Abstract
Short-chain fructooligosaccharides (scFOS) and other prebiotics are used to selectively stimulate the growth and activity of lactobacilli and bifidobacteria in the colon. However, there is little information on the mechanisms whereby prebiotics exert their specific effects upon such microorganisms. To study the genomic basis of scFOS metabolism in Lactobacillus plantarum WCFS1, two-color microarrays were used to screen for differentially expressed genes when grown on scFOS compared to glucose (control). A significant up-regulation (8- to 60-fold) was observed with a set of only five genes located in a single locus and predicted to encode a sucrose phosphoenolpyruvate transport system (PTS), a β-fructofuranosidase, a fructokinase, an α-glucosidase, and a sucrose operon repressor. Several other genes were slightly overexpressed, including pyruvate dehydrogenase. For the latter, no detectable activity in L. plantarum under various growth conditions has been previously reported. A mannose-PTS likely to encode glucose uptake was 50-fold down-regulated as well as, to a lower extent, other PTSs. Chemical analysis of the different moieties of scFOS that were depleted in the growth medium revealed that the trisaccharide 1-kestose present in scFOS was preferentially utilized, in comparison with the tetrasaccharide nystose and the pentasaccharide fructofuranosylnystose. The main end products of scFOS fermentation were lactate and acetate. This is the first example in lactobacilli of the association of a sucrose PTS and a β-fructofuranosidase that could be used for scFOS degradation.
Prebiotics are defined as nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon that can improve host health (19). Main targets for the prebiotic approach are bifidobacteria and lactobacilli, as these intestinal bacteria show several positive effects upon human well-being (18, 35).
Fructooligosaccharides are among the most extensively studied prebiotics and include a diverse family of fructose polymers which vary in length and can either be derivatives of simple fructose polymers or fructose moieties attached to a sucrose molecule. Most fructooligosaccharides marketed as ingredients for foods and nutritional supplements are either synthesized from sucrose using fructosyltransferases derived from Aspergillus niger (short chain fructooligosaccharides [scFOS]) or extracted from chicory roots (Cichoricum intybus) by a partial enzymatic hydrolysis of inulin (FOS). Their general structure can be represented by GFn or Fn, in which G and F are, respectively, a glucose or fructose unit and n is the number of fructosyl units. The different units are linked by β(2→1) bonds. The structures of these two sugar classes are similar. FOS and scFOS are resistant to digestion in the stomach and small intestine (3, 15) and thus are able to reach the colon, where they are selectively fermented by beneficial bacteria (7, 26).
While some limited insight has been provided for the manner by which bifidobacteria utilize fructooligosaccharides (33), mechanisms in lactobacilli are still unclear despite the commercial interest in this group of gram-positive bacteria (19). A recent study reported the presence of a fructofuranosidase responsible for the hydrolysis of fructooligosaccharides in L. acidophilus (4) encoded by an inducible operon that included a fructofuranosidase gene associated with the genes for an ATP-binding cassette (ABC) transport system. This study showed that L. acidophilus was able to use both scFOS and FOS. Similarly, an ATP-dependent transport system was implied in FOS utilization by L. paracasei (23).
L. plantarum is encountered in many food products and is a natural inhabitant of the human gastrointestinal tract (GIT) (16). It is also marketed as a probiotic (27). L. plantarum WCFS1 was originally isolated from human saliva (24). Its complete genome (3.3 Mb) has been analyzed in detail and is one of the largest known for lactic acid bacteria (24). One of its characteristics is the ability to ferment a large range of carbohydrates, such as monosaccharides (glucose, mannose, and galactose), disaccharides (sucrose, lactose, and trehalose), and oligosaccharides (raffinose and melezitose) (8). It has been also shown to survive passage through the stomach while remaining active and persisting for more than 6 days in the human GIT (43). The genetic basis for this persistence was investigated, and results showed that a number of genes encoded functions related to the metabolism of sugars were induced in situ in the GIT (9).
Metabolism of prebiotic fructooligosaccharides is an important feature for a Lactobacillus strain, and the mechanisms behind this have not been elucidated. Here, we describe a comparison of the expression of the genes of L. plantarum WCFS1 when grown on a chemically defined medium with scFOS or glucose using two-color oligonucleotide microarrays to screen differently expressed genes involved in metabolism. Analyses of the fractions of scFOS consumed were also performed to determine preference in the uptake of the different moieties.
MATERIALS AND METHODS
Growth with fructooligosaccharides in a chemically defined medium.
