Abstract
Salmonella spp. infection is a major cause of gastroenteritis, with many thousands of cases reported in the European Union every year. The use of probiotics offers the potential to improve this situation. Here, we investigate the effects of oral treatment of pigs with a defined lactic acid bacteria culture mixture on both clinical and microbiological signs of Salmonella enterica serovar Typhimurium infection. Fifteen weaned pigs blocked by sex and weight were administered control milk or a mixture of five probiotic strains as either a milk fermentate or milk suspension for a total of 30 days. The mixture consisted of two strains of Lactobacillus murinus and one strain each of Lactobacillus salivarius subsp. salivarius, Lactobacillus pentosus, and Pediococcus pentosaceous. Following probiotic administration for 6 days, animals were challenged orally with serovar Typhimurium; the health of the animals and the microbiological composition of their feces were monitored for 23 days postinfection. Animals treated with probiotic showed reduced incidence, severity, and duration of diarrhea. These animals also gained weight at a greater rate than control pigs administered skim milk. Mean fecal numbers of Salmonella were significantly reduced in probiotic-treated animals at 15 days postinfection (P = 0.01). The administered probiotic bacteria improved both the clinical and microbiological outcome of Salmonella infection. These strains offer significant benefit for use in the food industry and may have potential in human applications.
Probiotics, as usually defined, are “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (11). Probiotic properties have been ascribed to many microbial species, but those most commonly used are members of the lactic acid bacteria (LAB) group, particularly Lactobacillus spp. and Bifidobacterium spp. strains. Several studies have investigated the anti-Salmonella effects of potential probiotics by using in vitro procedures, particularly growth medium and tissue culture (8, 13).
The application of LAB probiotics has been linked by several authors with beneficial effects in models of gastrointestinal infection using small animals. Ogawa et al. reported that the use of Lactobacillus casei Shirota reduced colonization levels and decreased the severity of diarrhea in Escherichia coli O157:H7-infected infant rabbits (24). Using mice, Johnson-Henry et al. noted that a mixture of Lactobacillus spp. strains reduced gastric inflammation and bacterial colonization in Helicobacter pylori-infected animals (20). Varied results have been reported with the use of Salmonella spp. infection models. Pascual et al. noted complete exclusion by 21 days of S. enterica serovar Enteritidis when they used Lactobacillus salivarius in chickens (25), and Nisbet et al. observed a significant decrease in mortality due to S. enterica serovar Gallinarum infection in chicks treated with a commercial probiotic mixture (23). La Ragione et al. observed no beneficial effect on serovar Enteritidis fecal numbers or colonization of the intestine when they pretreated chickens with Lactobacillus johnsonii (21). The same authors did however note that E. coli numbers were reduced in the small intestine but not in the colon, cecum, or feces. They also claimed that the strain was very effective against Clostridium perfringens. Silva et al. observed improved survival for mice pretreated with Bifidobacterium longum during challenge with Salmonella spp. but no effect on numbers of the pathogen (27). They postulated that this may be due to a reduced inflammatory response mediated by the probiotic treatment but not population antagonism.
Reports of the efficacy of probiotic treatment in alleviating intestinal infection in large animals remain scarce. Zhao et al. claimed that the application of probiotic E. coli (no LAB) reduced the carriage of E. coli O157:H7 in cattle (29). Lema et al. observed that lambs infected with E. coli O157:H7 and then administered Lactobacillus acidophilus showed no beneficial effects (22). However, feeding the lambs a mixture of L. acidophilus and Streptococcus faecium, or the Streptococcus strain alone, significantly lowered numbers of the pathogenic strain. The greatest reduction in numbers was seen with the use of a mixture of L. acidophilus, S. faecium, L. casei, Lactobacillus fermentum, and Lactobacillus plantarum. Genovese et al. found that a competitive exclusion culture reduced the mortality and shedding of enterotoxigenic E. coli in neonatal pigs (18). The same group also observed that neonatal pigs treated with a similar culture shed significantly lower pathogen numbers after challenge with S. enterica serovar Choleraesuis and also exhibited reduced counts in the lower intestine (17). Whether or not symptoms of infection were alleviated was not described. Fedorka-Cray et al. reported that the application to serovar Choleraesuis-challenged piglets of a competitive exclusion culture of swine origin led to reduced Salmonella counts in their cecal contents and at the ileocolic junction. Reduced numbers of Salmonella-positive gut tissue samples were also seen; however, no clinical symptoms of infection were observed in any animals, including the controls (12).
