Abstract
A thorough understanding of the microorganisms and pathogens associated with the larval stage of the tropical ornate rock lobster, Panulirus ornatus, is required to overcome disease outbreaks that currently block aquaculture attempts. This study used microscopy in addition to culture and molecularly based microbiological techniques to characterize the bacterial community associated with cultured, developmental stage PI to PII P. ornatus phyllosomas. Scanning electron microscopy demonstrated colonization of phyllosomas by filamentous, rod-shaped, and coccus-shaped bacteria. A clone library constructed from dead phyllosomas sampled from the larval rearing tank on day 10 was dominated by Thiothrix-affiliated sequences (56% of clones). A comparable library from live phyllosomas also contained Thiothrix-affiliated sequences, though these only represented 19% of clones within the library. Fluorescent in situ hybridization (FISH) confirmed identification of the filamentous bacteria as Thiothrix sp., being present on dead phyllosomas. FISH also identified Leucothrix sp. and Vibrio sp., as well as a range of other rod- and coccus-shaped bacteria, colonizing both live and dead phyllosomas. The development of the microbial community associated with phyllosomas was monitored through a standard larval rearing run using denaturing gradient gel electrophoresis (DGGE). Vibrio sp.-affiliated bands dominated the profiles of live animals through the rearing period and dead phyllosomas sampled on selected days. The population of Vibrio sp. associated with phyllosomas was monitored with culture-based analysis on selective media and demonstrated to increase significantly on day 7, coinciding with the beginning of the larval molt. An isolated Vibrio harveyi strain demonstrated an identical 16S rRNA sequence with retrieved DGGE and clone library sequences. Colonization of phyllosomas with filamentous bacterial species potentially hinders the ability of the animals to molt and, combined with the added stress of the molt process, likely results in reduced immune function, allowing opportunistic pathogenic Vibrio sp. to cause larval mortalities.
Market forces have demanded an increased research investment into the culture of the tropical ornate rock lobster, Panulirus ornatus (1). However, a number of constraints, including an extended larval phase (34), a lack of suitable live and artificial feeds, and disease (5, 58, 73) have prevented the establishment of this aquaculture industry. During larval development, P. ornatus typically undergoes 20 or more molts over the extended larval period of approximately 160 days (33). This period is far greater than that of other commonly cultured crustaceans such as penaeid prawns, which typically undergo 12 molts over approximately 15 days before reaching postlarval stages (30). The long larval cycle of phyllosomas accentuates difficulties associated with controlling disease. Phyllosoma, like most crustacean larvae, are particularly susceptible to microbial infection, especially during the molt process, prior to the new shell hardening and subsequently providing some protection against pathogen invasion (41, 74). In addition, the culture environment tends to expose larvae to a variety of stresses that can compromise the larvae's immune system (70). Antibiotic and chemical treatments commonly used in aquaculture environments remove some but not all bacteria associated with phyllosomas, effectively selecting for certain organisms and accentuating the dynamics of the microbial community directly associated with phyllosomas (5).
Most studies examining the microbiological aspects of crustacean larval rearing have focused on the isolation and identification of pathogenic organisms, with Vibrio sp. being the most commonly reported pathogen (13, 21, 23, 24, 37, 39, 40, 51, 53, 62, 63). Previous research has questioned whether Vibrio spp. are in fact primary pathogens, instead opportunistically proliferating when the immune system of the host has become compromised as a result of other infectious agents or water quality issues (59, 61, 65, 68, 72). Particular species implicated in disease in larval and juvenile rock lobsters have included, V. alginolyticus, V. anguillarum, V. harveyi, and V. tubiashii (13, 23). Filamentous bacteria have also been associated with disease in aquaculture target species, including larval and juvenile rock lobsters. Bourne et al. (5) identified a Thiothrix-affiliated sequence in denaturing gradient gel electrophoresis (DGGE) analysis of early-stage P. ornatus phyllosomas. Diggles et al. (13) and Handlinger et al. (23) reported “Leucothrix-like” bacteria as disease causing organisms in Jasus sp. rock lobsters, while Johnson et al. (29) described an infestation of Leucothrix mucor on the surface of larvae of the lobster Homarus americanus.
Bourne et al. (5) examined the microbial diversity within a P. ornatus larval rearing system and reported a number of bacterial genera associated with phyllosoma larvae, including Alteromonas sp., Desulfobulbus mediterraneus, Pirulella sp., Pseudoalteromonas sp., an uncultured gammaproteobacterium clone, and a number of Vibrio sp. Vibrios were reported to be the dominant organisms with Vibrio parahaemolyticus being the dominant species. Further research reported the proliferation of Vibrio spp. within the hepatopancreas of P. ornatus phyllosomas and associated internal bacterial proliferation with larval mortality (73). In order to help establish P. ornatus as an aquaculture species, it is necessary to have a thorough understanding of the microbial composition and dynamics of the larval rearing system and how these impact on phyllosoma health. Such an understanding will allow a more effective microbial management regime to increase phyllosoma survival. The aim of the present study was to characterize the microbial community associated with cultured, early-stage P. ornatus phyllosomas and to determine how this microbial community changes over time in an attempt to identify bacteria that compromise the animal's health. The study focused on early-stage larval rearing since high mortality is frequently observed in this period.
MATERIALS AND METHODS
Larval rearing technology.
The larval rearing process, including broodstock source, treatment and spawning, tank design, stocking methods, feeding regimes, and water recirculation and treatment regimes, was conducted according to the methods of Bourne et al. (5). During standard larval rearing trials, molting from phyllosoma stages PI to PII occurs between days 7 and 12. Mass larval mortalities are observed during this period and commonly peak on day 10, being characterized by a larger than normal number of dead larvae accumulating on the bottom of the larval rearing tank. Typical rearing attempts result in surviving phyllosomas falling below 1% of the original population within 30 days of commencing the trial (5).
Sample collection.
Live phyllosomas were sampled from the top of the water column by focusing a light source into the larval rearing tank and collecting the attracted animals. Dead phyllosomas (1 to 4 h postmortem) were siphoned from “dead patches” on the bottom of the tank and sampled only on days where patches were observed. For clone library and fluorescence in situ hybridization (FISH) analysis, both live and dead samples were obtained on the day of a larval mass mortality event (day 10). For DGGE analysis, live phyllosomas were collected each day up until 5 days past a larval mass mortality event, and dead phyllosomas were collected on days where dead patches were observed. Live phyllosomas were collected 1 day prior to the PI-PII molt for scanning electron microscopy (SEM) and daily for culture-based analysis up until a larval mass mortality event, at which time dead phyllosomas were also collected. All collected phyllosomas were washed briefly in sterile artificial seawater (ASW) prior to sample processing.
Culture-based microbial analysis.
Vibrionaceae numbers associated with live phyllosomas were monitored daily through a larval rearing trial by collecting triplicate samples, each containing three larvae. Samples were homogenized with a pestle, resuspended in 1 ml of ASW, and serially diluted, and 100 μl from each dilution was inoculated onto thiosulfate-citrate-bile salt-sucrose (TCBS) agar (Difco) in triplicate. Plates were incubated overnight at 28°C, and individual colonies were counted, with the CFU counts standardized per phyllosoma. Plates affected by swarming cultures were not included in the analysis.
On the day of a larval mass mortality event (day 10), both live and dead phyllosomas were collected and processed separately as described above. Serial dilutions were inoculated onto Marine agar (Difco/ Becton Dickinson, Australia) and TCBS agar (Difco), followed by incubation overnight at 28°C. Morphologically distinct colonies were subcultured for later 16S rRNA analysis.
Extraction of genomic bacterial DNA.
Genomic DNA was extracted from isolated bacterial cultures by using a QIAGEN DNeasy tissue kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer's instructions for gram-positive bacteria. Similarly, total bacterial DNA associated with phyllosomas was extracted from approximately 10 homogenized phyllosomas by using a QIAGEN DNeasy tissue kit. All DNA was quantified on a 1.5% agarose gel stained with ethidium bromide (0.5 μg ml−1) and stored at −20°C.
PCR amplification of 16S rRNA genes.