L. plantarum strain WCFS1 (24) was grown anaerobically in 10 ml of chemically defined medium, the composition of which has been previously described (33), with the following modifications (in mg/liter): K2HPO4 (Merck, The Netherlands), 1,000; KH2PO4 (Merck), 5,000; tyrosine (Sigma, The Netherlands), 250; d-biotin (Sigma), 2.5; FeCl3·6H2O (Sigma), 3; MnCl2·4H2O (BDH, United Kingdom), 16; CuSO4·5H2O (BDH), 2.5; (NH4)6Mo7O24·4H2O (BDH), 2.5; cysteine-HCl (Sigma), 130. As the sole carbon source, 10 g/liter of the test sugars was added to the growth medium. Carbohydrates were either scFOS Actilight 950P (Beghin Meiji, France) or FOS Raftilose P95 (Orafti, Belgium). Glucose (BDH) was used as a control. Growth with the maximum quantity of residual sugars (0.5 g/liter), such as glucose, fructose (Sigma), or sucrose (Sigma,) contained in both scFOS and FOS was monitored in order to verify that this amount of sugar was negligible when growth occurred with fructooligosaccharides. scFOS had the following composition: >95% (wt/wt) fructooligosaccharides (trisaccharide [1-kestose] GF2, tetrasaccharide [nystose] GF3, and pentasaccharide [1F-fructosyfuranosylnystose] GF4) and <5% (wt/wt) residual sugars, such as glucose (G), fructose (F), and sucrose (S). FOS had the following composition: >95% (wt/wt) fructooligosaccharides (78% Fn-type molecules and 22% GFn-type molecules, with a degree of polymerization [DP] from 2 to 10) and <5% (wt/wt) residual sugars, such as glucose, fructose, and sucrose. Culture optical density at 600 nm (OD600) was measured using a spectrophotometer (Ultrospec 3000; Pharmacia Biotech) for 24 h.
Batch culture study: fermentations.
Four batch culture systems containing 500 ml of the chemically defined medium described above were prepared, two containing 1% (wt/vol) glucose and two containing 1% (wt/vol) scFOS as the sole carbon source. Media were gassed with O2-free N2 (15 ml/min) to keep the cultures anaerobic. The cultures were constantly stirred, and the temperature was kept constant at 37°C by means of a circulating water bath which pumped into a jacket surrounding the fermentors. Each fermentor was inoculated with 2% (vol/vol) with an overnight culture of L. plantarum WCFS1 grown in the same chemically defined medium with the same sugar as in the main culture, to generate approximately 108 cells/ml in the vessel. Every 30 min, 2.5 ml of culture fluid was removed from each batch culture system to measure pH and the OD600. When fermentation had reached an optical density whereby the bacterium was in the exponential phase of growth and where most of the residual sugars of scFOS had been depleted (i.e., an OD600 of 1 for fermentation with glucose and scFOS and an OD600 of 1.5 for scFOS), a 50-ml sample was removed. Forty milliliters of this sample was kept for further RNA extraction and immediately added to 160 ml of quenching buffer (60% [vol/vol] methanol, 66.7 mM HEPES, pH 6.5, −40°C) (32) and stored for at least 30 min at −40°C. The 10 ml left from the batch culture sampling was stored at −20°C for further analysis of end products of fermentation and sugars.
Sugar analysis. (i) Total sugar assay.
The general phenol-sulfuric assay was performed for determination of total carbohydrate contents (10, 13). The assay was calibrated with d-glucose standards or Actilight P950 using ultrapure water from 0 to 0.15 mg/ml. From each sample taken during the batch culture fermentations at an OD600 of 1 and an OD600 of 1.5, 1 ml of supernatant was used following centrifugation at 1,500 × g and diluted 50-fold with distilled water. The assay was performed in triplicate for each sample. Results were expressed as the percentage of sugar remaining in the samples (wt/vol) compared to the initial concentration of sugar present at the beginning of the fermentation.
(ii) Determination of disappearance of different fractions of scFOS by HPAEC-PAD.
In order to determine whether the DP of the different fractions of scFOS had an influence on the fermentation capability of L. plantarum WCFS1, high-performance anion exchange chromatography with pulse amperometric detection (HPAEC-PAD) was used to measure the concentration of the different moieties still present when batch culture samples with scFOS were taken. For each sample with scFOS, 1 ml of supernatant was kept after centrifugation at 1,500 × g for 10 min. Supernatants were then passed through a 0.2-μm filter and 20-fold diluted in distilled water. The assay was calibrated with scFOS (Actilight 950P) as a standard whose composition in scFOS content was known, i.e., 1-kestose GF2, 44% (wt/wt); nystose GF3, 46% (wt/wt); and 1F-β-fructofuranosylnystose GF4, 10% (wt/wt). Standards with glucose (BDH), fructose (Sigma), and sucrose (Sigma) were also included.
Samples were injected in 25-μl volumes for analysis by HPAEC-PAD using a pellicular anion-exchange resin-based column CarboPac PA1 (Dionex Ltd., United Kingdom). Carbohydrates were eluted at a flow rate of 1 ml/min using a gradient mobile phase of concentrations of sodium hydroxide and sodium acetate solutions at 20 ± 0.5°C. The four different mobile phases used were as follows: A (12.5 mM NaOH), B (125 mM NaOH), C (125 mM NaOH/500 mM Na-acetate), and D (8 mM Na-acetate). A cleaning and preconditioning step was initially carried out followed by a subsequent analysis period of 50 min where different concentrations of the different eluents were used to separate and elute carbohydrate in the mixtures injected. Results were expressed as the percentage of the concentration of the moiety left in the samples compared to the initial concentration at the start of the fermentation.