In this report, we investigate the effects of pretreatment of weaned pigs with a defined LAB culture mixture on both clinical and microbiological signs of Salmonella infection. The results of molecular analysis of the excreted cultures are also described.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Lactobacillus murinus DPC6002 and DPC6003, Lactobacillus pentosus DPC6004, Lactobacillus salivarius DPC6005, and Pediococcus pentosaceus DPC6006 are rifampin-resistant (Rifr) variants of cultures previously selected from a bank of porcine intestinal isolates on the basis of molecular typing and probiotic and technological properties, such as antimicrobial activity, bile tolerance, growth in milk, and survival in the porcine gastrointestinal tract (GIT) (2, 14, 15). These were routinely cultured at 37°C in MRS broth (Difco Laboratories, Detroit, MI) in anaerobic jars with CO2-generating kits (Anaerocult A; Merck, Darmstadt, Germany). For animal challenge, Salmonella enterica serovar Typhimurium PT12, a porcine isolate, was obtained from the culture collection of the Central Veterinary Research Laboratory and routinely cultured in tryptic soy broth (Merck, Darmstadt, Germany). To facilitate fecal enumeration, a nalidixic acid-resistant (Nalr) variant of the strain was generated by spread plating 100 μl of overnight culture onto brain heart infusion agar containing increasing concentrations of nalidixic acid (0 to 70 μg/ml). A single colony from a plate containing 50 μg/ml nalidixic acid was grown in broth and stocked. The randomly amplified polymorphic DNA (RAPD) banding pattern and growth characteristics of this variant were compared to those of the parent strain to ensure similarity.
Preparation of probiotic fermentate and suspension for pig feeding trial.
Individual cultures were grown in 10% reconstituted skim milk with yeast extract (RSM plus YE), with the exception of the L. salivarius strain, which was grown without YE, and mixed to obtain a fermentate mixture for pig feeding as described previously (15). For the suspension, 90-ml volumes of de Man Rogosa Sharpe broth (MRS; Merck, Darmstadt, Germany) were inoculated with the appropriate culture at 1% (vol/vol) and grown overnight in MRS broth at 37°C. Following incubation of each culture, the bacteria were harvested by centrifugation, resuspended in 900 ml of RSM or RSM plus YE, and mixed. This suspension was then aliquoted into 100-ml volumes, stored at 4°C, and used within 8 days. Bacterial numbers for each batch of fermentate and suspension were determined by plate counting on MRS agar to ensure consistency.
Pig challenge trial.
The pig feeding trial complied with all relevant legislation regarding the protection of animals and was approved by the Animal Research Ethics Committee at University College Dublin. Blood and fecal samples were collected from a total of 21 crossbred (Large White × Landrace) weaned pigs and analyzed as previously described to ensure the animals were negative for Salmonella carriage (1). The pigs were administered feed for 7 days postweaning in the Moorepark pig production facility and then trained to drink RSM while continuing to receive feed for a further week. Zinc oxide was included in feed for the first 7 days, as animals at this age are otherwise prone to illness. The zinc oxide was removed from their feed for the second week due to its antimicrobial properties. No antibiotic growth promoters were administered to the animals at any stage. Fifteen pigs which were Salmonella negative and drinking milk well were then transported to a pathogen challenge facility at the Central Veterinary Research Laboratory, Abbotstown, Dublin, Ireland, where they were blocked by sex and weight. Blood and fecal samples from all animals were again analyzed to confirm their Salmonella-negative status prior to challenge. Pigs within each block were assigned at random to one of three treatment groups (n = 5), as follows: control, probiotic fermentate, and probiotic suspension. Each animal was penned individually to prevent cross-contamination, and control animals were housed in an area separate from probiotic-treated animals. In addition to the cultures or skim milk administered throughout the trial as outlined below, all animals had unrestricted access to water and nonmedicated creep feed obtained from the Moorepark feed mill.
Once in the Central Veterinary Research Laboratory facility, the pigs were fed sterile RSM for 6 days, following which they were administered probiotic or control milk for 30 days. Pigs receiving probiotic culture were fed 100 ml daily of the appropriate mixture, providing a total dose of either ∼4 × 1010 CFU/day (fermentate) or ∼4 × 109 CFU/day (suspension). Preliminary experiments (data not shown) indicated that the numbers of each culture within the suspension and fermentate were approximately equal. Control animals received 100 ml of RSM plus YE daily. Following 6 days of probiotic administration, the animals in each treatment group were challenged orally with 1 × 108 CFU serovar Typhimurium PT12 Nalr daily for three consecutive days (i.e., days 0, 1, and 2 postinfection [p.i.]). Freshly voided fecal samples were collected from the pigs 4 days prior to Salmonella challenge (day −4) and at 2, 4, 8, 15, and 23 days p.i., i.e., following the first challenge. Probiotic counts were determined at days −4, 2, 8, 15, and 23; Salmonella organisms were enumerated at days 4, 8, 15, and 23 postinfection. All microbiological enumeration procedures are outlined below.
Pigs were weighed initially in Moorepark before their transport to Abbotstown and again at the end of the feeding period (day 23 p.i.). Their weights were recorded and growth rates (in g/day) calculated, with comparisons being made between the control and probiotic treatments.