The bacterial 16S rRNA gene was amplified from both isolates and extracted DNA from phyllosomas by using the conserved primers 27f and 1492r as described by Lane (35). For DGGE analysis, PCR on extracted bacterial DNA from phyllosomas was conducted as described by Ferris et al. (14), using the primers 1055f (conserved for domain Bacteria) and 1392r (universal conserved primer) (14). A 40-bp GC-clamp was attached to the 5′ end of the 1392r primer (14, 55). These primers amplify a 323-bp section of the 16S rRNA gene of members of the domain Bacteria, including the highly variable V9 region. All PCRs were performed in either an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany) or a PE Applied Biosystems GeneAmp PCR system 9700 (Perkin-Elmer, Maryland). PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide (0.5 μg ml−1) and stored at −20°C.
Clone library construction analysis.
Amplified bacterial 16S rRNA gene PCR products (27f/1492r) obtained from day 10 live and dead phyllosoma samples were cloned by using a TOPO TA cloning kit (version K2; Invitrogen, California) in accordance with the manufacturer's instructions. A total of 100 clones were randomly selected, and the 16S rRNA gene was reamplified. Clones were screened by restriction fragment length polymorphism (RFLP) analysis of PCR products using the restriction enzymes HhaI and HaeIII (Promega, Wisconsin). RFLP profiles were visualized on a 3% agarose gel and stained with ethidium bromide (1 μg ml−1), and clones were grouped into operational taxonomic units (OTUs). Representative clones of each OTU were selected and grown in Luria-Bertani (LB) ampicillin (50 μg ml−1) broth overnight. Plasmid DNA was extracted by using a QIAprep Spin Miniprep kit (QIAGEN) in accordance with the manufacturer's instructions, quantified, and directly sequenced.
DGGE.
We performed DGGE by using an INGENY phorU-2 (Ingeny International BV) DGGE system. PCR products were run on a 6.5% acrylamide gel with a 50 to 70% linear gradient of urea and formamide and electrophoresed at 60 V and 60°C for 20 h. Denaturing gradient gels were stained, destained, and photographed as described by Payne et al. (58). Distinct bands were excised, and DNA was eluted overnight at 4°C in 100 μl of nuclease-free water (Ambion) and reamplified by PCR as previously described. DGGE was performed on reamplified bands to check the mobility against original excised bands, and original PCR products of correct mobility were purified and directly sequenced.
Sequencing and phylogenetic analysis.
Partial and complete 16S rRNA gene sequences of bacterial clones were obtained by using the primers 27f, 339f, 732f, and 1492r (35). DGGE PCR products were sequenced with the primer 1055f (14). Automated sequencing was performed using either Amersham DYEnamic ET-terminator sequencing dye (Amersham Biosciences) or ABI BigDye terminator v3.1 sequencing dye (Applied Biosystems, California) in accordance with the manufacturer's instructions. Sequences were checked for chimera formation with the CHECK_CHIMERA software of the Ribosomal Database Project (48). Sequence data were aligned to the most similar sequence by using the BLAST database algorithm (3) and then further analyzed with the ARB software package (43). Tree topologies were evaluated by reconstructing phylogenies using evolutionary distance (PHYLIP distance method with the Jukes and Cantor model) analysis of aligned near full-length sequences (>1,000 bp) (42). Regions of ambiguous sequence were removed from the analysis. Aligned, partial 16S rRNA sequences (<1,000 bp) were subsequently inserted without changing the overall tree topology using the parsimony tool available within ARB. Bootstrap values were obtained for branching patterns by using the PHYLIP software package (version 3.65) (60), and values of ≥50% were included for the main nodes of the tree. The nucleotide sequence data of all isolates, DGGE bands, and clones appear in the GenBank nucleotide database under the accession numbers DQ831086 to DQ831095 and DQ831116 to DQ831122, DQ831072 to DQ831085 and DQ831109 to DQ831114, and DQ831043 to DQ831071 and DQ831096 to DQ831108, respectively.
SEM.
Day 9, PI phyllosomas were fixed in 3% glutaraldehyde diluted with 0.22-μm-pore-size-filtered ASW and left overnight at 4°C. The glutaraldehyde solution was removed and replaced with ASW twice, and fixed larvae were stored at 4°C. Prior to SEM analysis, larvae were washed in ASW three times at 10 min for each wash. Larvae were then fixed in 1% osmium tetroxide in 0.1 M cacodylate buffer (2 h at 4°C), washed in Milli-Q (Millipore) water (3 × 10 min) and dehydrated in an ethanol series (50% to 100%), followed by a 1:1 (vol/vol) ethanol-hexamethyldisilazane and 3 × 100% ethanol-hexamethyldisilazane series. Larvae were sputter coated with platinum at 25 mA for 150 s in a Balzars MFD 020 sputter coating unit. Specimens were examined in a JEOL 6400F scanning electron microscope at 10 kV.
FISH.
Day 10, PII phyllosomas were suspended in a 4% paraformaldehyde-phosphate-buffered saline solution and stored in darkness at 4°C overnight. After fixation, the larvae were washed three times in a 1:1 (vol/vol) phosphate-buffered saline-ethanol buffer and stored at 4°C until required. Larvae were placed onto a slide and hybridization solution containing the appropriate amount of formamide, and 50 ng of each probe was added, followed by incubation at 46°C for 2 to 3 h. Larvae were individually washed for 10 to 15 min in 50 ml of an appropriate wash buffer preheated to 48°C, rinsed with Milli-Q water, and filtered onto a 0.2 μM GTTP filter (Millipore) for orientation. Larvae were covered with fluorescence microscope oil (Zeiss), and a coverslip was placed on top and viewed and imaged on a Bio-Rad MRC-1024 confocal laser scanning microscope. The illumination source was a 15-mW argon-krypton laser (American Laser Corp.) with excitation peaks at 488 nm (blue), 568 nm (green), and 647 nm (red). The images were captured in three different photomultiplier tubes to distinguish between the probe conferred fluorescence and autofluorescence of the phyllosoma tissue. A 560-nm long-pass emission filter separated the green signal (fluorescein isothiocyanate) from the red signal, and a 640-nm short-pass emission filter separated the far-red signal (from Cy5) from the near-red signal (from Cy3). The confocal laser scanning microscope was controlled by an OS-2 PC running the Bio-Rad LaserSharp software package. Images were collected, and the final image evaluation was done in Adobe PhotoShop version 7.0. The green emission was presented in the green channel of the color image, the red emission was presented as the red channel and, by convention, the far-red emission (Cy5) was presented as the blue channel. The various probes, probe combinations, their respective formamide levels, and fluorochromes are provided in Table 1. The specificity of each probe was established by previous studies and checked against relevant controls with specificity references provided in Table 1.
TABLE 1.
Oligonucleotide probes used for FISH
| Probe | Sequence | rRNA target site (position) | Specificity | % Formamide | Source or reference |
|---|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | 16S (338-355) | Bacteria | 20 | 3a |
| GV | AGGCCACAACCTCCAAGTAG | 16S (841-822) | Vibrionaceae | 30 | 18a |
| G123T | CCTTCCGATCTCTATGCA | 16S (697-714) | Thiothrix sp. | 40 | 31 |
| LMU | CCCCTCTCCCAAACTCTA | 16S (652-669) | Leucothrix mucor | 35 | 72a |
| NonEUB338 | ACTCCTACGGGAGGCAGC | 16S (338-355) | Bacteria (negative control probe) | 20 | 3a |
Statistical analysis.
Culture-based bacterial counts were analyzed by using a one-way analysis of variance in SPSS version 10. Homogeneity of variances for the data set was not assumed. The data were transformed by using a log10 transformation, graphed on a histogram, and a normality curve was applied to check the normal distribution of the data. Where significant differences were recorded, data were further analyzed by using a Dunnett T3 post-hoc test.
Clone library data were analyzed by using various indices and models in order to give an indication of the variation of microbial diversity within the clone library (47). The freeware program, EstimateS, version 7.5 (10), was used to calculate the following indices: the Shannon-Weaver index (64), calculated as:
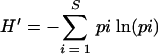 |
where pi is the proportion of clones belonging to the ith OTU and S is the total number of OTUs (52); the Fisher alpha log series richness index (15); and Chao1 (8, 9), calculated based on the number of species in a sample that are represented by one or two individuals (16). Rarefaction analysis was conducted on the clone library (25, 28, 67), and a rarefaction curve was produced by using the analytical approximation algorithm of Hurlbert (28). Calculations were performed on a personal computer with the freeware program aRarefact Win (26). Coverage (C) values were calculated by the equation C = [1 − (n/N)] × 100, where n is the number of unique clones, and N is the total number of clones examined (20).