End-product analysis.
The production of lactic, acetic, propionic, and butyric acids in the batch cultures was determined by high-performance liquid chromatography (HPLC). One milliliter of batch culture sample was centrifuged at 1,500 × g for 15 min, and 20 μl of the resulting supernatant was injected into a model 1050 UV HPLC (Hewlett Packard). The column was a prepacked Aminex HPX-87-H strong cation-exchange resin column (150 by 7.8 mm inner diameter), fitted with an ion-exclusive microguard refill cartridge (Bio-Rad). Degassed 0.005 M sulfuric acid was used as an eluent at a flow rate of 0.6 ml/min, and the operating temperature was 50°C. Organic acids were detected by UV at a wavelength of 220 nm and calibrated against standards of lactic, acetic, propionic, and butyric acids (Sigma) at concentrations between 10 and 100 mM. Initial concentrations of acetate present in the media were subtracted to quantify the amount of acetate produced during culture.
Transcriptome analysis. (i) Microarrays.
Microarrays containing in situ-synthesized 60-mer oligomers were produced by Agilent Technologies, according to a custom probe design based on the genome sequence of L. plantarum WCFS1. A total of 8,011 unique 60-mers having a theoretical melting temperature of approximately 82°C were selected. The melting temperature was calculated using nearest neighbor calculations (31), a Na+ concentration of 1 M, and an oligonucleotide concentration of 10−12 M. Of the unique probes, 1,596 were randomly selected and printed in duplicate on the array. The array contained a total of 9,607 custom 60-mers. Genes were represented by 1 (4.7%), 2 (6.8%), 3 (48.9%), 4 (32.5%), 5 (8.4%), or 6 probes (0.5%). A total of 260 putative genes were not represented on the array because no unique probe satisfying the selection criteria could be selected. Many of the putative genes not represented on the array were relatively short and encoded hypothetical proteins, transposase fragments, prophage proteins, ribosomal proteins, and tRNAs.
(ii) RNA extraction.
Following quenching, the cells were pelleted by centrifugation at 9,000 × g for 10 min at −20°C. The supernatant was then discarded, and the cell pellet was cooled in 60% (vol/vol) ethylene glycol. The cell pellet was transferred with a precooled spatula to a screw cap tube containing an extraction mixture (500 μl 1:1 phenol-chloroform, 30 μl of 10% [vol/vol] sodium dodecyl sulfate [SDS], 30 μl Na-acetate pH 5.2, 400 μl Tris-EDTA buffer [10 mM Tris(hydroxymethyl)amino methane, 1 mM EDTA, pH 7.4], and 500 mg of 75- to 150-μm glass beads). The cell pellet was thoroughly mixed with the extraction mixture, and the tube was immediately frozen in liquid nitrogen. Samples were stored at −80°C. The cells were then broken under frozen conditions in the Savant FastPrep FP120 (Qbiogen Inc., France) using two treatments of 40 seconds. The cells were then centrifuged at 4°C at 10,000 × g for 5 min. A 500-μl aliquot of the supernatant was transferred to a new tube, and 400 μl of cold chloroform was added. The tube was centrifuged at 4°C at 10,000 × g for 2 min. Isolation of RNA continued with the High Pure RNA isolation kit (Roche Diagnostics, Germany). Following isolation, 50 μl of RNA solution was obtained. The RNA concentration was measured at an OD of 260 nm with the ND-1000 spectrophotometer (NanoDrop Technologies Inc.). The A260/A280 ratio was measured to check purity of the RNA. Quality of the RNA obtained was analyzed with the 2100 Bioanalyser (Agilent Technologies). Samples whereby the 23S/16S RNA ratio was superior or equal to 1.6 were taken for further labeling.
(iii) Labeling and hybridization.
Five micrograms of RNA was used for each sample to be labeled. Each sample was labeled with cyanine 3 and cyanine 5. Indirect labeling was performed with the CyScribe first-strand cDNA labeling kit (Amersham, United Kingdom) according to the manufacturer's protocol. A 0.3-μg aliquot of Cydye-labeled cDNA was used for the hybridization for each sample. In total, six arrays (Agilent Technologies) were used. Each sample was hybridized twice, i.e., once for each dye. The arrays were hybridized for 17 h at 60°C. Slides were subsequently washed in solution 1 (700 ml MilliQ, 300 ml of 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.5 ml 10% [vol/vol] Triton X-102) for 10 min and in solution 2 (995 ml MilliQ, 5.0 ml 20× SSC, 0.5 ml 10% [vol/vol] Triton X-102; 4°C) for 5 min. Slides were then dried using a nitrogen-filled air gun to rapidly blow drops of solution from the slides. Slides were scanned with a ScanArray Express scanner (Perkin-Elmer), using a 10-nm resolution. Images were analyzed with the ImaGene 4.2 software (BioDicovery).
