Abstract
There are only two reports in the literature demonstrating the presence of Campylobacter spp. in marine mammals. One report describes the isolation of a new species, Campylobacter insulaenigrae sp. nov., from three harbor seals (Phoca vitulina) and a harbor porpoise (Phocoena phocoena) in Scotland, and the other describes the isolation of Campylobacter jejuni, Campylobacter lari, and an unknown Campylobacter species from northern elephant seals (Mirounga angustirostris) in California. In this study, 72 presumptive C. lari and unknown Campylobacter species strains were characterized using standard phenotypic methods, 16S rRNA PCR, and multilocus sequence typing (MLST). Phenotypic characterization of these isolates showed them to be variable in their ability to grow either at 42°C or on agar containing 1% glycine and in their sensitivity to nalidixic acid and cephalothin. Based on both 16S rRNA PCR and MLST, all but 1 of the 72 isolates were C. insulaenigrae, with one isolate being similar to but distinct from both Campylobacter upsaliensis and Campylobacter helveticus. Phylogenetic analysis identified two C. insulaenigrae clades: the primary clade, containing exclusively California strains, and a secondary clade, containing some California strains and all of the original Scottish strains. This study demonstrates the inability of phenotypic characterization to correctly identify all Campylobacter species and emphasizes the importance of molecular characterization via 16S rRNA sequence analysis or MLST for the identification of Campylobacter isolates from marine mammals.
Campylobacter spp. are gram-negative, motile spiral rods that can be zoonotic pathogens (15). Many species of Campylobacter, such as C. jejuni and C. lari, are oxidase and catalase positive and grow in thermophilic, microaerophilic conditions (15). Campylobacter spp. can cause gastroenteritis in humans, primates, birds, dogs, cats, cattle, and swine, with symptoms including fever, abdominal cramping, and mild to bloody diarrhea (5, 15, 16). The presence of Campylobacter in saltwater typically indicates recent fecal contamination, because these bacteria are not able to replicate or survive for prolonged periods in surface water (10, 18, 19). Sources of Campylobacter spp. in the environment are sewage treatment plants, agricultural runoff, freshwater outflow, and wildlife (17, 18).
Reports of Campylobacter species isolated from marine mammals are rare: to date, there have been only two reports in the literature. Campylobacter insulaenigrae sp. nov. (hereafter termed C. insulaenigrae) was isolated from three harbor seals (Phoca vitulina) and a harbor porpoise (Phocoena phocoena) in Scotland; the role of this bacterium in disease is not known (3). The isolation of C. jejuni, C. lari, and an unknown Campylobacter species from juvenile northern elephant seals (Mirounga angustirostris) in California was also reported (23).
The northern elephant seal is a large pinniped that spends most of the year in the Pacific Ocean, ranging as far as Alaska and Hawaii, with two periods spent annually on beaches and off-shore islands along the California and Mexican coast (12). Dense seal rookeries form during breeding and pupping (December-February) and molting (March-May) periods (12). Juvenile northern elephant seals are born on the rookeries and are weaned by their mothers after about a month. After fasting for a month, the juvenile seals begin venturing into the ocean in order to forage for food (12). Once juvenile seals must rely on their own abilities for food, they may become stressed and/or sick, making them potentially more susceptible to pathogens present in the environment. Sick juveniles are often found stranded along the California coastline and are then transported to marine mammal rehabilitation centers, such as The Marine Mammal Center (TMMC) in Sausalito, CA, for treatment.
Campylobacter-like organisms were isolated at TMMC from the feces of emaciated and dehydrated juvenile northern elephant seals stranded on nine beaches in northern California, between Bodega Bay and Monterey, during the months of March through May in 2000, 2001, and 2002 (unpublished data). This prompted a two-year study to investigate the prevalence of Campylobacter in both free-ranging and stranded northern elephant seals in northern California (23). C. jejuni, C. lari, and a novel Campylobacter sp. that was distinct from C. insulaenigrae when analyzed using biochemical techniques were identified. The purpose of this study was to provide phenotypic and molecular characterization of the C. lari and novel Campylobacter sp. isolates from free-ranging seals presented in our previous study, along with Campylobacter spp. isolated from seals during rehabilitation at TMMC. A novel multilocus sequence typing (MLST) method, similar to methods described for related Campylobacter species (2, 14), was developed to type the pinniped Campylobacter isolates.
MATERIALS AND METHODS
Bacterial isolates and phenotypic characterization.
A majority of the bacterial isolates used in the study were previously described in the work of Stoddard et al. (23). In this study, free-ranging seals were apparently healthy and stranded seals were compromised due to various conditions, with the top cause of stranding being emaciation with lung worm infection and bacterial infections being less-likely causes. All animals range between the ages of 2 and 7 months, but the exact ages cannot be known. There were 165 free-ranging seals and 196 stranded seals that were rectally swabbed. Swabs were placed in Cary-Blair transport medium (BD Diagnostics, Franklin Lakes, NJ) and held at 4°C until processed within 48 h of collection. Rectal swabs were streaked on a Brucella agar base with 5% sheep blood and vancomycin, amphotericin B, and cefoperazone (Campy-CVA agar) (Hardy Diagnostics, Santa Maria, CA) and incubated at 37°C for 48 to 96 h in 5.5-liter AnaeroPack containers (Mitsubishi Gas Chemical America, Inc., New York, NY) using CampyGen sachets (Oxoid, Inc., Ogdensburg, NY) to produce a microaerophilic environment. Three to five colonies were identified using standard procedures (15) and stored at −80°C in Microbank bead vials (Pro-Lab Diagnostics, Austin, TX).