RESULTS
SEM analysis.
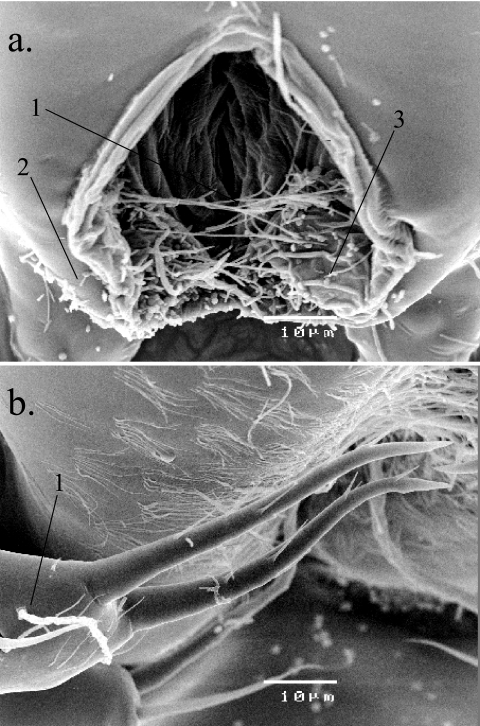
Analysis of live day 9 PI phyllosomas (1 day premolt) by SEM revealed colonization of the phyllosoma exterior surface by a range of bacteria. Filamentous, rod-shaped, and coccus-shaped bacteria were observed on the anus of the phyllosoma. Often, individual filaments had become entangled among one another as a result of the colonization (Fig. 1a). Colonization of PI phyllosoma mouthparts by filamentous bacteria was low (Fig. 1b); however, extensive colonization of the mouthparts of later-stage PIII phyllosomas (prior to molting) has been recently observed (4; results not shown). Similar extensive colonization of the carapace of later-stage phyllosomas by filamentous, rod-shaped, and coccus-shaped bacteria has also been commonly observed (4).
FIG. 1.
Colonization of phyllosomas by filamentous, rod-shaped and coccus-shaped bacteria. (a) Scanning electron micrograph of 1 day premolt, PI stage live phyllosoma anus showing colonization by filamentous (1), rod-shaped (2), and coccus-shaped (3) bacteria. (b) Scanning electron micrograph of 1 day premolt, PI stage live phyllosoma mouthparts showing colonization by filamentous bacteria (1).
Clone library analysis.
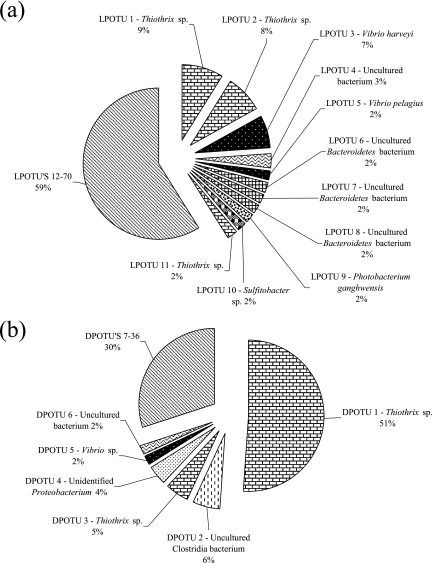
RFLP analysis of two clone libraries (100 clones each) constructed from live and dead phyllosomas collected on day 10 of a larval rearing trial identified 70 and 36 independent OTUs, respectively (Fig. 2). Representatives of OTUs that contained two or more clones were sequenced, and this consisted of 11 from the live phyllosoma library (LPL) and 6 from the dead phyllosoma library (DPL). The DPL was dominated by a sequence affiliated with a Thiothrix sp. that represented 51% of the library. A sequence affiliated with this same Thiothrix sp. also represented the largest OTU of the LPL (9%). OTUs 2 and 11 from the LPL and OTU 3 from the DPL were also affiliated with Thiothrix species, and each represented 8, 2, and 5% of their respective libraries. In addition, Vibrio sp.-affiliated sequences were present in both libraries, with OTU 3 from the LPL being closely affiliated with Vibrio harveyi (7% of the library) and OTU 5 affiliated with Vibrio pelagius (2% of the library). Within the DPL library, OTU 5, representing 2% of clones, was most closely affiliated with a Vibrio sp. Another retrieved sequence of interest, OTU 2, represented 6% of the DPL library and was affiliated with a strict anaerobe marine Clostridia bacterium. Within the LPL, OTU 10 represented 2% of the library and was affiliated with the sulfite-oxidizing bacterium, Sulfitobacter sp.
FIG. 2.
Frequencies of OTUs detected in 16S rRNA clone libraries derived from live (a) and dead (b) phyllosoma samples. Sequence affiliations are provided for OTUs representative of ≥2% of the total number of clones in the library. Calculations were made based upon the total number of clones associated with an OTU from which one or more representative clones had been sequenced.
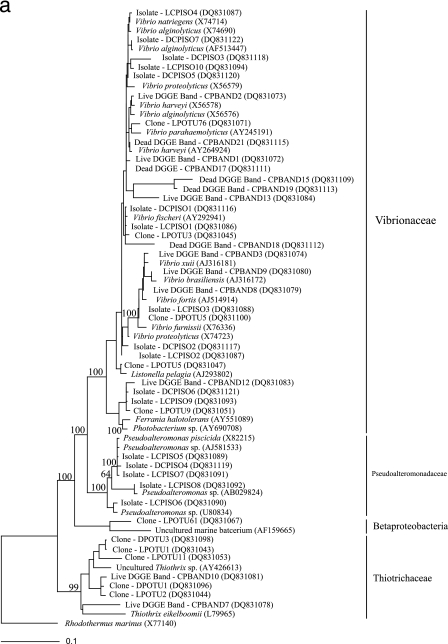
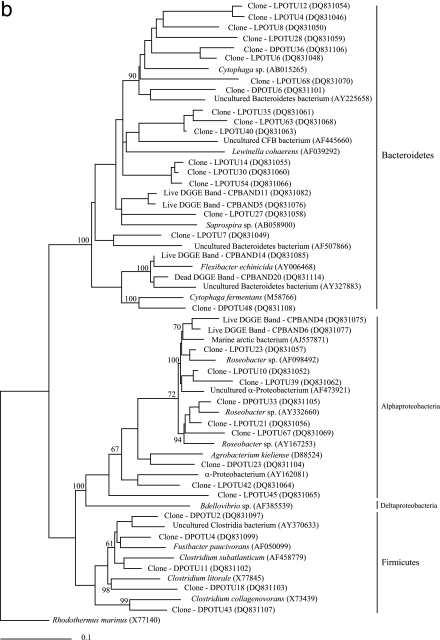
A further 18 clones from the LPL and 7 clones from the DPL, representing individual OTUs, were randomly selected and sequenced to provide additional phylogenetic information on the microbial community associated with live and dead phyllosomas. Within the DPL, three additional clones were affiliated with Clostridiales bacteria: Clostridium halophilum (OTU 11), Clostridium litorale (OTU 18), and Clostridium collagenovorans (OTU 43). A V. harveyi-affiliated sequence was retrieved from the LPL, along with several Alphaproteobacteria-related sequences, including Roseobacter pelophilus (OTU 21) and two Rhodobacteraceae bacteria (OTUs 23 and 39). Alphaproteobacteria-affiliated sequences were also retrieved from the DPL (OTU 23 [Mesorhizobium sp.] and OTU 33 [Roseobacter sp.]). Phylogenetic affiliations for all OTUs sequenced within these libraries are provided in Fig. 3, with most falling in the Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes, and Firmicutes.
FIG. 3.
Phylogenetic tree of 16S rRNA gene sequences recovered from bacterial isolates, clone libraries, and DGGE analysis of live and dead early stage P. ornatus phyllosomas taken from the larval rearing system. (a) Gammaproteobacteria; (b) sequences within the families Alphaproteobacteria, Betaproteobacteria, Epsilonproteobacteria, and the Bacteroidetes group. The scale bar represents 0.1 changes per nucleotide. GenBank accession numbers are provided for all isolate, clone, DGGE, and reference sequences. The proportion of each clone within the clone library is represented in brackets after the clone name. The locations of the DGGE bands are given in Fig. 4. Partial isolate, clone, and DGGE sequences were added to the phylogenetic tree by using the parsimony algorithm tool in the ARB software package (43). Rhodothermus marinus was used as an outgroup for the analysis. Bootstrap values of ≥50% are represented at the nodes of branching points.