Fecal samples collected from the pen of each animal between 3 and 7 days postinfection were examined, and the presence or absence of diarrhea was noted. Rectal temperature was recorded daily for 7 days postinfection. Observations of fecal consistency and the attitude of the animals, as recorded by one of the investigators, were combined to form a clinical scoring system (Table 1). These scores were recorded daily for the first 9 days postinfection and indicated the severity of illness observed in each animal over this period.
TABLE 1.
Clinical scores for pigs in different treatment groups for the first 9 days postinfection
| Pig no. | Total clinical scorea
|
||
|---|---|---|---|
| Control | Suspension | Fermentate | |
| 1 | 15 | 2 | 1 |
| 2 | 7 | 2 | 7 |
| 3 | 0 | 3 | 0 |
| 4 | 4 | 6 | 0 |
| 5 | 9 | 2 | 0 |
| Mean | 7.0 | 3.0 | 1.6 |
The scoring system is a combined scoring for attitude (0 = normal, 1 = must be stimulated to get up, 2 = gets up with help, 3 = cannot get up) and feces (0 = normal, 1 = soft, 2 = mild diarrhea, 3 = severe, watery diarrhea).
Microbiological analysis of pig fecal samples.
Fecal samples were stored at 4°C and analyzed within 24 h of collection; samples were homogenized in maximum recovery diluent (Lab M, United Kingdom) as 10-fold dilutions using a stomacher (Lab-Blender 400; Seward Medical, London, United Kingdom), further diluted in maximum recovery diluent, and appropriate dilutions were pour plated. The administered strains were enumerated following anaerobic incubation for 5 days at 37°C (as recommended by the manufacturer) on Lactobacillus selective agar (LBS agar; Becton Dickinson, Cockeysville, MD) containing 150 μg/ml of rifampin as a selective agent and 50 U/ml nystatin (Sigma) to inhibit yeasts and molds (LBS-RIF). In addition, 18 to 20 colonies selected from LBS-RIF plates from each probiotic-fed animal at day 8 and day 23 p.i. (i.e., up to 100 colonies in total for each treatment group) were cultured in MRS broth and fingerprinted by RAPD-PCR to determine the relative proportions of the individual probiotic strains. Genomic DNA was isolated from 1.5 ml of the overnight cultures according to the method outlined by Coakley et al. (5). The extracted DNA was then used as a template in PCR amplifications, which were performed using the random primer R2 (5′-GTGATGTGCTGGTGTTATGTTTA-3′; MWG Biotech, Ebersberg, Germany) as previously outlined (14), with the following modifications: PCR amplifications were performed in a total volume of 50 μl in an Eppendorf DNA thermal cycler (Eppendorf Scientific Inc., Westbury, NY) with 1.25 U of Taq DNA polymerase (Bioline, London, United Kingdom) added to the reaction mix. The PCR products (10 μl of each reaction mixture) were analyzed on a 1.5% (wt/vol) agarose (Sigma) gel, using a 100-bp ladder (New England Biolabs, Hitchin, Hertfordshire, United Kingdom) as a molecular weight standard. The fingerprints obtained from fecal isolates were compared with those of DNA from each of the cultures present in the administered probiotic mixtures, allowing for the identification of individual probiotic cultures in each fecal sample.
Preliminary experiments suggested that the numbers of Salmonella recovered from the pig feces would be very low and not suitable for counting by direct plating (data not shown). Fecal Salmonella numbers were therefore determined by a three-tube most probable number (MPN) method. Fecal samples were initially nonselectively enriched in buffered peptone water (BPW; Lab M). Ten grams of feces was added to 90 ml of BPW and homogenized for 4 minutes. The homogenate was then further diluted based on expected Salmonella numbers before being stored at 4°C. This second homogenate was then divided into three 50-ml, three 5-ml, and three 0.5-ml aliquots; the 0.5-ml aliquots were further diluted by the addition of 1 ml BPW. All tubes were incubated overnight at 37°C, after which samples were selectively enriched by adding 0.1 ml from each tube to 9.9 ml of Rappaport-Vassiliadis broth (Lab M) and incubating for 18 to 24 h at 42°C. A loopful of suspension from each tube was streaked on brilliant green agar (Lab M) containing 20 μg/ml of nalidixic acid (Sigma) and 25 μg/ml novobiocin (Sigma). The number of positive brilliant green agar plates was noted, and from this the number of Salmonella present in each sample was calculated using the MPN table of de Man (7). Where an end point for the MPN procedure was not reached (i.e., Salmonella numbers were either above or below the appropriate limit of detection [LOD]), the procedure was repeated using a further dilution of the original homogenate.
Statistical analysis.