This study included 5 presumptive C. lari strains and 2 unknown Campylobacter strains isolated from free-ranging seals, as well as 22 C. lari strains and 19 unknown Campylobacter strains isolated from stranded juvenile northern elephants that were brought to TMMC (23). In addition to the strains that were originally described by Stoddard et al. (23), this study included 18 C. lari strains and 6 unknown Campylobacter strains that were isolated from seals involved in the original study during rehabilitation at TMMC. Culture and identification of these isolates was the same as discussed above. Some of the isolates from the 2003-2004 study were not included due to contamination of or inability to resuscitate the original frozen stock. The C. lari strains were identified originally by phenotypic characterization; these strains were all gram-negative, curved rods that were oxidase positive, catalase positive, and urease negative, did not hydrolyze hippurate, reduce hydrogen sulfide (H2S), or reduce nitrate, were resistant to both 30 μg cephalothin and 30 μg nalidixic acid, and grew at 42°C.
Original Campylobacter sp. identification was done by Gram staining and determination of oxidase and catalase activity, nitrate reduction, hippurate hydrolysis, urease activity, H2S production on triple sugar iron (TSI) slants, growth at 42°C, and sensitivity to nalidixic acid and cephalothin. In addition to the parameters described above, each phenotype was confirmed for this study by measuring growth at 25°C and growth in the presence of 1% glycine or 3.5% NaCl, using procedures previously described (15).
DNA purification.
DNA for PCR and MLST was purified from bacterial strains grown on sheep blood agar plates (Hardy Diagnostics) using the boiling method (7). Briefly, a plateful of culture was suspended in 300 μl sterile distilled water and then boiled for 15 min. After boiling, the tubes were chilled immediately on ice for 5 min, followed by centrifugation at 15,000 × g for 5 min. The supernatant was then transferred to a new 1.5-ml tube.
16S rRNA gene PCR and sequence analysis.
The forward and reverse conserved 16S rRNA eubacterial primers used to amplify the 16S rRNA genes were designated 8FPL (5′ CTG CAG AGT TTG ATC CTG GCT CAG 3′) and 1492RPL (5′ CGG GTT ACC TTG TTA CGA CTT 3′). Each 50-μl sample amplified by PCR contained the following: 25 μl 2× FideliTaq PCR Master Mix (USB Corporation, Cleveland, OH), giving a final concentration of 200 μM (each) deoxynucleoside triphosphate and 1.5 mM MgCl2, 30 pmol (each) primer (6 μl), 60 to 80 μg genomic DNA (2 μl), and 17.0 μl water. Amplification was performed with a Mastercycler Gradient thermal cycler (Eppendorf North America, Westbury, NY) as follows: 94°C for 10 min, followed by 35 cycles of 94°C for 1 min, 63.1°C for 1 min, and 72°C for 2 min, followed by an elongation step at 72°C for 10 min. The 1,484-bp PCR product was separated on a 1.0% agarose gel by electrophoresis and visualized with ethidium bromide. The PCR products were then cleaned using the ExoSAP-IT system (USB Corporation), following the manufacturer's protocol by adding 8 μl of ExoSAP-IT to 20 μl of each reaction mixture. Each mixture was then run in the Mastercycler Gradient thermal cycler as follows: 37°C for 15 min followed by heating at 80°C for 15 min. Forward and reverse sequencing reactions were performed by the DNA Sequencing Facility, Division of Biological Sciences, University of California, Davis. The DNA sequences were corrected using Chromas (Technelysium Pty. Ltd., Tewantin, Qld, Australia) and compared to known sequences using the NCBI BLAST database (http://www.ncbi.nlm.nih.gov/).
MLST.
Scottish C. insulaenigrae isolates were purchased from the NCTC (London, United Kingdom). MLST primer sets are listed in Table 1. Each MLST amplification mixture contained: 50 ng genomic DNA, 1× MasterAmp PCR buffer (Epicentre, Madison, WI), 1× MasterAmp PCR enhancer (Epicenter), 2.5 mM MgCl2, 250 μM (each) deoxynucleoside triphosphates, 50 pmol (each) primer, and 1 U Taq polymerase (New England Biolabs, Beverly, MA). PCRs for MLST were performed with a Tetrad thermocycler (Bio-Rad, Hercules, CA) with the following settings: 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 2 min. Amplicons were purified on a BioRobot 8000 workstation (QIAGEN, Valencia, CA). Cycle sequencing reactions were performed with a Tetrad thermocycler, using the ABI PRISM BigDye terminator cycle sequencing kit (version 3.1; Applied Biosystems, Foster City, CA) and standard protocols. Extension products were purified using 96-well DyeEx plates (QIAGEN). DNA sequencing was performed on an ABI PRISM 3130XL genetic analyzer (Applied Biosystems), using POP-7 polymer and the ABI PRISM Genetic Analyzer Data Collection and ABI PRISM Genetic Analyzer Sequencing Analysis software.