Calculated diversity indices (LPL/DPL [Shannon-Weaver H′, 3.97/2.32; Fisher alpha, 103.64/20.17; and Chao-1, 283.87/181]), the percentage of library coverage (LPL, 31%; DPL, 64%), and the absence of a visible asymptote in the LPL rarefaction analysis curve (figure not shown) confirmed higher bacterial diversity within the LPL than within the DPL, supporting the larger number of RFLP patterns detected within this library. All indices for the LPL confirmed that analysis of a larger number of clones would be required to obtain a better representative picture of the bacterial diversity within this sample.
DGGE analysis.
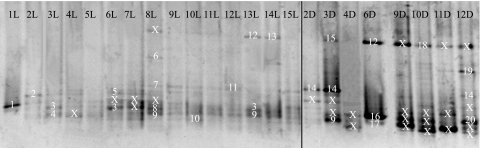
Vibrio sp.-affiliated sequences dominated DGGE profiles of both live phyllosoma (LP) and dead phyllosoma (DP) (LP bands 1 to 3, 8, 9, 12, and 13; DP bands 15 to 18 and 20) (Fig. 4), being present on most days and constituting ca. 54% of retrieved sequences obtained from LP bands and 60% of retrieved sequences obtained from DP bands. Interestingly, a sequence was retrieved from the day 10 LP profile and affiliated with a Thiothrix sp. (band 10) (Fig. 4), demonstrating high sequence identity with clone sequences that dominated the LP and DP libraries. Based upon bands of similar mobility for which no sequence information was obtained, its appearance coincided with the day prior to the start of the PI to PII molt (day 6) and was then present for the remainder of the larval rearing trial. Bands of similar mobility were present in the DP profile, although the sequence information for these bands was not obtained. Further sequences retrieved from the LP profile affiliated with Methylobacter marinus (band 7) (Fig. 4) and an alphaproteobacterial sequence related to the Rhodobacteriaceae bacterium (band 4) (Fig. 4) and closely affiliated with retrieved sequences from the LP clone library. Other sequence affiliations obtained from the DGGE analysis of LP and DP were related to the Sphingobacteria, Alphaproteobacteria, and Flavobacteria groups (Table 2 and Fig. 3). Interestingly, in both LP and DP DGGE profiles there were no distinct changes in the microbial community observed between days 9 and 10, when high larval mortality was observed within the larval rearing tank, although a new band appeared on day 8 but was absent from the profile the next day. No sequence information could be obtained for this band.
FIG. 4.
DGGE profile of 16S rRNA gene fragments of live (L) and dead (D) early-stage P. ornatus phyllosomas taken over the duration of a larval rearing trial. The numbers at the top of the figure (in black) represent days. The numbers in the main body of the figure (in white) represent the bands that were cut from the gel and sequenced. These sequences are present in the phylogenetic tree in Fig. 3. Bands marked with the symbol “X” represent bands for which sequence data could not be obtained.
TABLE 2.
| Band no. | DNA source | Closest relative (database accession no.) | Alignment (bp) | Similarityb (%) | Taxonomic description |
|---|---|---|---|---|---|
| 1 | Live phyllosoma | Vibrio corallilyticus, strain LMG 21349 (AJ440004.1) | 327/327 | 100 | Gammaproteobacteria |
| 2 | Live phyllosoma | Vibrio harveyi, strain LB4 (DQ146935.1) | 332/334 | 99 | Gammaproteobacteria |
| 3 | Live phyllosoma | Vibrio campbellii, strain 90-69B3 (AY738129.1) | 323/325 | 99 | Gammaproteobacteria |
| 4 | Live phyllosoma | Rhodobacteraceae bacterium 183, strain 183 (AJ810844.1) | 158/163 | 96 | Alphaproteobacteria |
| 5 | Live phyllosoma | Saprospira sp., strain SS03-4 (AB191040.1) | 203/222 | 91 | Sphingobacteria |
| 6 | Live phyllosoma | Roseivivax sp. strain NT N52 (AB166990.1) | 165/169 | 97 | Alphaproteobacteria |
| 7 | Live phyllosoma | Methylobacter marinus A45 (AF304197.1) | 252/265 | 95 | Gammaproteobacteria |
| 8 | Live phyllosoma | Vibrio natriegens, strain 01/252 (AJ874353.1) | 315/315 | 100 | Gammaproteobacteria |
| 9 | Live phyllosoma | Vibrio harveyi, strain S35 (AY750578.1) | 321/321 | 100 | Gammaproteobacteria |
| 10 | Live phyllosoma | Uncultured Thiothrix sp. clone UP23b (AY426613.1) | 308/315 | 97 | Gammaproteobacteria |
| 11 | Live phyllosoma | Saprospira sp. SS03-4 (AB191040.1) | 246/264 | 93 | Sphingobacteria |
| 12 | Live phyllosoma | Vibrio olivaceus (AY827492.1) | 71/73 | 97 | Gammaproteobacteria |
| 13 | Live phyllosoma | Vibrio hispanicus, strain LMG 13213 (AY254042.2) | 256/269 | 95 | Gammaproteobacteria |
| 14 | Dead phyllosoma | Flexibacter echinicida, strain F11 (AY006470.1) | 315/315 | 100 | Sphingobacteria |
| 15 | Dead phyllosoma | Vibrio vulnificus (AY264936.1) | 184/198 | 92 | Gammaproteobacteria |
| 16 | Dead phyllosoma | Vibrio proteolyticus (AF513463.1) | 323/323 | 100 | Gammaproteobacteria |
| 17 | Dead phyllosoma | Vibrio shilonii strain MP-3 (AY911392.1) | 285/300 | 95 | Gammaproteobacteria |
| 18 | Dead phyllosoma | Vibrio vulnificus, strain 1003(O) (AY676129.1) | 193/202 | 95 | Gammaproteobacteria |
| 19 | Dead phyllosoma | Psychroserpens mesophilus, strain KOPRI 13650 (DQ001321.1) | 301/315 | 95 | Flavobacteria |
| 20 | Dead phyllosoma | Vibrio harveyi, strain LB4 (DQ146935.1) | 315/315 | 100 | Gammaproteobacteria |
FISH analysis.
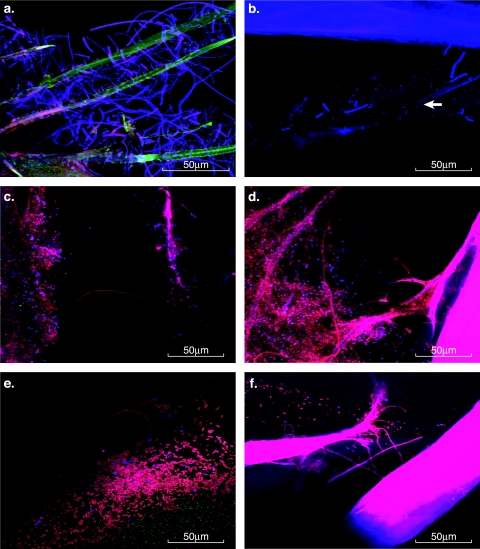
Probing of whole phyllosomas with the fluorescently labeled domain Bacteria probe EUB338 demonstrated extensive colonization by filamentous bacteria. Based upon the large percentage of retrieved clone sequences, a fluorescently labeled oligonucleotide probe targeting Thiothrix species (G123T) was used to probe whole animals and confirmed that the extensive colonization by filamentous bacteria on the carapace and appendages (Fig. 5a) of dead phyllosomas sampled on day 10 were Thiothix-related organisms. Surprisingly, analysis of live PII phyllosomas sampled on day 10 did not reveal Thiothrix sp. filaments, despite the presence of retrieved sequences in the corresponding clone library. A Leucothrix mucor specific probe (LMU) demonstrated the presence of sporadic numbers of L. mucor filaments on the carapace and appendages of dead phyllosomas (Fig. 5b). FISH analysis of live phyllosomas with an LMU probe did not detect the presence of L. mucor filaments on live, day 10, PII phyllosomas. Probing of whole animals with a Vibrionaceae-specific probe (GV) identified large numbers of Vibrio sp. cells externally on both live (Fig. 5c) and dead (Fig. 5d) phyllosomas. Unidentified bacteria, comprised largely of rods and to a lesser extent cocci, were also present on the eyestalks (Fig. 5e), limbs, and carapaces of dead phyllosomas and the appendages and carapaces of live phyllosomas. Analysis of the limbs of some live phyllosomas with a EUB338 probe demonstrated low numbers of filamentous bacteria (Fig. 5f), although since these specific animals were not probed with the Thiothrix-specific probe (G123T), it is unknown whether these filaments are indeed Thiothrix-related species consistent with sequences retrieved from the clone library analysis.
