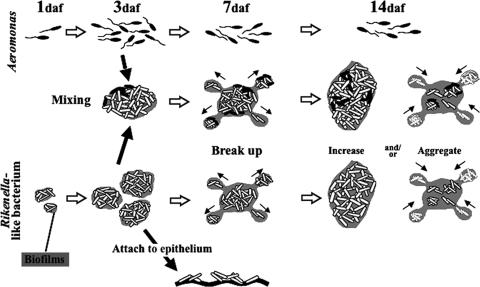
FIG. 5.
Model of the spatial-temporal dynamics of A. veronii and Rikenella-like bacteria in the leech crop. A. veronii increases in number until 3 daf and subsequently gradually decreases. The Rikenella-like bacteria increase in concentration and reach a plateau 7 daf. By 3 daf, Rikenella-like bacteria were detected that attached to the leech epithelium. The microcolonies are encased in an N-acetylglucosamine-containing matrix and may form a biofilm in the ILF, either a free-floating granular biofilm or one associated with erythrocytes. The apparent biofilms break apart at 7 daf, and subsequently the colony size increases by growth and/or colony aggregations. Mixed microcolonies were detected from 3 daf onwards, suggesting the presence of two-species biofilms. The number of cells in the mixed microcolony was consistently greater than in the single-species microcolonies.
Acknowledgments
We thank R. Koga from AIST Tsukuba, Japan, for technical advice for FISH, D. Gage from the University of Connecticut for the use of the microtome and technical advice, and A. Silver (University of Connecticut) and M. Yokoyama (University of Connecticut) for helpful comments.
This research was supported by Japan Society for the Promotion of Science fellowships for young scientists to Y.K. and the NSF Career Award MCB 0448052 to J.G.
Footnotes
Published ahead of print on 2 February 2007.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angenent, L. T., S. Sung, and L. Raskin. 2004. Formation of granules and Methanosaeta fibres in an anaerobic migrating blanket reactor (AMBR). Environ. Microbiol. 6:315-322. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Braschler, T. R., S. Merino, J. M. Tomas, and J. Graf. 2003. Complement resistance is essential for colonization of the digestive tract of Hirudo medicinalis by Aeromonas strains. Appl. Environ. Microbiol. 69:4268-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick, N. A., K. F. Raffa, R. M. Goodman, and J. Handelsman. 2004. Census of the bacterial community of the gypsy moth larval midgut by using culturing and culture-independent methods. Appl. Environ. Microbiol. 70:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 7.Brune, A., D. Emerson, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune, A., E. Miambi, and J. A. Breznak. 1995. Roles of oxygen and the intestinal microflora in the metabolism of lignin-derived phenylpropanoids and other monoaromatic compounds by termites. Appl. Environ. Microbiol. 61:2688-2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colwell, R. R., and M. T. MacDonell. 1986. Proposal to recognize the family Aeromonadaceae fam. nov. Int. J. Syst. Bacteriol. 36:473-477. [Google Scholar]
- 10.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eroglu, C., M. Hokelek, E. Guneren, S. Esen, A. Pekbay, and O. A. Uysal. 2001. Bacterial flora of Hirudo medicinalis and their antibiotic sensitivities in the Middle Black Sea Region, Turkey. Ann. Plast. Surg. 47:70-73. [DOI] [PubMed] [Google Scholar]
- 12.Fenollar, F., P. E. Fournier, and R. Legre. 1999. Unusual case of Aeromonas sobria cellulitis associated with the use of leeches. Eur. J. Clin. Microbiol. Infect. Dis. 18:72-73. [DOI] [PubMed] [Google Scholar]
- 13.Gavîn, R., S. Merino, M. Altarriba, R. Canals, J. G. Shaw, and J. M. Tomâs. 2003. Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol. Lett. 224:77-83. [DOI] [PubMed] [Google Scholar]
- 14.Gavin, R., A. A. Rabaan, S. Merino, J. M. Tomas, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383-397. [DOI] [PubMed] [Google Scholar]
- 15.Graf, J. 1999. Symbiosis of Aeromonas veronii biovar sobria and Hirudo medicinalis, the medicinal leech: a novel model for digestive tract associations. Infect. Immun. 67:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graf, J. 2002. The effect of the symbionts on the physiology of Hirudo medicinalis, the medicinal leech. Invertebr. Reprod. Dev. 41:269-275. [Google Scholar]