TABLE 1.
Campylobacter insulaenigrae MLST primer sets
| Locus | Forward (5′-3′)
|
Reverse (5′-3′)
|
Amplicon size (bp) | ||
|---|---|---|---|---|---|
| Primer | Sequence | Primer | Sequence | ||
| aspA | aspAF2 | GAAGCWAAAGCWAAAGAATAYAAAGAT | aspAR2 | GAGTTTTTTGCAWGCTTCWGGATT | 690 |
| atpA | atpAF | GWCAAGGDGTTATYTGTATWTATGTTGC | atpAR | TTTAADAVYTCAACCATTCTTTGTCC | 700 |
| glnA | glnAF | TGATAGGMACTTGGCAYCATATYAC | glnAR | ARRCTCATATGMACATGCATACCA | 751 |
| glyA | insglyF1 | GCTTTTTAATTGTMYTTTTGCTAATGT | insglyR1 | GCTARATCTGCATCTTTRCCRCTAAA | 750 |
| pgi | inspgiF | GTTTTAGTAGGTATGGGTGGATCAAGC | inspgiR | TATTTGGAATCAAAGGAGCTTTATAGC | 760 |
| pgm | inspgmF2 | AATTCTTTCCCTAAAAATTTAACTCTT | inspgmR2 | AGTAAGCGAATTAAATTTTYAGTTCCT | 764 |
| tkt | tktF1 | GCAAAYTCAGGMCAYCCAGGTGC | instktR1 | TTTACATCTTCAGGAATTTCAAAAGTT | 791 |
Assignment of allele numbers and STs and phylogenetic analysis.
The Perl program MLSTparser (14) was modified to accept the novel locus composition of the C. insulaenigrae sequence types (STs). Allele sequences and STs were extracted from the FASTA-formatted forward and reverse reads using MLSTparser, as described previously (14); each unique allele and ST was assigned an arbitrary number. Each allele also was queried against the MLST databases of the related Campylobacter species, C. lari, C. upsaliensis, and C. helveticus (http://pubmlst.org/); however, none of the alleles identified for C. insulaenigrae were identified previously for these three other Campylobacter species. Allele and ST data for C. insulaenigrae were deposited in the PubMLST database and are available online (http://pubmlst.org/cinsulaenigrae/).
Variable sites and calculation of the dn/ds ratios were performed using START2 (http://pubmlst.org/software/analysis/). Allele sequences for each strain were concatenated in the order aspA-atpA-glnA-glyA-pgi-pgm-tkt for a final composite length of 3,363 bp. The concatenated sequences were aligned using CLUSTALX, and a dendrogram was constructed using the neighbor-joining method with the Kimura two-parameter distance estimation method. Phylogenetic analyses were performed using MEGA version 2.1 (11).
RESULTS
Phenotypic characterization.
As described above, 45 presumptive C. lari strains and 27 unknown Campylobacter strains from both free-ranging and stranded northern elephant seals from California were characterized. All Campylobacter spp. had the following phenotypic characteristics: they were oxidase positive with the ability to reduce nitrate but not hydrolyze hippurate, urease negative, did not produce H2S on TSI slants, did not grow at 25°C, and were not able to grow in the presence of 3.5% sodium chloride (Table 2). All but one isolate was catalase positive (strain 3413).
TABLE 2.
Differential biochemical and phenotypic characteristics of Campylobacter insulaenigrae isolates from seals in California
| Pattern no. or speciesa | No. of isolates with pattern | Presence of:
|
Growth at/in:
|
Presence of:
|
Hippurate hydrolysis | Sensitivity tod:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Catalase | Oxidase | 25°C | 42°C | 3.5% NaCl | 1.0% Glycine | Nitrate | H2S in TSI | Urease | Cephalothin | Nalidixic Acid | |||
| 1 | 22 | + | + | − | + | − | + | − | − | R | R | ||
| 2 | 6 | + | + | − | − | − | + | + | − | − | − | R | R |
| 3 | 13 | + | + | − | + | − | − | + | − | − | − | R | R |
| 4 | 2 | + | + | − | − | − | − | + | − | − | − | R | R |
| 5 | 11 | + | + | − | + | − | + | + | − | − | − | S | R |
| 6 | 3 | + | + | − | − | − | + | + | − | − | − | S | R |
| 7 | 12 | + | + | − | + | − | − | + | − | − | − | S | R |
| 8 | 2 | + | + | − | − | − | − | + | − | − | − | S | R |
| 9 | 1 | − | + | − | − | − | + | + | − | − | − | S | S |
| C. insulaenigrae | 4 | + | + | − | − | − | + | + | − | − | − | R | R |
| C. lari | + | + | −b | + | − | + | + | − | +/− | − | R | Rc | |
| C. upsaliensis | − | + | − | +c | − | + | + | − | − | − | S | S | |
| C. helveticus | − | + | − | + | − | +/− | + | − | − | − | S | S | |
Patterns 1 to 8 represent C. insulaenigrae, and pattern 9 represents a C. upsaliensis/C. helveticus-like strain. Biochemical and phenotypic characteristics of the related Campylobacter species C. lari, C. upsaliensis, and C. helveticus are provided for comparison. C. lari, C. upsaliensis, and C. helveticus characteristics are taken from the work of Lastovica and Skirrow (11a) and On (20). C. insulaenigrae characteristics are taken from the work of Foster et al. (3).