FIG. 5.
FISH micrographs of whole animal mounts of P. ornatus phyllosomas. (a) FISH image of a dead phyllosoma limb probed with a Cy3-labeled domain Bacteria-specific probe (EUB338) and a Cy5-labeled Thiothrix sp.-specific probe (G123T). Bacteria on the surface of the animal appear pink; Thiothrix sp. bacteria protruding from the appendages of the animal appear blue. (b) FISH image of a dead phyllosoma carapace probed with a Cy5-labeled Leucothrix mucor-specific probe. L. mucor filaments protruding from the appendages of the animal appear blue. (c) FISH image of a live phyllosoma limb probed with a Cy3-labeled domain Bacteria-specific probe (EUB338) and a Cy5-labeled Vibro sp.-specific probe. Bacteria on the surface of the animal appear pink; Vibrio sp. appear blue. (d) FISH image of a dead phyllosoma eyestalk probed with a Cy3-labeled domain Bacteria-specific probe (EUB338) and a Cy5-labeled Vibro sp.-specific probe. Bacteria on the surface of the animal appear pink; Vibrio sp. appear blue. (e) FISH image of a dead phyllosoma eyestalk probed with a Cy3-labeled domain Bacteria-specific probe (EUB338) and a Cy5-labeled Vibro sp.-specific probe. Bacteria on the surface of the animal appear pink; Vibrio sp. appear blue. (f) FISH image of a live phyllosoma limb probed with a Cy3-labeled domain Bacteria-specific probe (EUB338) and a Cy5-labeled Leucothrix mucor-specific probe. Bacteria on the surface of the animal appear pink; Leucothrix mucor filaments would appear blue.
Culture-based analysis.
CFU counts on TCBS agar for bacteria associated with phyllosomas detected an increase in Vibrionaceae numbers during the early stages of a larval rearing trial. Numbers increased 14% from days 1 to 2 (244 to 284 CFU phyllosoma−1), 29% from days 2 to 4 (284 to 398 CFU phyllosoma−1), and 39% from days 6 to 7 (388 to 635 CFU phyllosoma−1). A large increase of 71% occurred in Vibrionaceae CFU counts from days 7 to 8 (635 to 2,220 CFU phyllosoma−1), coinciding with the beginning of the PI-PII larval molt. Vibrionaceae CFU counts then increased a further 3% from day 8 to day 9 (2,220 to 2,288 CFU phyllosoma−1). Statistical analysis of the data demonstrated a significant difference in CFU counts for days 8 and 9 compared to CFU counts on all other days of the time series (one-way analysis of variance, P = 0.00 and P < 0.05). After day 9, swarming bacterial colonies precluded any further statistically valid CFU data from being obtained.
To provide an indication of the microbial diversity and potential pathogen(s) associated with both live and dead phyllosomas on the day of a larval mass mortality event (day 10), morphologically different bacterial strains were isolated on Marine and TCBS agar (Fig. 3a and Table 3). A total of 10 strains were isolated from live phyllosomas, whereas 7 strains were isolated from dead phyllosomas. The dominant genera isolated were identified as Vibrio sp. (nine isolates), with isolates closely affiliated with V. harveyi (LCPISO1 and DCPISO1) and V. campbellii (LCPISO2 and DCPISO2) being recovered from both live and dead phyllosomas and sharing nearly identical sequence identity. Other isolated Vibrio sp. included V. alginolyticus (LCPISO11), V. natriegens (DCPISO3 and DCPISO5), and V. parahaemolyticus (DCPISO7). Isolates affiliated with the genus Pseudoalteromonas were also prevalent (five isolates), particularly in live phyllosomas (four of five isolates). Other isolated cultures were affiliated with the genus Photobacterium (three isolates).
TABLE 3.
Affiliation of sequences retrieved from bacteria isolated from live and dead cultured phyllosomas based upon a partial (>250-bp) 16S rRNA sequence
| Isolate no.a | Mediumb | Closest relative (database accession no.) | Alignment (bp) | Similarityc (%) | Taxonomic description |
|---|---|---|---|---|---|
| LCPISO1 | TCBS | Vibrio harveyi, strain S35 (AY750578.1) | 936/938 | 99 | Gammaproteobacteria |
| LCPISO2 | TCBS | Vibrio campbellii, strain 90-69B3 (AY738129.1) | 943/944 | 99 | Gammaproteobacteria |
| LCPISO3 | TCBS | Vibrio sp., strain V794 (DQ146993.1) | 937/940 | 99 | Gammaproteobacteria |
| LCPISO4 | MA | Photobacterium phosphoreum (AY292916.1) | 935/939 | 99 | Gammaproteobacteria |
| LCPISO5 | MA | Pseudoalteromonas piscicida (AF081498.1) | 944/945 | 99 | Gammaproteobacteria |
| LCPISO6 | MA | Pseudoalteromonas sp., strain S9 (U80834.1) | 906/915 | 99 | Gammaproteobacteria |
| LCPISO7 | MA | Pseudoalteromonas piscicida (AF297959.1) | 921/923 | 99 | Gammaproteobacteria |
| LCPISO8 | MA | Pseudoalteromonas sp., strain S511-1 (AB029824.1) | 826/826 | 100 | Gammaproteobacteria |
| LCPISO9 | MA | Photobacterium ganghwensis, strain FR1311 (AY960847.1) | 834/835 | 99 | Gammaproteobacteria |
| LCPISO10 | MA | Vibrio alginolyticus, strain CIP 70.65 (X74691.1) | 878/884 | 99 | Gammaproteobacteria |
| DCPISO1 | TCBS | Vibrio harveyi, strain S35 (AY750578.1) | 953/954 | 99 | Gammaproteobacteria |
| DCPISO2 | TCBS | Vibrio campbellii, strain 90-69B3 (AY738129.1) | 963/972 | 99 | Gammaproteobacteria |
| DCPISO3 | MA | Vibrio natriegens, strain 01/097 (AJ874352.1) | 951/954 | 99 | Gammaproteobacteria |
| DCPISO4 | MA | Pseudoalteromonas piscicida (AB090233.1) | 922/928 | 99 | Gammaproteobacteria |
| DCPISO5 | MA | Vibrio natriegens, strain 01/097 (AJ874352.1) | 907/908 | 99 | Gammaproteobacteria |
| DCPISO6 | MA | Photobacterium ganghwensis, strain FR1311 (AY960847.1) | 781/783 | 99 | Gammaproteobacteria |
| DCPISO7 | MA | Vibrio parahaemolyticus, strain MP-2 (AY911391.1) | 831/835 | 99 | Gammaproteobacteria |
LCP, live phyllosoma; DCP, dead phyllosoma.
MA, marine agar; TCBS, thiocitrate, bile salt, sucrose agar.
Sequences were aligned to the closest relative by using BLAST (3). The similarity was calculated with gaps not taken into account.
DISCUSSION
High mortality during early larval stages, particularly upon initial stocking and around molt periods, is commonly associated with larval rearing of the tropical rock lobster, Panulirus ornatus (5). To date, opportunistic bacterial pathogens, particularly Vibrio sp., have been implicated as a potential cause of larval mortalities (4, 5, 73). The results from the present study demonstrated colonization by filamentous bacteria as a contributing detrimental factor to phyllosoma survival. SEM identified filamentous bacteria on phyllosomas, and FISH confirmed heavy infestation of dead animals on day 10 by Thiothrix sp. bacteria. Clone library analysis supported Thiothrix identification with 56% of retrieved sequences from the library of dead phyllosomas sampled on day 10 affiliating with this organism. In addition, 19% of clones from the live phyllosoma library also sampled on day 10 demonstrated sequence identity to Thiothrix sp. and a partial 16S rRNA sequence closely affiliated with a Thiothrix sp. was retrieved from the DGGE analysis of live phyllosomas on this day. Due to the detection limit of DGGE, it is believed that only predominant species present in a microbial community can be detected (18, 45, 54, 55, 57), suggesting that Thiothrix sp. are also a dominant organism associated with live phyllosomas. Recent work by Bourne et al. (4) used SEM to monitor the progressive colonization of filamentous bacteria in later-stage PIII phyllosomas. Extensive colonization and entanglement of mouth parts were observed and believed to render the animal unable to feed adequately. Heavy epibiont growth has also been attributed to increased phyllosoma mortality due to a reduction of respiratory effectiveness as demand for oxygen increases during molt periods (5, 11, 12, 23). In addition, such colonization has compromised animal health in other crustacean species with reports of penaeid prawns affected by the filamentous bacteria, Leucothrix mucor (38).