The microarray hybridization scheme consisted of a loop design with six arrays with the following samples hybridized on one array and labeled with cyanine3 and cyanine5, respectively: G1 and F1, F1 and F2, F2 and G2, G2 and F3, F3 and F4, and F4 and G1. Here, G1 and G2 represent biological duplicates taken when L. plantarum WCFS1 was grown on glucose at an OD600 of 1, F1 and F2 represent biological duplicates on scFOS at an OD600 of 1, and F3 and F4 represent biological duplicates on scFOS taken at an OD600 of 1.5. Samples F1 and F3 were taken from one culture at different optical densities, whereas F2 and F4 were taken from the duplicate culture.
Data analysis.
After blank spots had been removed, array measurements were normalized by local fitting of an M-A plot applying the loess algorithm (46), using the Limma package (39) in R (http://www.r-project.org). Spot intensities in both channels (I1 and I2) were thus converted to M-A coordinates, where M = log2(I1/I2) and A = log2(I1/I2)/2. Normalized intensities were used for further analysis.
First, it was confirmed that there were no systematic differences between the biological duplicates (G1/G2, F1/F2, and F3/F4) by comparing the error between replicates to the within-array error between duplicate spots. Subsequently, biological replicates were treated as duplicate measurements, and their averages were compared using linear modeling functions from the Limma package. The statistical significance of differences was calculated from variation in biological duplicates, using the eBayes function in Limma (cross-probe variance estimation) and false discovery rate (FDR) adjustment of the P values (38). The list of differentially expressed genes was projected onto the metabolic pathways known for L. plantarum (41) using the Sympheny software (Genomatica, Inc., San Diego, Calif.) to visualize which pathways were particularly affected. Only genes with FDR-adjusted P values less than 0.1, corresponding to at most 10% false positives, were displayed.
Database searches were performed using nonredundant sequence accessible at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) by using the BlastP program (1) for genes strongly upregulated with scFOS. To predict the location of the proteins, the biological software SignalP (http://www.cbs.dtu.dk/services/SignalP) was exploited.
Microarray accession numbers.
The microarray platform and microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession numbers GPL4318 and GSE5882, respectively.
RESULTS
Preferential growth of L. plantarum WCFS1 with glucose and scFOS.
L. plantarum WCFS1 was found to grow in a chemically defined medium under anaerobic conditions only with glucose or scFOS and reached, respectively, OD600 of 5.2 and 2.2 after 24-h incubation (data not shown). In contrast, L. plantarum WCFS1 did not grow well with FOS, and the maximum OD reached was 0.7 in the same period. When grown with 0.5 g/liter of residual sugars such as glucose, fructose, or sucrose, the OD600 never exceeded 0.4 after 24 h of fermentation, while in the medium without any carbohydrate the OD600 never exceeded 0.2. Hence, it was concluded that scFOS was a preferred substrate and was therefore chosen to be compared with growth of L. plantarum WCFS1 on glucose in controlled and duplicate batch cultures whereby the OD and pH where determined (Fig. 1). Growth of L. plantarum WCFS1 was more rapid in the medium with glucose added compared to scFOS, which was supported by a rapid acidification as an indication of bacterial fermentation.
FIG. 1.
OD600 and pH in batch cultures with L. plantarum WCFS1 in a chemically defined medium with 1% (wt/vol) glucose or scFOS as the sole carbon source. Curves are the averages of values from two biological duplicates ± the standard deviations. ▪, OD600 with glucose; □, OD600 with scFOS; ▴, pH with glucose; ▵, pH with scFOS.
Sugar consumption and uptake of different fractions of scFOS.
To assess the quantity of sugars consumed by L. plantarum WCFS1, the total sugar content was measured at an OD600 of 1 for glucose and scFOS and an OD600 of 1.5 for scFOS (Table 1). Results indicated that glucose and scFOS were partially consumed at the time of sampling.
TABLE 1.
Percentage of total carbohydrate contents remaining when L. plantarum WCFS1 was grown in a chemically defined medium with 1% (wt/vol) glucose or scFOS as the sole carbon source, compared to T0a
| Carbon source and time of sampling | Carbohydrate (% wt/vol) in medium compared to T0 |
|---|---|
| Glucose, OD600 = 1 | 69.4 (±1.9) |
| scFOS, OD600 = 1 | 63.4 (±8.6) |
| scFOS, OD600 = 1.5 | 46.7 (±2.3) |
Determined by phenol-sulfuric assay. Values are the averages of two biological duplicates (± standard deviation).