Some strains positive/resistant.
Some strains negative/sensitive.
S, sensitive; R, resistant.
There was variability in the results for growth at 42°C or in the presence of 1% glycine and in the sensitivity to nalidixic acid and cephalothin (Table 2). The most common phenotypic and biochemical pattern that was observed was pattern number 1 (n = 22), which is consistent with C. lari (Table 2). The second-most-common phenotypic and biochemical pattern was pattern number 3 (n = 13) (Table 2). There were six isolates with pattern 2, which is consistent with the originally described C. insulaenigrae phenotype. Pattern number 9 was found with only one isolate, and this pattern could be compatible with several different Campylobacter spp., including C. upsaliensis and C. helveticus.
16S rRNA gene PCR analysis.
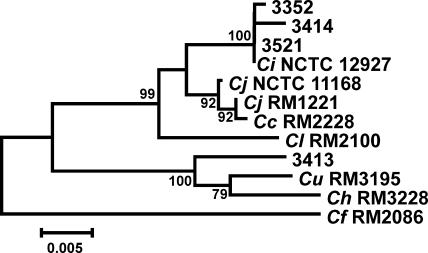
All strains except strain 3413, which was found to have phenotype pattern 9, were identified as C. insulaenigrae based on both forward and reverse sequence analysis of the 1,484 base pairs of the 16S rRNA gene (Fig. 1). Strain number 3413 was identified as being closely related to C. upsaliensis after both forward and reverse 16S sequence analysis (Fig. 1).
FIG. 1.
Dendrogram of Campylobacter 16S rRNA gene sequences, including four representative sequences from this study. The dendrogram was constructed using the neighbor-joining algorithm and the Kimura two-parameter distance estimation method. Bootstrap values of >75%, generated from 500 replicates, are shown at the nodes. The scale bar represents substitutions per site. The remaining C. insulaenigrae 16S sequences were not substantially different from the 16S sequences of the three strains (3352, 3414, and 3521) represented in the tree. Additional 16S sequences from Campylobacter sequenced or type strains are included for comparison. Ci, C. insulaenigrae; Cj, C. jejuni; Cc, C. coli; Cl, C. lari; Cu, C. upsaliensis; Ch, C. helveticus; Cf, C. fetus.
Multilocus sequence typing.
Preliminary phylogenetic analysis of C. insulaenigrae using 303 Campylobacter 16S rRNA sequences indicated that the closest relative of C. insulaenigrae was C. lari and that C. insulaenigrae was a member of the thermophilic subgroup of campylobacters (data not shown). A multilocus sequence typing method for C. lari and for thermophilic Campylobacter spp. in general was described by Miller et al. (14). Since C. insulaenigrae was similar to C. lari, our initial MLST primer sets included the C. lari aspAF2-aspAR2, pgmF3-pgmR3, and tktF2-tktR primer sets and the thermophilic atpAF-atpAR, glnAF-glnAR, gltAF-gltAR, and glyAF-glyAR primer sets (14).
PCR analysis of the 75 strains showed that, as with C. lari (14), the gene encoding citrate synthase, gltA, is not present and may be absent in C. insulaenigrae. Therefore, the pgi locus was substituted for gltA. Additionally, the glyA, pgm, and tkt loci were amplified inconsistently using the original primer sets; therefore, new C. insulaenigrae-specific primer sets had to be designed. Using the aligned sequences of the glyA, pgi, pgm, and tkt loci extracted from the available Campylobacter and Arcobacter genome sequences, degenerate primers were designed for each locus 300 bp upstream and downstream of the allelic endpoints. These primers were used to sequence the four NCTC C. insulaenigrae strains, and the resultant sequences were then used to design C. insulaenigrae-specific primers. The finalized primer sets are described in Table 1.
Genetic diversity of C. insulaenigrae.
Within the 75 C. insulaenigrae strains typed in this study, a large number of alleles were found at each of the 7 loci, ranging from 9 at the aspA and glnA loci to 14 at the atpA and pgm loci (Table 3). The percentage of variable sites was relatively consistent from locus to locus. Analysis of the 80 alleles identified in this study, using the Campylobacter PubMLST databases, did not identify any alleles potentially transferred laterally from the thermophilic Campylobacter spp. The ratio of nonsynonymous to synonymous base substitutions (dn/ds) ranged from 0 (atpA) to 0.011 (aspA), indicating that the C. insulaenigrae MLST loci were not subject to positive selection.
TABLE 3.