Data pertaining to animal weight, temperature, presence or absence of diarrhea, and clinical score were studied in one of two ways. For investigation of data regarding individual probiotic groups, one-way analysis of variance (ANOVA) was used to analyze normally distributed data. Data exhibiting a nonnormal pattern of distribution were analyzed by the nonparametric Kruskal-Wallis one-way ANOVA on ranks test, with post hoc comparison by Dunnett's method. Where data were considered on the basis of a single probiotic grouping (i.e., suspension and fermentate combined), Student's t test was used for data satisfying assumptions of normality; where nonparametric analysis was required, the Mann-Whitney U test was used. Where serovar Typhimurium MPN counts were not obtained due to the result falling below a detectable level, the value of the applicable LOD was substituted; this value was also added to all other data obtained at that time point. Where MPN counts were not obtained due to the result being above a detectable level, the value of the applicable LOD was again substituted; this generally applied where there was insufficient fecal sample available to perform a repeated MPN procedure. Where sufficient fecal sample was available to perform repeated MPN analyses but results were still above detectable levels 14 days after initial sample collection, values were left blank due to the uncertainty inherent in using samples outside this time frame. Salmonella counts were transformed to log base 10 before analysis by one-way ANOVA, with post hoc multiple comparison procedures performed using the Holm-Sidak method. Differences were considered significant if P was <0.05. Data are reported as the mean ± standard error of the mean. All calculations were performed using the program SigmaStat 3.1 (Systat Software, Erkrath, Germany).
RESULTS
Physical indicators.
The weights of the animals in the control group increased by a mean of 500.6 ± 57.9 g/day, a figure similar to that normally seen for weaners in Ireland. Higher mean weight increases were seen in animals treated with either the suspension (636.4 ± 21.7 g/day) or fermentate (632.4 ± 34.5 g/day), although these differences were not significant when using a P value of <0.05 (P = 0.06). Thirteen of 25 individual fecal samples collected between days 3 and 7 p.i. from animals fed control milk were positive for the presence of diarrhea. This compared with only 6 positive samples (of 50 overall) collected from animals fed probiotic, three each from animals fed the probiotic as suspension or as fermentate. Statistical analysis indicated that the feeding of probiotic to animals was associated with a significantly lower risk of diarrhea (P < 0.001). Of the five pigs in the control group, four exhibited diarrhea during the 5-day period. Of the animals fed probiotic, two of those fed the probiotic suspension gave diarrhea-positive stool samples, with only one fermentate-fed animal showing diarrhea during the study. Statistical analysis of the mean clinical score of animals in the control group (7.0) and animals fed probiotic when taken as a single group (suspension and fermentate scores combined; 2.1) resulted in a significantly lower score for the probiotic-fed animals. When the probiotic treatments were compared separately (suspension versus control and fermentate versus control) against the control group, their mean clinical scores, while lower, were not significantly so (P = 0.11) (Table 1). Significant increases in rectal temperature were not detected in animals from any group postinfection.
Fecal excretion of administered cultures.
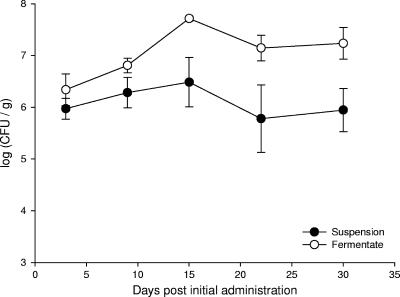
Total numbers of excreted probiotic cultures rose in all treated animals to a peak of between 6.48 and 7.71 log10 CFU/g feces at 15 days after the first administration (8 days postinfection). Compared to suspension-treated animals, higher total counts were observed in all cases from pigs fed the fermentate (Fig. 1); this is in keeping with the higher probiotic count delivered in the fermentate mixture.
FIG. 1.
Mean total numbers of rifampin-resistant colonies isolated from fecal samples from pigs administered a probiotic mixture as either fermentate or suspension.
Effects on fecal excretion of S. enterica serovar Typhimurium.
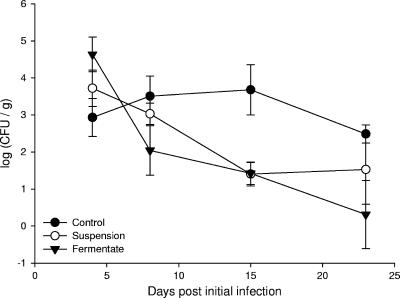
Salmonella spp. in fecal samples of control and treatment groups were enumerated by the MPN technique at regular intervals postinfection. No statistical differences were observed between control and probiotic treatment groups at 4 days postinfection (P = 0.09) (Fig. 2). At 8 days postinfection, the mean Salmonella numbers of the suspension and fermentate groups (3.03 and 2.04 log10 CFU/g feces, respectively) were lower than that of the control group (3.51 log10 CFU/g), although this difference was not shown to be statistically significant (P = 0.18). At 15 days postinfection, the numbers of Salmonella detected in the control samples were significantly higher than those of the probiotic grouping (P = 0.01); analyzed individually, the mean values of the suspension and fermentate treatment groups (1.4 and 1.42 log10 CFU/g feces, respectively) were also shown to be statistically significantly different from that of the control (3.68 log10 CFU/g) (Fig. 2).
FIG. 2.
Numbers of Salmonella enumerated by MPN from feces of pigs administered either control milk or probiotic and subsequently challenged with S. enterica serovar Typhimurium PT12 on three consecutive days.