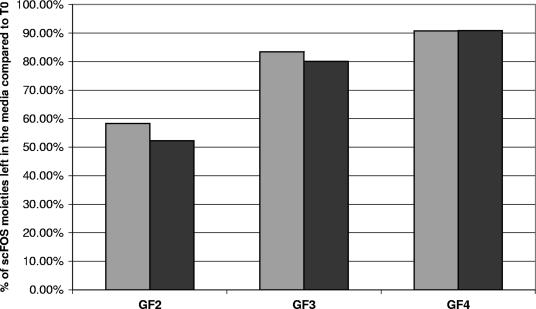
A disappearance in the medium of different fractions (GF2, GF3, and GF4) of scFOS was measured by HPAEC-PAD (Fig. 2). This revealed a preferential utilization of the GF2 moiety in scFOS compared to the GF3 and GF4 moieties. At an OD600 of 1 and 1.5, 58.3% (wt/vol) and 52.3% (wt/vol) of the initial GF2 content, respectively, was still present in the medium. This proportion was higher for GF3 at an OD600 of 1 and 1.5, with 83 and 80% (wt/vol), respectively. The GF4 moiety was hardly used, as 90.8% (wt/vol) was still present in the medium at OD600 of both 1 and 1.5. No accumulation of mono- or disaccharides was observed in the medium with scFOS, and most of the residual sugars were depleted at these time points. Traces of sucrose were detected in the medium with scFOS at the beginning of the fermentation (1% [wt/vol] of total sugars at maximum) and at an OD600 of 1 but not at an OD600 of 1.5.
FIG. 2.
Percentages of the different moieties, GF2, GF3, and GF4, left when L. plantarum WCFS1 was grown in a chemically defined medium with 1% scFOS (wt/vol) as the sole carbon source to an OD600 of 1 (light gray bars) or an OD600 of 1.5 (dark gray bars), compared to T0. Values are the averages of two biological duplicates.
Lactate and acetate, main end products of fermentation.
Lactate and to a lower extent acetate were the main end products of sugar fermentation under anaerobic conditions (Fig. 3). There was a slightly higher concentration of these compounds with scFOS at an OD600 of 1 and a higher concentration with scFOS at OD600 of 1.5 compared to glucose at an OD600 of 1. These results were in accordance with pH values recorded in the media of the batch cultures, which were of 4.10 for scFOS at an OD600 of 1, 3.80 for scFOS at an OD600 of 1.5, and 4.17 for glucose at an OD600 of 1.
FIG. 3.
Concentration of the end products lactate (light gray bars) and acetate (dark gray bars) when L. plantarum WCFS1 was grown in a chemically defined medium with 1% (wt/vol) glucose or scFOS as the sole carbon source. G, medium with glucose at an OD600 of 1; FOD1, scFOS at an OD600 of 1; FOD1.5, scFOS at an OD600 of 1.5. Values are the averages of two biological duplicates.
Transcriptome results.
To relate differences in the physiological response of L. plantarum WCFS1 to scFOS compared to glucose, the global transcriptome response was determined from a whole-genome microarray. Hence, RNA was isolated from samples taken from the batch cultures for physiological analysis and, after labeling, hybridized to the microarrays using a loop design. No significant differences were found between the biological duplicates grown on glucose or scFOS and harvested at an OD600 of 1 (samples G1 and G2 or F1 and F2). However, a cluster of genes involved in pyrimidine metabolism (lp_2697, lp_2698, lp_2700, and lp_2702) was found to be differentially expressed in the biological duplicates grown on scFOS and harvested at an OD600 of 1.5 (samples F3 and F4). Nevertheless, globally the biological duplicates were similar, and their average was taken in subsequent data analyses.
A growth phase effect was seen in one culture growing on scFOS when comparing samples F2 and F4 for a limited number of genes which were significantly differentially expressed (lp_2697, lp_2698, lp_2699, lp_2700, lp_2702, and lp_2371). These genes are predicted to participate in the synthesis of purine and pyrimidine. This effect was not apparent when comparing samples F1 and F3 from the duplicate cultures.
Using the average of biological duplicates, the effect of carbohydrate source (glucose or scFOS) on the transcriptome of L. plantarum WCFS1 was investigated. A total of 185 genes were found to be significantly differentially expressed when comparing glucose to scFOS at an OD600 of 1, and 191 were found when comparing glucose and scFOS at an OD600 of 1.5, which represents approximately 5% of the genome of L. plantarum WCFS1. Ninety-seven of these genes were differentially expressed both for scFOS at OD600 levels of 1 and 1.5 compared to glucose. Functionality of the genes differentially expressed was mostly related to carbohydrate metabolism or coding for transport binding proteins. Approximately half were up-regulated, and the remainder were down-regulated (Tables 2 and 3). Clustered genes (lp_0184 to lp_0189) could be distinguished which were strongly up-regulated with scFOS (8- to 60-fold). This set consisted of a fructokinase, a sucrose PTS EIIBCA, a β-fructofuranosidase, a sucrose operon repressor, and an α-glucosidase (Fig. 4). Some of these genes contained conserved domains, such as a domain of the repressor ORF kinase (ROK) family for the fructokinase, a periplasmic binding protein, and a sugar binding domain of the LacI family (pfam00532) as well as a helix-turn-helix motif of the lactose operon repressor (smart00354) for the sucrose operon repressor. Similarities with the products of the gene encoding a sucrose PTS (lp_0185) in Lactobacillus spp. strains other than L. plantarum strains were low. The first proteins showing a degree of similarity were a sucrose PTS from L. sakei (61% identity) and a sucrose PTS from L. acidophilus (57% identity). However, strong similarity with a sucrose PTS from a Pediococcus pentosaceus strain (97% identity) was identified. Similarities with the products of the gene encoding a β-fructofuranosidase (lp_0187) in Lactobacillus spp. strains other than L. plantarum strains were also low. The first enzyme showing a certain degree of similarity was a sucrose-6-phosphate hydrolase from L. sakei (50% identity) or sucrose-6-phosphate hydrolase from L. acidophilus (45% identity). Strong similarities with sucrose-6-phosphate hydrolases were obtained with a Pediococcus acidilactici and Pediococcus pentosaceus strain (98 and 97% identity).