Diversity at the Campylobacter insulaenigrae MLST locia
| Locus | No. of alleles (% of isolatesb) | Variable sites (%)c | dn/dsd |
|---|---|---|---|
| aspA | 9 (12.0) | 7 | 0.110 |
| atpA | 14 (18.7) | 13 | 0.000 |
| glnA | 9 (12.0) | 12 | 0.015 |
| glyA | 11 (14.7) | 13 | 0.100 |
| pgi | 13 (17.3) | 10 | 0.066 |
| pgm | 14 (18.7) | 17 | 0.035 |
| tkt | 10 (13.3) | 14 | 0.011 |
Includes results for C. insulaenigrae strains from Scotland.
No. of alleles/strains typed.
No. of polymorphic sites/allele size (nucleotides).
Ratio of nonsynonymous to synonymous sites.
STs and clonal complexes.
Using the 7 loci, a total of 40 STs were identified among the 75 C. insulaenigrae strains typed in this study, which also included the original C. insulaenigrae isolates from Scotland (see Table S1 in the supplemental material). Among the California strains, 33.8% (n = 24) had unique STs, with the most common STs being ST-25 (n = 12) and ST-5 (n = 8). A comparison of the biochemical and phenotypic patterns with the assigned STs showed no correlation, although all of the isolates in ST-5 showed phenotypic pattern 1 (see Table S1 in the supplemental material). Campylobacter insulaenigrae isolated from the same seals (ES2171, ES2235, ES2275, ES2284, and ES2297) both at stranding and during their stay at TMMC showed different STs at these two time points and were therefore kept in the study (see Table S1 in the supplemental material). In a comparison of free-ranging seals, stranded seals, and seals at TMMC, there was substantial overlap in the STs.
Approximately half (22/40) of the STs identified in this study could be organized into 6 clonal complexes, although none of the clonal complexes contained a predicted founder ST (data not shown). Four of the complexes contain only 2 members, the fifth contains 4 members (ST-9, ST-27, ST-31, and ST-33), and the sixth contains 10 members (ST-6, ST-7, ST-13, ST-16, ST-18, ST-21, ST-22, ST-28, ST-36, and ST-40).
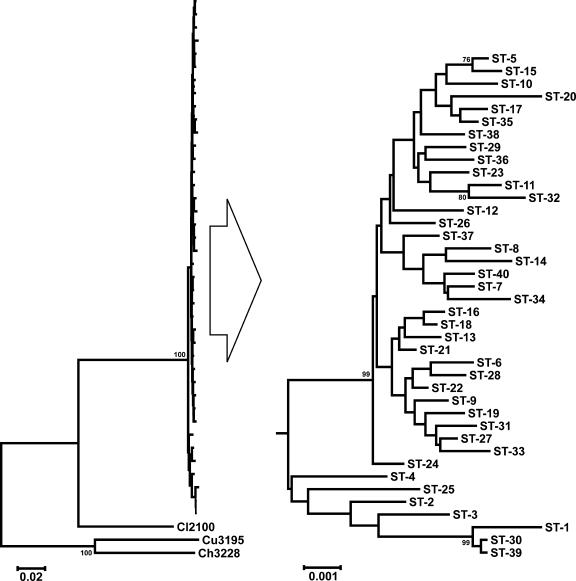
Pairwise comparisons of the 3,363-bp concatenated allele sequences indicated that those from the C. insulaenigrae strains isolated from northern elephant seals in California were 85%, 73%, and 73% similar to the cognate concatenated allele sequences from C. lari, C. upsaliensis, and C. helveticus, respectively. The C. insulaenigrae strains from Scotland all possessed unique STs (ST-1 through ST-4), which were distinct from those of the C. insulaenigrae strains isolated from the California seals. Although the Scottish C. insulaenigrae strains were found by phylogenetic analysis to be members of a secondary clade (containing both the “Scottish” STs and STs ST-25, ST-30, and ST-39) which was distinct from the clade containing the majority of California C. insulaenigrae isolates (Fig. 2), pairwise comparisons of concatenated allele sequences indicated ≤1.4% sequence divergence between members of the primary and secondary clades. Nonetheless, it is noteworthy that 83% (15/18) of the strains isolated at TMMC during 2004 were members of the secondary clade, compared to 8% (2/26) of strains isolated from free-ranging seals during the same time period; no members (0/27) of the secondary clade were isolated from California seals, free-ranging or stranded, during 2003.
FIG. 2.
Dendrogram of Campylobacter insulaenigrae STs, including original isolates from the United Kingdom (ST-1 through ST-4). The dendrogram was constructed using the neighbor-joining algorithm and the Kimura two-parameter distance estimation method. Bootstrap values of >75%, generated from 500 replicates, are shown at the nodes. The scale bar represents substitutions per site. Three representative C. lari (Cl2100), C. upsaliensis (Cu3195), and C. helveticus (Ch3228) STs (using similar concatenated MLST alleles) are included for comparison. The C. insulaenigrae portion of the tree on the left is expanded on the right (large arrow) to improve clarity, since the scale of this portion of the tree with the related Campylobacter spp. included is too small to readily distinguish the C. insulaenigrae STs (note different scale bars).