Analysis of the final fecal samples from the animals, taken 23 days after the initial Salmonella infection, indicated no significant difference in numbers of Salmonella between the control and probiotic groupings (P = 0.2) (Fig. 2).
Excretion of administered probiotic cultures.
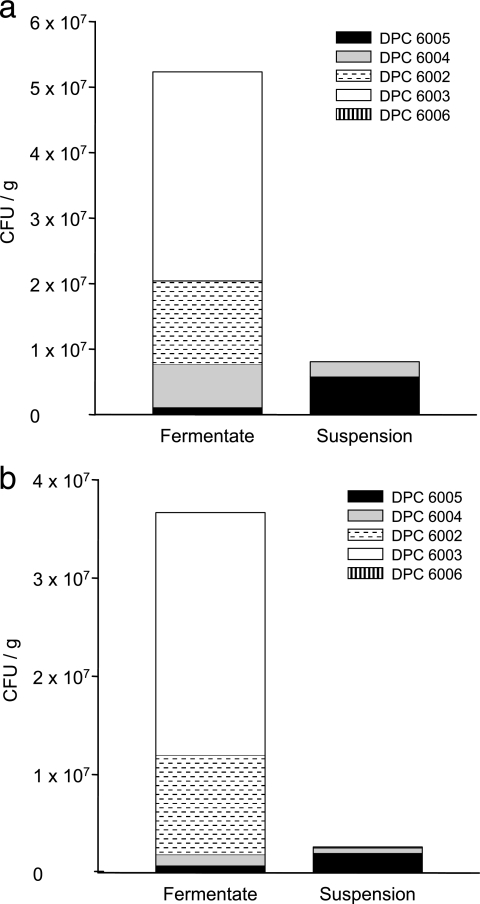
Fecal samples taken at days 8 and 23 postinfection from animals fed probiotic as suspension or fermentate were analyzed by RAPD-PCR, and the composition of individual probiotic cultures within the mixture was identified (Fig. 3).
FIG. 3.
Numbers and distribution of individual probiotic cultures isolated from fecal samples from pigs administered a probiotic mixture as either a fermentate or suspension. (a) Eight days post-initial infection; (b) 23 days p.i.
At 8 and 23 days postinfection, L. murinus DPC 6003 was by far the most prevalent culture recovered from animals receiving the fermentate treatment, accounting for 60% of colonies analyzed at day 8 and 67% at day 23. Relatively large numbers of L. murinus DPC 6002 were also recovered, accounting for 24% of colonies at day 8 and approximately 28% at day 23. At 8 days postinfection, 12.5% of colonies taken from fermentate samples were identified as L. pentosus DPC 6004, while this culture only accounted for 3% of colonies from the same animals at day 23 postinfection. Two percent of colonies taken from fermentate samples at both 8 and 23 days postinfection were identified as L. salivarius DPC 6005. Only one colony of Pediococcus pentosaceus DPC 6006 was recovered at day 8 p.i. and did not account for any of the isolates at day 23 (Fig. 3).
The proportions of colonies recovered from fecal samples of animals administered the suspension treatment differed considerably from those observed in fermentate-fed animals. L. salivarius DPC 6005 accounted for 72% and 77% of colonies recovered at day 8 and day 23 p.i., respectively. The remaining 28% of colonies at day 8 were all identified as L. pentosus DPC 6004; this probiotic represented 19% of the total at day 23. Low numbers of both L. murinus DPC 6002 (2.5%) and L. murinus DPC 6003 (1%) were also identified at day 23. No colonies of P. pentosaceus DPC 6006 were identified in either case (Fig. 3).
When the overall numbers of Rifr probiotic colonies were enumerated, it was observed that total numbers recovered from fermentate samples were 2 to 20 times higher than those from suspension samples (Fig. 1); this is in keeping with the higher initial numbers in the fermentate administered. Numbers at day 23 were slightly lower than those at day 8. Combining these data with the relative proportions for individual cultures obtained above gives a good indication of the actual numbers represented by each culture. While large numbers of L. murinus strains DPC 6002 and 6003 were recovered from fermentate samples but not from suspension samples, the combined numbers (CFU/g) of DPC 6005 and DPC 6004 were very similar in both the fermentate and suspension samples at both day 8 (8.0 × 106 CFU/g and 8.08 × 106 CFU/g, respectively) and day 23 (1.9 ×106 CFU/g and 1.0 × 106 CFU/g, respectively) (Fig. 3).
DISCUSSION
Salmonella spp. infection remains a major cause of food-borne gastroenteritis, with an estimated 160,000 cases of human salmonellosis reported annually in the European Union (9, 10). Serovar Typhimurium and serovar Enteritidis are the serotypes most commonly associated with cases of food-borne salmonellosis in humans (9). In the United States, the FoodNet program has noted that the estimated incidence of Salmonella infection has declined at a slower rate than other major food-borne illnesses (3). Consumption of pork products containing salmonellae leads to many cases of food-borne illness each year (3, 28). Serovar Typhimurium remains the serovar most commonly isolated from pigs in Ireland and elsewhere (1, 6, 26).