As phyllosomas molt from stage PI to PII they shed the outer shell allowing colonization organisms to also be discarded. Dead phyllosomas sampled on day 10 are stage PI animals that have failed to successfully molt with heavy bacterial colonization likely contributing to mortality. In contrast, live animals sampled on day 10 have successfully shed the outer shell resulting in lower colonization. This contributed to the inability to detect Thiothrix sp. during FISH analysis despite the retrieval of sequences in the corresponding clone library. FISH analysis was also limited to a small number of individual animals (∼5), whereas clone library analysis was a pooled sample of multiple phyllosomas (∼10), allowing a greater representative view of bacterial diversity. Some phyllosomas analyzed with FISH and the EUB338 probe detected filamentous bacteria associated with live phyllosomas, although unfortunately these animals were not probed with the Thiothrix-specific probe to confirm phylogenetic identification of these filaments. Many unknown filamentous bacteria which did not hybridize to either Thiothrix sp. or Leucothrix mucor specific probes were also detected on both live and dead sampled phyllosomas. Although a large number of bacteria have a filamentous morphology it is acknowledged the probes used in the present study do not cover all Thiothrix species with a search of Ribosomal Database Project database showing only 80 out of 97 Thiothrix sp.-related sequences are specific for the G123T probe. Although molt shedding reduces the microbial colonization load, it appears to maintain microbial diversity associated with the phyllosomas since indices and rarefaction analysis demonstrated greater microbial diversity associated with live phyllosomas compared to dead animals. Although appearing counterintuitive it is likely that dominant bacteria, in this case, Thiothrix sp., proliferate on phyllosoma postmortem. In addition, high bacterial diversity may promote phyllosoma health by preventing the establishment of pathogenic organisms through competition for nutrients and space. Such phenomenon has been previously reported in the gastrointestinal tract of chickens where the mature microbiota confer resistance to infection by Salmonella enterica (50, 75).
Vibrio-affiliated sequences were commonly retrieved from both live and dead phyllosoma samples with clone library and DGGE analysis, while FISH confirmed the presence of Vibrio sp. cells on the external surfaces of phyllosomas on day 10. Vibrio sp. dominated isolated bacterial strains from culture-based studies, whereas specific Vibrio counts demonstrated a significant increase in Vibrionaceae-related organisms from days 7 to 8 correlating with the beginning of the larval molt from PI-PII. This is a time where phyllosomas are particularly susceptible to infection (41) as a result of the phyllosoma's external barriers to pathogen invasion being compromised (73, 74). Previous studies have implicated Vibrio sp. as potential pathogens associated with P. ornatus and Jasus verreauxi phyllosoma mortalities (5, 13, 22, 23, 73). Vibrio sp. derived sequences from each analysis method demonstrated a high degree of identity, with one Vibrio harveyi-affiliated organism isolated from both live and dead phyllosomas and sequences retrieved from live phyllosoma DGGE analysis and represented as 7% of the live phyllosoma clone library. V. harveyi has been implicated as a pathogen in phyllosomas of the New Zealand spiny lobster, Jasus verreauxi (13) and also as a pathogen among prawn larvae (24, 37, 39, 40, 51, 53). In addition, sequences affiliated with another known crustacean pathogen, Vibrio campbellii, previously associated with disease in the pacific oyster (66), abalone (44), and in various penaeid prawn species (7, 21, 27, 62, 63), were identified from isolates recovered from both live and dead phyllosomas and in live phyllosoma DGGE analysis. Bands showing similar mobility to the live phyllosoma-V. campbellii-affiliated band were also present in dead phyllosoma DGGE analysis.
Numerous clone sequences were retrieved from phyllosomas that affiliated with marine microbial strains involved in nutrient cycling processes, including the methanotrophic bacteria Methylobacter marinus (LP DGGE band 7) and the nitrogen-fixing root nodule bacterium Mesorhizobium sp. (DP OTU 23). Sulfur cycling appears to be an important process within the aquaculture rearing environment, as indicated by the dominance of retrieved Thiothrix sp. sequences, as well as the presence of clostridium-related sequences and a Sulfitobacter sp. sequence. Thiothrix spp. are sulfur-oxidizing bacteria that have been described in a number of environments, including activated-sludge wastewater treatment plants (31, 56), and as epibionts on a number of aquatic invertebrates (6, 17, 36, 71). Thiothrix spp. oxidize sulfide compounds, such as hydrogen sulfide (H2S), to sulfate (SO4). The presence of Sulfitobacter sp. indicates the presence of sulfite (SO2), which is a nutrient requirement of this organism, being oxidized to produce SO4 (32, 69). Environments that contain sulfite may have oxygen-deficient zones that are likely to be toxic to aquatic species (19). Clostridium spp. are strict anaerobes commonly found in soil environments (46) but also isolated from marine systems (2, 49) and able to utilize a range of compounds as energy sources, such as cellulose, sugars, starch, and pectin (46), but of interest to the present study are those that obtain their energy by utilizing amino acids. Clostridium spp. ferment individual amino acids or amino acid pairs, which results in the production of a range of foul-smelling substances, including H2S (46), potentially acting as a substrate for Thiothrix sp. No sequences were obtained that affiliated with sulfate-reducing bacteria (SRB), a group widespread in aquatic and terrestrial environments that become anoxic through microbial decomposition processes (46). SRB are an important group of bacteria in sulfur cycling that reduce the sulfate produced by such organisms as Thiothrix sp. and Sulfitobacter sp. back to sulfide, allowing the cycle to continue. It is likely that SRB, such as Desulfovibrio sp., are present within the larval rearing system, but closer analysis showed that this group of bacteria was poorly represented by the primers used in the present study. Further analysis of the larval rearing system using primers that target the SRB is required to gain an understanding of the genera and species present that represent this group.
Phyllosomas sampled in the present study were defined as “living” as opposed to “healthy” since it is unknown whether the animals are compromised even though they still possess the ability to swim in the water column. Because phyllosomas are small and transparent, high numbers of “dead” phyllosomas could only be sampled from the large 5,000-liter larval rearing tanks by siphoning accumulated dead patches. Although microbial turnover is possible on the dead phyllosomas, dead animals were sampled as soon as possible postmortem (1 to 4 h), and only selected phyllosomas that were not showing obvious signs of decay were incorporated into the analysis. As a result, microbial community changes postmortem are expected to be minimal, and dominant members of the community will be maintained.
One challenge for successful aquaculture of the tropical rock lobster is to establish larval rearing technology on a sufficient scale to make it commercially viable. As a result, larval rearing trials are undertaken in 5,000-liter tanks which, although good for commercial applications, make replication difficult. The results presented in the present study are from one trial, although representative of results seen from other larval rearing runs. This is confirmed by previous published studies that have found both Thiothrix- and Vibrio-affiliated organisms within the larval rearing environment (4, 5, 73). The present study is unique in that it has provided an in-depth understanding of the community dynamics associated with the phyllosomas, whereas other studies have focused on broader microbial changes within the whole system.
Extensive Thiothrix sp. colonization appears to have a negative effect on the health of early-stage phyllosomas by hindering their ability to molt from stage PI to stage PII. Studies of later-stage phyllosomas (PIII to PIV) have recently also confirmed heavy epibiont fouling compromising animal health and supporting the findings of the present study (4). The period around the molt is a time when phyllosomas are particularly susceptible to infection by pathogenic bacteria such as Vibrio sp. (41, 73) as a result of the new shell of the phyllosomas being soft after the shedding of the old carapace (73, 74). This is also a time when Vibrio sp. numbers have been reported to dramatically increase within phyllosomas, resulting in subsequent mass larval mortalities (73). In order to control Thiothrix sp. proliferation, sulfide compounds within the larval rearing system need to be lowered or removed. Ozonation of all incoming larval rearing water may serve as an effective Thiothrix sp. control mechanism by reducing sulfide compounds through the removal of bacteria that produce such compounds. Isolated V. harveyi and V. campbellii strains will also be tested to determine whether they exhibit a significant pathogenic effect on phyllosomas based upon the satisfaction of Koch's postulates.