DISCUSSION
In 2003 and 2004, presumptive strains of C. lari and an unknown Campylobacter species were isolated from northern elephant seals in California, based on phenotypic identification techniques. In this study, the exact identities of all but one of these strains were determined, based on 16S rRNA sequence analysis and MLST, to be Campylobacter insulaenigrae (3). Although the Campylobacter strains isolated from seals in California are C. insulaenigrae, they show variation from the Scottish strains both phenotypically and genotypically with regard to biochemical characteristics and MLST sequence types (3). Identification of the Campylobacter sp. isolates from these California seals using phenotypic characterization was found often to lead to an incorrect identification of some strains as C. lari.
The original characterization of C. insulaenigrae isolated from marine mammals in Scotland showed this species to be a nonthermophilic Campylobacter species that is able to grow on medium containing 1% glycine and is resistant to both nalidixic acid and cephalothin (Table 2) (3). In the present study of 71 California C. insulaenigrae isolates, we found variable growth at 42°C or on medium containing 1% glycine, as well as variation in sensitivity to cephalothin. The major phenotypic profile (phenotypic pattern 1) was consistent with an identification of C. lari; the misidentification of strains as C. lari has been reported previously (22). The ability to hydrolyze indoxyl acetate was not tested in this study; however, of the 29 original strains isolated from stranded northern elephant seals brought to TMMC between 2000 and 2002, all were found to be negative (unpublished data). The inability to hydrolyze indoxyl acetate is consistent with both the original C. insulaenigrae isolates from Scotland and C. lari and therefore would not aid in differentiating these species (3).
Phenotypic testing can be problematic due to the lack of standardization for tests that are used and the fact that variable methodology can affect the outcome (20). Another problem is that interpretation of tests can be subjective when they are used for identification. Additionally, it is apparent that, similar to the eight phenotypic patterns identified for C. insulaenigrae, multiple phenotypic subgroups are present for each of the three related Campylobacter species: C. lari, C. upsaliensis, and C. helveticus (Table 2); thus, there is no one phenotypic pattern that could be used to identify unambiguously any of these four phenotypically variable species. Even C. jejuni strains, identified typically through the use of hippuricase assays (13, 24), can be misidentified due to the presence of a relatively uncommon hippuricase-negative subgroup. The growing trend of resistance to nalidixic acid among Campylobacter species further confounds the identification of species using phenotypic testing (1). Interpretation of strain sensitivity to nalidixic acid and cephalothin has been determined to be a common mistake in species identification (22).
Because phenotypic methodology commonly results in misidentification of Campylobacter species isolated from marine mammals, it is recommended that a molecular method be included in the identification scheme for Campylobacter. A simple genotypic method of identification of Campylobacter isolates is 16S rRNA amplification and sequence analysis (9, 20, 22). Although phenotypic methods were unable to speciate unambiguously the Campylobacter strains isolated from marine mammals in this study, 16S rRNA PCR and sequence analysis correctly identified all 71 strains as C. insulaenigrae. Another identification method which would be rapid and would not require phenotypic characterization is the use of multiplex PCR. A multiplex PCR utilizing the lipid A gene lpxA has been developed to differentiate between C. coli, C. jejuni, C. lari, and C. upsaliensis (8) and could be modified to include C. insulaenigrae.
The present study helped to establish a multilocus sequence typing method that is valid for use with C. insulaenigrae, since the previously established methods (2, 14) were not successful in characterizing this species. Of the previously described thermophilic MLST primer sets (14), only the primer sets for three loci (aspA, atpA, and glnA) were amplified for C. insulaenigrae strains consistently without modification of the primers. The glyA, pgi, pgm, and tkt primer sets also had to be redesigned de novo, since the original primer sets, used in the MLST methods for other thermophilic Campylobacter spp., inconsistently amplified DNA from the strains in this study. Although the addition of these C. insulaenigrae-specific primer pairs would make it more difficult to obtain a complete ST from any given strain of Campylobacter isolated from a marine mammal, the common primer sets for the atpA and glnA loci could be used at minimum to identify such strains to the species level, since these primer pairs will amplify DNA from all known thermophilic Campylobacter species. Indeed, allelic sequences at these loci were used to identify the novel strain 3413.
The citrate synthase (gltA) gene could not be amplified for any of the C. insulaenigrae strains, suggesting strongly that the gltA gene is absent from this species. In a previous study developing an MLST method for multiple Campylobacter spp., the gltA gene of C. lari also could not be amplified (14). Analysis of the recently completed C. lari genome indicated that in strain RM2100, the gltA gene specifically and multiple tricarboxylic acid cycle genes in general were absent (unpublished data). Pairwise comparisons of the concatenated C. insulaenigrae allele sequences indicated that the closest relative to C. insulaenigrae is C. lari (85% similarity). Additionally, the four original Scottish C. insulaenigrae strains were 98.3% similar to C. lari, based on pairwise sequence comparisons of 16S rRNA gene sequences (3).
Campylobacter lari is a thermophilic Campylobacter species which was first isolated from healthy gulls and is often found in the feces of other healthy bird species (15, 21). One could hypothesize that C. insulaenigrae evolved from C. lari based on the presence of both of these Campylobacter species in marine birds and mammals. Gulls could shed the bacteria in the marine environment, and they could then have been picked up by seals and other marine mammals with concomitant host adaptation. Alternatively, C. lari and C. insulaenigrae could share a common ancestor. Other factors in support of these theories are their genetic relatedness and the fact that both Campylobacter species lack the citrate synthase gene.