The probiotic bacteria used in this study were selected based upon their ability to survive simulated gastrointestinal environments and to prevent Salmonella invasion of intestinal epithelial cells (2). We have also examined the ability of the five strains to survive transit through the pig GIT in healthy animals and highlighted the advantages of using the strains in combination (15). In the current study, we examined whether these procedures were effective in isolating bacteria with in vivo anti-Salmonella activity by investigating the efficacy of the five strains in improving the outcome of Salmonella infection in weaned pigs.
Application of either of the probiotic treatments in this study resulted in reduced numbers of fecal Salmonella. While lower mean counts were observed for probiotic-treated animals at a number of sampling points, these differences were significant at 15 days postinfection, with the control animals exhibiting a mean Salmonella count approximately 200-fold higher than either of the probiotic groups. As the number of salmonellae shed by the pigs was sometimes higher than expected, some samples were not sufficiently diluted in order to obtain an endpoint in the MPN technique. This made comparison at day 4 p.i. particularly difficult, as low sample quantities resulted in only two counts being obtained for control animals. The lack of observed probiotic effect at this point may be due to high numbers of administered Salmonella still being excreted after the third consecutive day of inoculation, i.e., “flushing through.” Salmonella counts rose slightly for pigs in the suspension group at 23 days p.i. We believe that this may be due to reinfection of animals from their surroundings. Such environmental infection has been shown to be an important factor in Salmonella carriage in pigs (16, 19). A number of reports have been published describing probiotic-mediated reduction in intestinal Salmonella numbers (12, 17); these studies have, however, utilized serovar Choleraesuis, which has not been implicated as a major cause of human disease. The culture used by these groups is a complicated mix of different bacteria, including species of the genera Clostridium and Enterococcus; it is possible that this composition could lead to pathogen transmission and may not be suitable for human application. Those studies also described the administration of cultures to neonatal pigs which had not yet established a stable gut flora, in contrast to our use of older, weaned animals.
While the candidate cultures proved effective in the primary goal of reducing intestinal Salmonella numbers, we also investigated their effects on several disease indicators. Administration of either form of probiotic mixture clearly led to alleviation of physical signs of illness in the animals, although not always significantly so. In all cases, no significant differences were seen between results for the fermentate and suspension groups. Perhaps the most obvious sign of gastrointestinal infection is the presence of diarrhea. While over 50% of stool samples from control animals were diarrhea positive, the corresponding figure for probiotic-treated pigs was significantly lower at 12% in both cases. Ogawa et al. have demonstrated a reduced severity of diarrhea in E. coli O157:H7-infected rabbits (24); we are not aware of any reports of probiotic-mediated reduction in incidence of diarrhea in any model of Salmonella infection to date. Clinical scoring systems are used in an attempt to quantify objectively the severity of disease between groups of animals; such a system was devised, with higher scores indicating more severe illness. While both probiotic treatment groups yielded lower mean scores than the control group, these were not significantly so. However, investigation of the culture-fed animals as a single group (based on results detailed below) did result in significantly lower clinical scores. In addition to the differences recorded in clinical scores between treated and control groups, the veterinarian responsible for animal welfare noted a marked difference between the control and probiotic groups in overall demeanor, with treated groups being consistently brighter and more alert. The weight gain of the animals in the individual probiotic groups across the trial period, while greater than that of control animals, was not significantly so based on a P level of <0.05. Improved growth performance in pigs due to the administration of probiotics has previously been demonstrated by Chang (4).
RAPD-PCR analysis of excreted probiotic cultures indicated that the two L. murinus strains dominated in animals administered the probiotic fermentate. This is in keeping with our previous study (15), during which pigs were also fed probiotic-fermented milk. However, examination of colonies isolated from animals treated with a milk suspension containing the same strains in similar relative proportions demonstrated that the two L. murinus strains comprised only a small proportion of excreted probiotics. An examination of the composition of the milk fermentate showed that the numbers of the individual cultures present remained approximately the same during storage (data not shown); it is thus likely that the milk fermentation process gives the L. murinus strains a selective advantage in subsequent intestinal conditions. Investigation of the numbers of excreted individual cultures (Fig. 3) revealed similar combined levels of L. pentosus DPC6004 and L. salivarius DPC 6005 in both fermentate- and suspension-fed animals at day 8 p.i. and day 23 p.i. Due to the lack of differences observed in the results for both probiotic groupings, it may be possible that any probiotic effect can be ascribed to the common factor(s) in the two treatments, i.e., L. pentosus DPC6004 and L. salivarius DPC 6005; whether either DPC6004 or DPC6005 alone or the combination of both strains is the source of the beneficial effects seen is not yet clear. This hypothesis is supported by our earlier results (15) showing no decrease in numbers of Enterobacteriaceae isolated from the feces of pigs fed either of the L. murinus strains but substantial reductions in animals fed any of strains DPC6004, DPC6005, or DPC6006 or a mixture of all five strains. The results therefore also suggest that the dominant bacteria in probiotic mixtures may not be responsible for the observed beneficial effects. However, as we cannot say with certainty that the fecal microbiological composition mirrors that of the intestine, further investigations will be necessary to establish the accuracy of this theory. This observation of a potentially similar “active ingredient” in both probiotic groups led us to combine the data from both groups in some statistical tests, as described earlier.