Acknowledgments
We thank Matt Kenway, Don Booth, Jane Gioffre, Grant Milton, and Justin Hochen (Australian Institute of Marine Science) for their assistance in the larval rearing of phyllosomas. We thank Lone Høj (Australian Institute of Marine Science) for helpful scientific advice and discussion.
Financial support for this research was received from Jeff McCloy.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.ABARE. 2003. Australian Fisheries Statistics 2002 Canberra. ABARE, Canberra, Australia.
- 2.al Saif, N., and J. S. Brazier. 1996. The distribution of Clostridium difficile in the environment of South Wales. J. Med. Microbiol. 45:133-137. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaeffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourne, D., N. Webster, M. Payne, L. Hoj, M. Skindersoe, and M. Hall. Towards the development of rock lobster aquaculture: aspects of the microbiology of phyllosoma rearing of the ornate rock lobster Panulirus ornatus. Aquaculture, in press.
- 5.Bourne, D. G., N. Young, N. Webster, M. Payne, M. Salmon, S. Demel, and M. Hall. 2004. Microbial community dynamics in a larval aquaculture system of the tropical rock lobster, Panulirus ornatus. Aquaculture 242:31-51. [Google Scholar]
- 6.Brigmon, R. L., and C. De Ridder. 1998. Symbiotic relationship of Thiothrix spp. with an echinoderm. Appl. Environ. Microbiol. 64:3491-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgents, J. E., K. G. Burnett, and L. E. Burnett. 2005. Effects of hypoxia and hypercapnic hypoxia on the localization and the elimination of Vibrio campbellii in Litopenaeus vannamei, the Pacific White Shrimp. Biol. Bull. 208:159-168. [DOI] [PubMed] [Google Scholar]
- 8.Chao, A. 1987. Estimating the population-size for capture recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 9.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 10.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples. User's guide and application, version 7.5. [Online.] http://purl.oclc.org/estimates.
- 11.Crear, B. J., and G. N. R. Forteath. 1998. A physiological investigation into methods of improving post capture survival of both the southern rock lobster Jasus edwardsii, and the western rock lobster Panulirus cygnus. FRDC final report, FRDC project no. 1999/315. University of Tasmania, Launceston, Tasmania.
- 12.Diggles, B. K. 1999. Diseases of spiny lobsters in New Zealand, p. 18-34. In L. H. Evans and J. B. Jones (ed.), Conference proceedings of the International Symposium on Lobster Health Management. Curtin University of Technology, Perth, Australia.
- 13.Diggles, B. K., G. A. Moss, J. Carson, and C. D. Anderson. 2000. Luminous vibriosis in rock lobster Jasus verreauxi (Decapoda: Palinuridae) phyllosoma larvae associated with infection by Vibrio harveyi. Dis. Aquat. Org. 43:127-137. [DOI] [PubMed] [Google Scholar]
- 14.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, R. A., A. S. Corber, and C. B. Williams. 1943. The relation between the number of species and the number of individuals in a random sample of an animal population. J. Anim. Ecol. 12:42-58. [Google Scholar]
- 16.Foggo, A., S. D. Rundle, and D. T. Bilton. 2003. The net result: evaluating species richness extrapolation techniques for littoral pond invertebrates. Freshw. Biol. 48:1756-1764. [Google Scholar]
- 17.Ford, P. L., and N. J. Scott. 1996. Thiothrix sp. (Beggiatoaceae) from tadpoles in Western Mexico. Southwest. Nat. 41:328-331. [Google Scholar]
- 18.Gelsomino, A., A. C. Keijzer-Wolters, G. Cacco, and J. D. van Elsas. 1999. Assessment of bacterial community structure in soil by polymerase chain reaction and denaturing gradient gel electrophoresis. J. Microbiol. Methods 38:1-15. [DOI] [PubMed] [Google Scholar]
- 18a.Giuliano, L., M. De Domenico, E. De Domenico, M. G. Hofle, and M. M. Yakimov. 1999. Identification of culturable oligotrophic bacteria within naturally occuring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb. Ecol. 37:77-85. [DOI] [PubMed] [Google Scholar]
- 19.Gobble, M. G., C. M. West, and R. L. Kratz. 2004. Removal of sulfite from incinerator air pollution control. IT3 '04 Conference, May 10-14, 2004, Phoenix, Arizona. RMT, Inc., Madison, WI. http://www.rmtinc.com/public/docs/Sulfite_Oxid_04.pdf.
- 20.Good, I. J. 1953. The population frequencies of species and the estimation to the population parameters. Biometrika 40:237-264. [Google Scholar]
- 21.Hameed, A. S. S., and P. V. Rao. 1994. Studies on the chemical control of a Vibrio campbellii-like bacterium affecting hatchery-reared Penaeus indicus larvae. Aquaculture 127:1-9. [Google Scholar]
- 22.Handlinger, J., J. Carson, A. Ritar, and B. Crear. 2000. A study of diseases in cultured phyllosoma larvae and juveniles of southern rock lobster (Jasus edwardsii). J. Shellfish Res. 19:676. [Google Scholar]
- 23.Handlinger, J., J. Carson, A. J. Ritar, B. J. Crear, D. P. Taylor, and D. Johnston. 1999. Disease conditions of cultured phyllosoma larvae and juveniles of the southern rock lobster (Jasus edwardsii, Decapoda; Palinuridae), p. 75-87. In L. H. Evans and J. B. Jones (ed.), Proceedings of the International Symposium on Lobster Health Management. Curtin University of Technology, Perth, Australia.
- 24.Harris, L. J., and L. Owens. 1999. Production of exotoxins by two luminous Vibrio harveyi strains known to be primary pathogens of Penaeus monodon larvae. Dis. Aquat. Org. 38:11-22. [Google Scholar]
- 25.Heck, K. L. J., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurements and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 26.Holland, S. H. 1988. A rarefactwin program, version 1.2. [Online.] http://www.uga.edu/strata/Software.html.
- 27.Holman, J. D., K. G. Burnett, and L. E. Burnett. 2004. Effects of hypercapnic hypoxia on the clearance of Vibrio campbellii in the Atlantic Blue Crab, Callinectes sapidus Rathbun. Biol. Bull. 206:188-196. [DOI] [PubMed] [Google Scholar]
- 28.Hurlbert, S. H. 1971. The non-concept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, P. W., J. M. Sieburth, A. Sastry, C. R. Arnold, and M. S. Doty. 1971. Leucothrix mucor infestation of benthic crustacea, fish eggs, and tropical algae. Limnol. Oceanogr. 16:962-969. [Google Scholar]
- 30.Jory, D., and T. Cabrera. 2003. Marine shrimp, p. 382-419. In J. S. Lucas and P. C. Southgate (ed.), Aquaculture: farming aquatic animals and plants. Blackwell Publishing, Ltd., London, Great Britain.
- 31.Kanagawa, T., Y. Kamagata, S. Aruga, T. Kohno, M. Horn, and M. Wagner. 2000. Phylogenetic analysis of and oligonucleotide probe development for eikelboom type 021N filamentous bacteria isolated from bulking activated sludge. Appl. Environ. Microbiol. 66:5043-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kappler, U., and C. Dahl. 2001. Enzymology and molecular biology of prokaryotic sulfite oxidation. FEMS Microbiol. Lett. 203:1-9. [DOI] [PubMed] [Google Scholar]
- 33.Kittaka, J. 2000. Culture of the spiny lobster. Blackwell Scientific Publications, Oxford, England.
- 34.Kittaka, J., and F. A. Abrunhosa. 1997. Characteristics of palinurids (Decapoda: Crustacea) in larval culture. Hydrobiologia 358:305-311. [Google Scholar]
- 35.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. S. M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, England.