For the C. insulaenigrae strains isolated from northern elephant seals in California, there were a large number of alleles but not much variation at the nucleotide level among the alleles. Nevertheless, phylogenetic analysis identified two C. insulaenigrae clades: the primary clade, containing exclusively California strains, and a secondary clade, containing some California strains and all of the original Scottish strains. As described above, the California strains present in the secondary clade were almost entirely those isolated at TMMC during 2004 and comprised sequence types ST-25, ST-30, and ST-39. Further analysis of strains isolated at TMMC indicates the presence of other predominant STs, namely, ST-5 and ST-23, during the previous year. This suggests that in each year a set of distinct C. insulaenigrae strains was propagated laterally through the facility, probably fairly early in the season, infecting multiple seals in rehabilitation. Further evidence for this is provided by the five seals from which C. insulaenigrae was isolated at the time of admission and again during rehabilitation. Strains with a distinct and with one exception unique ST were isolated from each of the five seals upon admission, but each was infected with one of the prevailing STs during rehabilitation. Interestingly, the prevailing STs differed between 2003 and 2004, indicating that the source was probably an infected seal introduced to the facility and not an endogenous source, such as the water supply. The transmission of isolates between seals is probably due to the fact that multiple seals (up to five) can be housed together in one pen and the pens are connected, allowing for contact with seals in the same pen and neighboring pens. Also, the presence of secondary clade STs only in 2004 may indicate a population shift among the C. insulaenigrae strains infecting marine mammals. Typing of additional strains, isolated during the subsequent years, will be necessary to answer some of these questions.
The absence of substantial sequence variation among the MLST alleles could be explained by the fact that these strains are isolated from the same species of seal within a similar geographic location in California. We have additional Campylobacter isolates from northern elephant seals and California sea lions (Zalophus californianus) in southern California that will be characterized and compared to the current strains in a future study in order to further elucidate the variability within C. insulaenigrae. We also hope to find more strains from these animals that are similar to the one C. upsaliensis/C. helveticus-like organism. This isolate (strain 3413) was identified as C. upsaliensis using 16S rRNA PCR, and preliminary MLST results indicate that this strain is similar to but distinct from (approximately 10% sequence divergence) both C. upsaliensis and C. helveticus. Campylobacter upsaliensis and C. helveticus are species that are associated with healthy cats and dogs but are sometimes isolated from animals with diarrhea (4, 21). In addition to further characterization of C. insulaenigrae and the C. upsaliensis-like organism, it will be important to determine if these Campylobacter species have virulence genes, since it is not known if they can cause disease in marine mammals.
The isolation of C. insulaenigrae from northern elephant seals in California has helped to further shed light on this complex and highly variable species. Campylobacter insulaenigrae is a phenotypically diverse species that may have evolved from or at least shares a common ancestor with C. lari. The identification of C. insulaenigrae by phenotypic methods often can be misleading; therefore, it is important to develop a standardized methodology for identification of Campylobacter species from marine mammals which does not rely entirely on phenotypic identification but also includes a molecular characterization using methods such as 16S rRNA PCR and/or multiplex PCR. A novel multilocus sequence typing method was developed for the identification of C. insulaenigrae and has proven to be useful for comparison within and between Campylobacter species. Finally, sequence data obtained by MLST of these C. insulaenigrae strains provide further evidence that C. insulaenigrae sp. nov. is a new Campylobacter species, which may be restricted to marine mammals but is genetically distinct from related thermophilic Campylobacter species, such as C. lari. Sequence typing and characterization of additional strains will only serve to solidify its position within the genus.
Supplementary Material
Acknowledgments
We thank Guilin Wang, S. Joy Milan, Spencer Jang, Anita Wong, Juliet Herrara, Eileen Samitz, and Joann Yee for technical assistance. We also thank Michelle Caudle, Denise Greig, the volunteers at The Marine Mammal Center, Pat Morris, Dan Costa (MMPA permit 87-1593-01), Sarah Allen (MMPA permit 373-1575), Brent Stewart (MMPA permit 486-1506), Brian Hatfield, the University of California Reserve System, and the rangers of Año Nuevo State Reserve, Point Reyes National Seashore, and the National Park Service for assistance with sampling of northern elephant seals.
This publication was supported in part by the West Coast Center for Oceans and Human Health as part of the NOAA Oceans and Human Health Initiative, WCCOHH publication no. 8. The WCCOHH is part of the National Marine Fisheries Service's Northwest Fisheries Science Center, Seattle, WA. This project was also supported in part by the Wildlife Health Center, School of Veterinary Medicine, University of California, Davis.
This publication made use of the Campylobacter MultiLocus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley at the University of Oxford (6).
Footnotes
Published ahead of print on 26 January 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altwegg, M., A. Burnens, J. Zollinger-Iten, and J. L. Penner. 1987. Problems in identification of Campylobacter jejuni associated with acquisition of resistance to nalidixic acid. J. Clin. Microbiol. 25:1807-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39: 14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster, G., B. Holmes, A. G. Steigerwalt, P. A. Lawson, P. Thorne, D. E. Byrer, H. M. Ross, J. Xerry, P. M. Thompson, and M. D. Collins. 2004. Campylobacter insulaenigrae sp. nov., isolated from marine mammals. Int. J. Syst. Evol. Microbiol. 54:2369-2373. [DOI] [PubMed] [Google Scholar]
- 4.Fox, J. G. 2006. Enteric bacterial infections, p. 339-368. In C. E. Greene (ed.), Infectious diseases of the dog and cat. Saunders Elsevier, St. Louis, MO.