The data presented in this report show that the probiotic mixtures used led to an amelioration of diarrhea in S. enterica serovar Typhimurium-infected pigs early in the course of infection and reduced pathogen counts over a longer time frame. This demonstrates the validity of using commensal LAB strains in the prevention of gastrointestinal infection and underlines the usefulness of the in vitro and in vivo procedures used to isolate and select the bacteria (2, 15).
Acknowledgments
We acknowledge the assistance of all the staff at the Central Veterinary Research Laboratory, Dublin, and at the Pig Production Development Unit, Moorepark Research Centre, Fermoy, Ireland. We are also grateful to Michael Cronin for assistance with statistical analysis.
This work was funded by the Irish Government under National Development Plan 2000-2006 and by Science Foundation Ireland.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Casey, P. G., D. Butler, G. E. Gardiner, M. Tangney, P. Simpson, P. G. Lawlor, C. Stanton, R. P. Ross, C. Hill, and G. F. Fitzgerald. 2004. Salmonella carriage in an Irish pig herd: correlation between serological and bacteriological detection methods. J. Food Prot. 67:2797-2800. [DOI] [PubMed] [Google Scholar]
- 2.Casey, P. G., G. D. Casey, G. E. Gardiner, M. Tangney, C. Stanton, R. P. Ross, C. Hill, and G. F. Fitzgerald. 2004. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Lett. Appl. Microbiol. 39:431-438. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2006. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, United States, 2005. Morb. Mortal. Wkly. Rep. 55:392-395. [PubMed] [Google Scholar]
- 4.Chang, Y. H., J. K. Kim, H. J. Kim, W. Y. Kim, Y. B. Kim, and Y. H. Park. 2001. Selection of a potential probiotic Lactobacillus strain and subsequent in vivo studies. Antonie Leeuwenhoek 80:193-199. [DOI] [PubMed] [Google Scholar]
- 5.Coakley, M., R. P. Ross, and D. Donnelly. 1996. Application of the polymerase chain reaction to the rapid analysis of brewery yeast strains. J. Inst. Brew. 102:349-354. [Google Scholar]
- 6.Davies, R., G. Paiba, S. Evans, and B. Dalziel. 2000. Surveys for Salmonella in pigs, cattle and sheep at slaughter in Great Britain. Vet. Rec. 147:695. [PubMed] [Google Scholar]
- 7.de Man, J. C. 1983. MPN tables, corrected. Eur. J. Appl. Microbiol. 17:301-305. [Google Scholar]
- 8.Drago, L., M. R. Gismondo, A. Lombardi, C. de Haen, and L. Gozzini. 1997. Inhibition of in vitro growth of enteropathogens by new Lactobacillus isolates of human intestinal origin. FEMS Microbiol. Lett. 153:455-463. [DOI] [PubMed] [Google Scholar]
- 9.European Commission. 2002. Trends and sources of zoonotic agents in animals, feeding stuffs, food and man in the European Union and Norway in 2000. European Commission document SANCO/927/2002. http://europa.eu.int/comm/food/fs/sfp/mr/mr08_en.pdf.
- 10.European Commission. 2002. Directive of the European Parliament and of the Council on the monitoring of zoonoses and zoonotic agents, amending Council Decision 90/424/EEC and repealing Council Directive 92/117/EEC. http://europa.eu.int/eur-lex/pri/en/lip/latest/doc/2002/com2002_0684en01.doc.
- 11.FAO/WHO. 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Joint FAO/WHO expert consultation. Cordoba, Argentina, 1-4 October 2001. http://www.who.int/foodsafety/publications/fs_management/probiotics/en/.