- 36.Larkin, J. M., M. C. Henk, and S. D. Burton. 1990. Occurrence of a Thiothrix sp. attached to mayfly larvae and presence of parasitic bacteria in the Thiothrix sp. Appl. Environ. Microbiol. 56:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, K. K., S. R. Yu, F. R. Chen, T. I. Yang, and P. C. Liu. 1996. Virulence of Vibrio alginolyticus isolated from diseased tiger prawn, Penaeus monodon. Curr. Microbiol. 32:229-231. [DOI] [PubMed] [Google Scholar]
- 38.Lightner, D. V. 1983. Diseases of cultured penaeid shrimp, p. 289-320. In J. P. McVey and J. R. Moore (ed.), CRC handbook of mariculture, vol. 1. CRC Press, Boca Raton, FL. [Google Scholar]
- 39.Liu, P. C., K. K. Lee, and S. N. Chen. 1996. Pathogenicity of different isolates of Vibrio harveyi in tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 22:413-416. [Google Scholar]
- 40.Liu, P. C., and K. K. Lee. 1999. Cysteine protease is a major exotoxin of pathogenic luminous Vibrio harveyi in the tiger prawn, Penaeus monodon. Lett. Appl. Microbiol. 28:428-430. [DOI] [PubMed] [Google Scholar]
- 41.Liu, C. H., S. T. Yeh, S. Y. Cheng, and J. C. Chen. 2004. The immune response of the white shrimp Litopenaeus vannamei and its susceptibility to Vibrio infection in relation with the moult cycle. Fish Shellfish Immunol. 16:151-161. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 43.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, J., Q. Wang, F. Ma, and M. Liu. 1996. A pathogen of septicopyaemia in the abalone Haliotis discus hannai Ino. Dalian, China. J. Fish. China/Shuichan Xuebao 20:332-336. [Google Scholar]
- 45.Maarit Niemi, R., I. Heiskanen, K. Wallenius, and K. Lindstrom. 2001. Extraction and purification of DNA in rhizosphere soil samples for PCR-DGGE analysis of bacterial consortia. J. Microbiol. Methods 45:155-165. [DOI] [PubMed] [Google Scholar]
- 46.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Biology of microorganisms. Prentice-Hall International, Inc., New York, NY.
- 47.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 48.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1996. The Ribosomal Database Project (RDP). Nucleic Acids Res. 24:82-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, W. A., M. A. Miller, I. A. Gardner, E. R. Atwill, B. A. Byrne, S. Jang, M. Harris, J. Ames, D. Jessup, D. Paradies, K. Worcester, A. Melli, and P. A. Conrad. 2006. Salmonella spp., Vibrio spp., Clostridium perfringens, and Plesiomonas shigelloides in marine and freshwater invertebrates from coastal California ecosystems. Microb. Ecol. 52:198-206. [DOI] [PubMed] [Google Scholar]
- 50.Milner, K. C., and M. F. Schaffer. 1952. Bacteriological studies of experimental Salmonella infections in chicks. J. Infect. Dis. 90:81-96. [DOI] [PubMed] [Google Scholar]
- 51.Montero, A. B., and B. Austin. 1999. Characterization of extracellular products from an isolate of Vibrio harveyi recovered from diseased post-larval Penaeus vannamei (Bonne). J. Fish Dis. 22:377-386. [Google Scholar]
- 52.Moyer, C. B., F. Dobbs, and D. Karl. 1994. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 60:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munro, J., J. Oakey, E. Bromage, and L. Owens. 2003. Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Org. 54:187-194. [DOI] [PubMed] [Google Scholar]
- 54.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 55.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nielsen, P. H., M. A. de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 57.Nikolausz, M., R. Sipos, S. Revesz, A. Szekely, and K. Marialigeti. 2005. Observation of bias associated with re-amplification of DNA isolated from denaturing gradient gels. FEMS Microbiol. Lett. 244:385-390. [DOI] [PubMed] [Google Scholar]
- 58.Payne, M. S., M. R. Hall, R. Bannister, L. Sly, and D. G. Bourne. 2006. Microbial diversity within the water column of a larval rearing system for the ornate rock lobster (Panulirus ornatus). Aquaculture 258:80-90. [Google Scholar]
- 59.Rengpipat, S., S. Rukpratanporn, S. Piyatiratitivorakul, and P. Menasaveta. 2000. Immunity enhancement in black tiger shrimp (Penaeus monodon) by a probiont bacterium (Bacillus S11). Aquaculture 191:271-288. [Google Scholar]
- 60.Retief, J. D. 2000. Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132:243-258. [DOI] [PubMed] [Google Scholar]
- 61.Rosemark, R., and W. S. Fisher. 1988. Vibriosis of lobster, p. 240-243. In C. J. Sindermann and D. V. Lightner (ed.), Disease diagnosis and control in North American marine aquaculture, 2nd rev. Elsevier Science Publishers, B.V., Amsterdam, The Netherlands.
- 62.Sahul Hameed, A. S. 1995. Susceptibility of three Penaeus species to a Vibrio campbellii-like bacterium. J. World Aquacult. Soc. 26:315-319. [Google Scholar]
- 63.Sahul Hameed, A. S., P. V. Rao, J. J. Farmer, F. W. Hickman-Brenner, and G. R. Fanning. 1996. Characteristics and pathogenicity of a Vibrio campbellii-like bacterium affecting hatchery-reared Penaeus indicus (Milne Edwards, 1837) larvae. Aquacult. Res. 27:853-863. [Google Scholar]
- 64.Shannon, C. E., and W. Weaver. 1963. The mathematical theory of communication. University of Illinois Press, Urbana.
- 65.Shao, Z. L. 2001. Aquaculture pharmaceuticals and biologicals: current perspectives and future possibilities. Adv. Drug Deliv. Rev. 50:229-243. [DOI] [PubMed] [Google Scholar]
- 66.Shen, X., Y. Cai, and W. Fang. 2005. Identification of Vibrio campbellii isolated from cultured pacific oyster. Acta Microbiol. Sin. 45:177-180. [PubMed] [Google Scholar]
- 67.Simberloff, D. 1978. Use of rarefaction and related methods in ecology, p. 150-165. In K. L. Dickson, J. J. Cairns, and R. J. Livingston (ed.), Biological data in water pollution assessment: quantitative and statistical analyses. American Society for Testing and Materials, Philadelphia, PA.
- 68.Smith, V. J., J. H. Brown, and C. Hauton. 2003. Immunostimulation in crustaceans: does it really protect against infection? Fish Shellfish Immunol. 15:71-90. [DOI] [PubMed] [Google Scholar]
- 69.Sorokin, D. Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov.: a new heterotrophic bacterium from the black sea specialized on sulfite oxidation. Microbiology 64:295-305. [Google Scholar]
- 70.Takahashi, Y., T. Itami, and M. Kondo. 1995. Immunodefense system of crustacea. Fish Pathol./Gyobyo Kenkyu 30:141-150. [Google Scholar]
- 71.Temara, A., C. De Ridder, J. C. Kuenen, and L. A. Robertson. 1993. Sulfide-oxidizing bacteria in the burrowing echinoid, Echinocardium cordatum (Echinodermata). Mar. Biol. 115:179-185. [Google Scholar]
- 72.Vandenberghe, J., Y. Li, L. Verdonck, J. Li, P. Sorgeloos, H. S. Xu, and J. Swings. 1998. Vibriosis associated with Penaeus chinensis (Crustacea: Decapoda) larvae in Chinese shrimp hatcheries. Aquaculture 169:121-132. [Google Scholar]
- 72a.Wagner, M., R. Amann, H. Lemmer, W. Manz, and K. H. Schleifer. 1994. Probing activated sludge with fluorescently labeled rRNA targeted oligonucleotides. Water Sci. Technol. 29:15-23.
- 73.Webster, N. S., D. G. Bourne, and M. Hall. 2006. Vibrionaceae infection in phyllosomas of the tropical rock lobster Panulirus ornatus as detected by fluorescence in situ hybridization. Aquaculture 255:173-178. [Google Scholar]
- 74.Wibowo, S., S. Putro, S. Sukarto, and S. Wardoyo. 1992. Bacterial load of normal moulting and delayed moulting farmed tiger shrimp (Penaeus monodon Fab.). FAO, Rome, Italy.
- 75.Zhu, X. Y., T. Zhong, Y. Pandya, and R. D. Joerger. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124-137. [DOI] [PMC free article] [PubMed] [Google Scholar]