- 17.Graf, J., Y. Kikuchi, and R. V. Rio. 2006. Leeches and their microbiota: naturally simple symbiosis models. Trends Microbiol. 14:365-371. [DOI] [PubMed] [Google Scholar]
- 18.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 19.Hongoh, Y., H. Yuzawa, M. Ohkuma, and T. Kudo. 2003. Evaluation of primers and PCR conditions for the analysis of 16S rRNA genes from a natural environment. FEMS Microbiol. Lett. 221:299-304. [DOI] [PubMed] [Google Scholar]
- 20.Indergand, S., and J. Graf. 2000. Ingested blood contributes to the specificity of the symbiosis of Aeromonas veronii biovar Sobria and Hirudo medicinalis, the medicinal leech. Appl. Environ. Microbiol. 66:4735-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 22.Kämpfer, P., R. Erhart, C. Beimfohr, J. Böhringer, M. Wagner, and R. Amann. 1996. Characterization of bacterial communities from activated sludge: culture-dependent numerical identification versus in situ identification using group- and genus-specific rRNA-targeted oligonucleotide probes. Microb. Ecol. 32:101-121. [DOI] [PubMed] [Google Scholar]
- 23.Kaneuchi, C., and T. Mistoka. 1978. Bacteroides microfusus, a new species from the intestines of calves, chickens, and Japanese quails. Int. J. Syst. Bacteriol. 28:478-481. [Google Scholar]
- 24.Kikuchi, Y., X. Y. Meng, and T. Fukatsu. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lettinga, G., A. Van Velsen, S. Hobma, and W. Zeeuw. 1980. Use of the upflow sludge blanket (USB) reactor concept for biological wastewater treatment, especially for anaerobic treatment. Biotech. Bioeng. 22:699-734. [Google Scholar]
- 26.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindh, J. M., O. Terenius, and I. Faye. 2005. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl. Environ. Microbiol. 71:7217-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 29.Mackay, D. R., E. K. Manders, G. C. Saggers, D. R. Banducci, J. Prinsloo, and K. Klugman. 1999. Aeromonas species isolated from medicinal leeches. Ann. Plast. Surg. 42:275-279. [DOI] [PubMed] [Google Scholar]
- 30.MacLeod, F. A., S. R. Guiot, and J. W. Costerton. 1990. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl. Environ. Microbiol. 56:1598-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 32.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 34.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 35.Rawls, J. F., B. S. Samuel, and J. I. Gordon. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 101:4596-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Development Core Team. 2005. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 37.Sakurai, M., R. Koga, T. Tsuchida, X. Y. Meng, and T. Fukatsu. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer, R. T. 1986. Leech biology and behavior. Clarendon Press, Oxford, United Kingdom.
- 39.Sonnenburg, J. L., J. Xu, D. D. Leip, C. H. Chen, B. P. Westover, J. Weatherford, J. D. Buhler, and J. I. Gordon. 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307:1955-1959. [DOI] [PubMed] [Google Scholar]
- 40.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trontelj, P., and S. Y. Utevsky. 2005. Celebrity with a neglected taxonomy: molecular systematics of the medicinal leech (genus Hirudo). Mol. Phylogenet. Evol. 34:616-624. [DOI] [PubMed] [Google Scholar]
- 42.Worthen, P. L., C. J. Gode, and J. Graf. 2006. Culture-independent characterization of the digestive-tract microbiota of the medicinal leech reveals a tripartite symbiosis. Appl. Environ. Microbiol. 72:4775-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 100:10452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, H., D. Schmitt-Wagner, U. Stingl, and A. Brune. 2005. Niche heterogeneity determines bacterial community structure in the termite gut (Reticulitermes santonensis). Environ. Microbiol. 7:916-932. [DOI] [PubMed] [Google Scholar]