- 12.Fedorka-Cray, P. J., J. S. Bailey, N. J. Stern, N. A. Cox, S. R. Ladely, and M. Musgrove. 1999. Mucosal competitive exclusion to reduce Salmonella in swine. J. Food Prot. 62:1376-1380. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez, M. F., S. Boris, and C. Barbes. 2003. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 94:449-455. [DOI] [PubMed] [Google Scholar]
- 14.Gardiner, G., R. P. Ross, J. K. Collins, G. Fitzgerald, and C. Stanton. 1998. Development of a probiotic cheddar cheese containing human-derived Lactobacillus paracasei strains. Appl. Environ. Microbiol. 64:2192-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardiner, G. E., P. G. Casey, G. Casey, P. B. Lynch, P. G. Lawlor, C. Hill, G. F. Fitzgerald, C. Stanton, and R. P. Ross. 2004. Relative ability of orally administered Lactobacillus murinus to predominate and persist in the porcine gastrointestinal tract. Appl. Environ. Microbiol. 70:1895-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gebreyes, W. A., P. R. Davies, P.-K. Turkson, W. E. Morrow, J. A. Funk, and C. Altier. 2004. Salmonella enterica serovars from pigs on farms and after slaughter and validity of using bacteriologic data to define herd Salmonella status. J. Food Prot. 67:691-697. [DOI] [PubMed] [Google Scholar]
- 17.Genovese, K. J., R. C. Anderson, R. B. Harvey, T. R. Callaway, T. L. Poole, T. S. Edrington, P. J. Fedorka-Cray, and D. J. Nisbet. 2003. Competitive exclusion of Salmonella from the gut of neonatal and weaned pigs. J. Food Prot. 66:1353-1359. [DOI] [PubMed] [Google Scholar]
- 18.Genovese, K. J., R. C. Anderson, R. B. Harvey, and D. J. Nisbet. 2000. Competitive exclusion treatment reduces the mortality and fecal shedding associated with enterotoxigenic Escherichia coli infection in nursery-raised neonatal pigs. Can. J. Vet. Res. 64:204-207. [PMC free article] [PubMed] [Google Scholar]
- 19.Hurd, H. S., J. K. Gailey, J. D. McKean, and M. H. Rostagno. 2001. Rapid infection in market-weight swine following exposure to a Salmonella typhimurium-contaminated environment. Am. J. Vet. Res. 62:1194-1197. [DOI] [PubMed] [Google Scholar]
- 20.Johnson-Henry, K. C., D. J. Mitchell, Y. Avitzur, E. Galindo-Mata, N. L. Jones, and P. M. Sherman. 2004. Probiotics reduce bacterial colonization and gastric inflammation in H. pylori-infected mice. Dig. Dis. Sci. 49:1095-1102. [DOI] [PubMed] [Google Scholar]
- 21.La Ragione, R. M., A. Narbad, M. J. Gasson, and M. J. Woodward. 2004. In vivo characterization of Lactobacillus johnsonii FI9785 for use as a defined competitive exclusion agent against bacterial pathogens in poultry. Lett. Appl. Microbiol. 38:197-205. [DOI] [PubMed] [Google Scholar]
- 22.Lema, M., L. Williams, and D. R. Rao. 2001. Reduction of fecal shedding of enterohemorrhagic Escherichia coli O157:H7 in lambs by feeding microbial feed supplement. Small Rumin. Res. 39:31-39. [DOI] [PubMed] [Google Scholar]
- 23.Nisbet, D. J., G. I. Tellez, V. K. Lowry, R. C. Anderson, G. Garcia, G. Nava, M. H. Kogut, D. E. Corrier, and L. H. Stanker. 1998. Effect of a commercial competitive exclusion culture (Preempt) on mortality and horizontal transmission of Salmonella gallinarum in broiler chickens. Avian Dis. 42:651-656. [PubMed] [Google Scholar]
- 24.Ogawa, M., K. Shimizu, K. Nomoto, M. Takahashi, M. Watanuki, R. Tanaka, T. Tanaka, T. Hamabata, S. Yamasaki, and Y. Takeda. 2002. Protective effect of Lactobacillus casei strain Shirota on Shiga toxin-producing Escherichia coli O157:H7 infection in infant rabbits. Infect. Immun. 69:1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pascual, M., M. Hugas, J. I. Badiola, J. M. Monfort, and M. Garriga. 1999. Lactobacillus salivarius CTC2197 prevents Salmonella enteritidis colonization in chickens. Appl. Environ. Microbiol. 65:4981-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quirke, A.-M., N. Leonard, G. Kelly, J. Egan, P. B. Lynch, T. Rowe, and P. J. Quinn. 2001. Prevalence of Salmonella serotypes on pig carcasses from high- and low-risk herds slaughtered in three abattoirs. Berl. Munch. Tierarztl. 114:360-362. [PubMed] [Google Scholar]
- 27.Silva, A. M., F. H. Barbosa, R. Duarte, L. Q. Vieira, R. M. Arantes, and J. R. Nicoli. 2004. Effect of Bifidobacterium longum ingestion on experimental salmonellosis in mice. J. Appl. Microbiol. 97:29-37. [DOI] [PubMed] [Google Scholar]
- 28.Swanenburg, M., H. A. Urlings, J. M. Snijders, D. A. Keuzenkamp, and F. van Knapen. 2002. Salmonella in slaughter pigs: prevalence, serotypes and critical control points during slaughter in two slaughterhouses. Int. J. Food Microbiol. 70:243-254. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, T., M. P. Doyle, B. G. Harmon, C. A. Brown, P. O. Mueller, and A. H. Parks. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J. Clin. Microbiol. 36:641-647. [DOI] [PMC free article] [PubMed] [Google Scholar]