TABLE 2.
Genes showing mainly up-regulation in L. plantarum WCFS1 grown on scFOS at an OD600 of 1 or 1.5 versus glucose
| Function group and ORF | Description | M FOD1a | M FOD1.5b |
|---|---|---|---|
| Energy metabolism and transport | |||
| lp_0184 | Fructokinase | 5.8 | 4.9 |
| lp_0185 | Sucrose PTS, EIIBCA | 6.0 | 4.6 |
| lp_0187 | β-Fructofuranosidase | 3.7 | 3.0 |
| lp_0189 | α-Glucosidase | 3.7 | 3.2 |
| lp_0193 | α-Glucosidase | 0.8 | NSc |
| lp_0280 | Transport protein | 0.8 | NS |
| lp_0286 | Cellobiose PTS, EIIC | 1.0 | NS |
| lp_0329 | Acetaldehyde dehydrogenase | 1.0 | NS |
| lp_0393 | Galactoside O-acetyltransferase | 0.7 | NS |
| lp_0764 | Phosphoglucomutase | 0.7 | NS |
| lp_0849 | Pyruvate oxidase | 0.8 | NS |
| lp_1101 | l-Lactate dehydrogenase | 1.0 | NS |
| lp_1102 | Transport protein | 1.0 | NS |
| lp_1105 | Malic enzyme, NAD-dependent | 0.9 | NS |
| lp_1106 | [Citrate (pro-3S)-lyase] ligase | 1.0 | NS |
| lp_1107 | Citrate lyase, acyl carrier protein | 1.0 | 1.0 |
| lp_1108 | Citrate lyase, β chain | 1.0 | 1.0 |
| lp_1109 | Citrate lyase, α chain | 1.1 | 1.0 |
| lp_1112 | Fumarate hydratase | 0.8 | NS |
| lp_1273 | Phosphocarrier protein Hpr | 0.8 | NS |
| lp_1274 | Phosphoenolpyruvate-protein phosphotransferase | 0.8 | NS |
| lp_1398 | β-Glucosides PTS, EIIA | 0.8 | NS |
| lp_1481 | Nitrite extrusion | 0.8 | NS |
| lp_1498 | Nitrate reductase, β chain | 0.8 | NS |
| lp_1499 | Nitrate reductase, delta chain | 0.7 | NS |
| lp_2096 | 1-Phosphofructokinase | 1.1 | NS |
| lp_2097 | Fructose PTS, EIIABC | 1.5 | NS |
| lp_2151 | Pyruvate dehydrogenase complex, E3 component dehydrogenase | 1.6 | 1.2 |
| lp_2152 | Pyruvate dehydrogenase complex, E2 component | 1.4 | 1.5 |
| lp_2153 | Pyruvate dehydrogenase complex, E1 component, β subunit | 1.6 | 1.1 |
| lp_2154 | Pyruvate dehydrogenase complex, E1 component, α subunit | 1.7 | NS |
| lp_2371 | Uracil transport protein | NS | 2.2 |
| lp_2757 | Glucan 1,4-α-maltohydrolase | 0.8 | NS |
| lp_2684 | Aromatic amino acid specific aminotransferase | 0.7 | 1.5 |
| lp_2776 | d-Serine dehydratase | NS | 1.0 |
| lp_2794 | Flavodoxin | 0.7 | NS |
| lp_2920 | Amino acid transport protein | NS | 1.0 |
| lp_2969 | N-Acetylglucosamine PTS, EIICBA | 0.9 | NS |
| lp_3010 | Cellobiose PTS, EIIC | 1.1 | NS |
| lp_3011 | 6-Phospho-β-glucosidase | NS | 1.0 |
| lp_3045 | Short-chain dehydrogenase/oxidoreductase | 0.8 | NS |
| lp_3092 | Succinate-semialdehyde dehydrogenase (NAD(P)+) | 0.8 | 1.2 |
| lp_3170 | Phosphoglycerate mutase | 1.2 | NS |
| 1p_3279 | Potassium uptake protein | NS | 1.0 |
| lp_3284 | Quaternary ammonium compound-resistance protein | NS | 1.3 |
| lp_3313 | Formate C-acetyltransferase | 0.9 | NS |
| lp_3314 | Formate acetyltransferase activating enzyme | 0.9 | NS |
| lp_3418 | Phosphoenolpyruvate carboxykinase (ATP) | NS | 1.1 |
| lp_3420 | Glutamate decarboxylase | 0.8 | NS |
| lp_3483 | β-Galactosidase, large subunit | 0.9 | NS |
| lp_3484 | β-Galactosidase, small subunit | 0.7 | NS |
| lp_3487 | Aldose 1-epimerase | NS | 1.0 |
| lp_3525 | 6-Phospho-β-glucosidase | 0.8 | NS |
| lp_3589 | Pyruvate oxidase | 1.0 | NS |
| lp_3605 | Myo-inositol 2-dehydrogenase | 0.9 | NS |