- 5.Hirsh, D. C. 2004. Spiral-curved organisms III: Campylobacter, Arcobacter, Lawsonia, p. 134-140. In D. C. Hirsh, N. J. MacLachlan, and R. L. Walker (ed.), Veterinary microbiology. Blackwell Science, Inc., Ames, IA.
- 6.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinform. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josefsen, M. H., P. S. Lubeck, F. Hansen, and J. Hoorfar. 2004. Towards an international standard for PCR-based detection of foodborne thermotolerant campylobacters: interaction of enrichment media and pre-PCR treatment on carcass rinse samples. J. Microbiol. Methods 58:39-48. [DOI] [PubMed] [Google Scholar]
- 8.Klena, J. D., C. T. Parker, K. Knibb, J. C. Ibbitt, P. M. Devane, S. T. Horn, W. G. Miller, and M. E. Konkel. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koene, M. G., D. J. Houwers, J. R. Dijkstra, B. Duim, and J. A. Wagenaar. 2004. Simultaneous presence of multiple Campylobacter species in dogs. J. Clin. Microbiol. 42:819-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korhonen, L. K., and P. J. Martikainen. 1991. Survival of Escherichia coli and Campylobacter jejuni in untreated and filtered lake water. J. Appl. Bacteriol. 71:379-382. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 11a.Lastovica, A. J., and M. B. Skirrow. 2000. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and C. coli, p. 89-120. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter. ASM Press, Washington, DC.
- 12.Le Boeuf, B. J., and R. M. Laws. 1994. Elephant seals: an introduction to the genus, p. 1-26. In B. J. Le Boeuf and R. M. Laws (ed.), Elephant seals: population ecology, behavior, and physiology. University of California Press, Berkeley, CA.
- 13.Lucey, B., F. O'Halloran, and S. Fanning. 2004. Molecular-based identification and typing of Campylobacter jejuni and C. coli. Methods Mol. Biol. 268:33-47. [DOI] [PubMed] [Google Scholar]
- 14.Miller, W. G., S. L. On, G. Wang, S. Fontanoz, A. J. Lastovica, and R. E. Mandrell. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nachamkin, I. 2003. Campylobacter and Arcobacter, p. 902-914. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology. ASM Press, Washington, DC.
- 16.Nielsen, E. M. 2002. Occurrence and strain diversity of thermophilic campylobacters in cattle of different age groups in dairy herds. Lett. Appl. Microbiol. 35:85-89. [DOI] [PubMed] [Google Scholar]
- 17.Obiri-Danso, K., and K. Jones. 1999. The effect of a new sewage treatment plant on faecal indicator numbers, campylobacters and bathing water compliance in Morecambe Bay. J. Appl. Microbiol. 86:603-614. [DOI] [PubMed] [Google Scholar]
- 18.Obiri-Danso, K., and K. Jones. 1999. Distribution and seasonality of microbial indicators and thermophilic campylobacters in two freshwater bathing sites on the River Lune in northwest England. J. Appl. Microbiol. 87:822-832. [DOI] [PubMed] [Google Scholar]
- 19.Obiri-Danso, K., N. Paul, and K. Jones. 2001. The effects of UVB and temperature on the survival of natural populations and pure cultures of Campylobacter jejuni, Camp. coli, Camp. lari and urease-positive thermophilic campylobacters (UPTC) in surface waters. J. Appl. Microbiol. 90:256-267. [DOI] [PubMed] [Google Scholar]
- 20.On, S. L. 1996. Identification methods for campylobacters, helicobacters, and related organisms. Clin. Microbiol. Rev. 9:405-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Songer, J. G., and K. W. Post. 2005. The genera Campylobacter, Helicobacter, and Arcobacter, p. 223-231. In J. G. Songer and K. W. Post (ed.), Veterinary microbiology: bacterial and fungal agents of animal disease. Saunders Elsevier, St. Louis, MO.
- 22.Steinhauserova, I., J. Ceskova, K. Fojtikova, and I. Obrovska. 2001. Identification of thermophilic Campylobacter spp. by phenotypic and molecular methods. J. Appl. Microbiol. 90:470-475. [DOI] [PubMed] [Google Scholar]
- 23.Stoddard, R. A., F. M. D. Gulland, E. R. Atwill, J. Lawrence, S. Jang, and P. A. Conrad. 2005. Salmonella and Campylobacter spp. in northern elephant seals, California. Emerg. Infect. Dis. 11:1967-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wainø, M., D. D. Bang, M. Lund, S. Nordentoft, J. S. Andersen, K. Pedersen, and M. Madsen. 2003. Identification of campylobacteria isolated from Danish broilers by phenotypic tests and species-specific PCR assays. J. Appl. Microbiol. 95:649-655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.