| Purines, pyrimidines, nucleosides, and nucleotides | |||
| lp_0242 | Nucleoside-diphosphate kinase | 1.2 | NS |
| lp_2374 | Uracil phosphoribosyltransferase | NS | 1.1 |
| lp_2591 | Purine nucleosidase | 0.9 | 1.0 |
| lp_2697 | Orotate phosphoribosyltransferase | −0.9 | 2.6 |
| lp_2698 | Orotidine-5′-phosphate decarboxylase | −0.9 | 2.3 |
| lp_2699 | Dihydroorotate oxidase | −0.8 | 2.2 |
| lp_2700 | Carbamoyl-phosphate synthase, pyrimidine-specific, large chain | −1.1 | 2.6 |
| lp_2702 | Dihydroorotase | −1.2 | 3.2 |
| lp_2703 | Aspartate carbamoyltransferase | NS | 2.7 |
| lp_2719 | Phosphoribosylamine-glycine ligase | 0.7 | 1.3 |
| lp_2720 | Bifunctional protein:phosphoribosylaminoimidazolecarboxamide | 0.8 | 1.2 |
| lp_2721 | Phosphoribosylglycinamide formyltransferase | 0.7 | 1.0 |
| lp_2722 | Phosphoribosylformylglycinamidine cyclo-ligase | 0.9 | 1.2 |
| lp_2723 | Amidophosphoribosyltransferase precursor | 0.9 | NS |
| lp_2724 | Phosphoribosylformylglycinamidine synthase II | 0.9 | NS |
| lp_2725 | Phosphoribosylformylglycinamidine synthase I | 0.7 | NS |
| lp_2726 | Conserved purine biosynthesis cluster protein | 1.0 | NS |
| lp_2728 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | 0.8 | NS |
| lp_3194 | IMP dehydrogenase (fragment) | NS | 1.0 |
| lp_3269 | Adenylosuccinate lyase | 1.9 | 1.1 |
| lp_3270 | Adenylosuccinate synthase | 2.6 | 1.4 |
| lp_3271 | GMP reductase | 3.4 | 1.8 |
| Regulatory function | |||
| lp_0188 | Sucrose operon repressor | 2.8 | 1.9 |
| lp_0836 | Regulatory protein Spx | 0.8 | 1.1 |
| lp_1153 | Transcription regulator (putative) | NS | 1.2 |
| lp_1700 | Sensory protein | NS | 1.0 |
| lp_2095 | Transcription regulator of fructose operon | 0.9 | NS |
| lp_2228 | Regulatory protein Spx | NS | 1.1 |
| lp_2704 | Pyrimidine operon regulator | NS | 2.0 |
| lp_2800 | Transcription regulator (putative) | NS | 1.0 |
| Hypothetical protein | |||
| lp_0058 | Unknown | 0.8 | NS |
| lp_0137 | Oxidoreductase | NS | 1.0 |
| lp_0155 | Unknown | 0.7 | NS |
| lp_0156 | Unknown | 0.9 | NS |
| lp_0240 | Unknown | 0.8 | NS |
| lp_0244 | Oxidoreductase | NS | 1.3 |
| lp_0260 | Unknown | NS | 1.3 |
| lp_0320 | Unknown | 0.9 | 1.1 |
| lp_0459 | Unknown | NS | 1.0 |
| lp_0509 | Unknown | 1.0 | 1.7 |
| lp_0524 | Unknown | NS | 1.0 |
| lp_0865 | Unknown | 0.7 | NS |
| lp_0899 | Unknown | 0.8 | 1.3 |
| lp_0927 | Unknown | 0.8 | 1.8 |
| lp_0928 | Unknown | 0.8 | 1.9 |
| lp_0934 | Unknown | NS | 1.3 |
| lp_0935 | Unknown | NS | 1.3 |
| lp_0995 | Unknown | NS | 1.3 |
| lp_1163 | Nucleotide-binding protein, universal stress protein UspA family | NS | 1.0 |
| lp_1168 | Unknown | 1.0 | 1.0 |
| lp_1395 | Unknown | NS | 1.1 |
| lp_1521 | Oxidoreductase | NS | 1.4 |
| lp_1527 | Hydrolase, HAD superfamily | 0.8 | 1.2 |
| lp_1566 | Unknown | 0.8 | NS |
| lp_1577 | Unknown | 1.3 | 1.7 |
| lp_1590 | Integral membrane protein | NS | 1.9 |
| lp_1704 | Integral membrane protein | NS | 1.2 |
| lp_1706 | Integral membrane protein | NS | 1.2 |
| lp_1708 | Unknown | 1.0 | 1.3 |
| lp_1834 | Unknown | 1.3 | 1.1 |
| lp_1872 | Unknown | 0.7 | NS |
| lp_1880 | Unknown | 1.1 | 1.1 |
| lp_1972 | Unknown | 0.7 | 1.1 |
| lp_2755 | Integral membrane protein (putative) | 0.7 | 1.2 |
| lp_2113 | Unknown | 0.9 | 1.2 |
| lp_3243 | Unknown | 0.7 | 1.3 |
| lp_3250 | Unknown | 1.2 | 1.2 |
| lp_3318 | Oxidoreductase | 1.2 | NS |
| lp_3433 | Unknown | NS | 1.1 |
| lp_3489 | Oxidoreductase | 0.7 | NS |
| lp_3577 | Integral membrane protein | NS | 1.6 |
| Cellular processes | |||
| lp_0129 | Small heat shock protein | NS | 1.0 |
| lp_0355 | Cell division protein Suf1 | 0.8 | 1.1 |
| lp_0929 | Alkaline shock protein | 0.8 | 1.8 |
| lp_0930 | Alkaline shock protein | 0.7 | 2.1 |
| lp_1701 | Nucleotide-binding protein, universal stress protein UspA family | NS | 1.0 |
| lp_1747 | Nucleotide-binding protein, universal stress protein UspA family | NS | 1.3 |
| lp_2323 | Thiol peroxidase | NS | 1.1 |
| lp_2544 | NADH peroxidase | 0.9 | 1.1 |
| lp_2668 | Small heat shock protein | 0.7 | 1.6 |
| lp_3128 | Stress induced DNA binding protein | 1.0 | 2.1 |
| lp_3352 | Small heat shock protein | NS | 1.2 |
| lp_3578 | Catalase | NS | 1.1 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | |||
| lp_2270 | Thioredoxin | NS | 1.1 |
| lp_2633 | Thioredoxin H-type | NS | 1.2 |
| lp_2771 | Nicotinate phosphoribosyltransferase | 1.0 | 2.8 |
| Protein synthesis | |||
| lp_0737 | Ribosomal protein S30EA | 1.4 | 1.5 |
| lp_1973 | Ribosomal protein S21 | 0.8 | 1.2 |
| lp_2216 | Ribosomal protein S14-2 | 0.7 | 1.1 |
| lp_2126 | Ribosomal protein S20 | 0.9 | NS |
| Cell envelope | |||
| lp_0304 | Extracellular protein | 0.9 | NS |
| lp_1697 | Extracellular protein | NS | 1.2 |
| lp_1812 | Lipoprotein precursor (putative) | NS | 1.0 |
| lp_1819 | Teichoic acid biosynthesis protein | 0.7 | NS |
| lp_2200 | Penicillin binding protein 2B | NS | 1.0 |
| lp_2845 | Extracellular protein | 0.9 | NS |
| Phage and prophage related functions | |||
| lp_0660 | Prophage P1 protein 37 | NS | 1.1 |
| lp_0860 | Transposase, fragment | 0.8 | NS |
| lp_2441 | Prophage P2a protein 16 | 0.8 | NS |
| lp_2443 | Prophage P2a protein 14 | NS | 1.3 |
| lp_2445 | Prophage P2a protein 12 | 0.8 | 1.1 |
| lp_2446 | Prophage P2a protein 11 | 0.8 | 1.2 |
| lp_2447 | Prophage P2a protein 10 | 0.8 | 1.3 |
| lp_2457 | Prophage P2b protein 24 | NS | NS |
| lp_3383 | Prophage P3 protein 7, DNA replication | NS | NS |
| RNA | |||
| lp_RNA01 | 0.7 | 1.8 |
M FOD1, log2(intensity of signal with scFOS at an OD600 of 1/intensity of signal with glucose at an OD600 of 1).
M FOD1.5, log2(intensity of signal with scFOS at an OD600 of 1.5/intensity of signal with glucose at an OD600 of 1).
NS, nonsignificantly differentially expressed compared to expression with glucose at an OD600 of 1.
TABLE 3.
Genes showing mainly a down-regulation in L. plantarum WCFS1 grown on scFOS at an OD 1 or OD 1.5 versus glucose
| Function group and ORF | Description | M FOD1a | M FOD1.5b |
|---|---|---|---|
| Energy metabolism and transport | |||
| lp_0230 | Mannitol PTS, EIICB | −0.9 | −1.1 |
| lp_0329 | Acetaldehyde dehydrogenase | −0.7 | NSc |
| lp_0349 | Ammonium transport protein | −1.2 | NS |
| lp_0436 | Cellobiose PTS, EIIC | −0.9 | −1.3 |
| lp_0576 | Mannose PTS, EIIC | −5.4 | −5.5 |
| lp_0575 | Mannose PTS, EIIAB | −4.3 | −4.6 |
| lp_0577 | Mannose PTS, EIID | −4.2 | −3.7 |
| lp_0587 | Mannose PTS, EIIB | −0.7 | NS |
| lp_0822 | Glutamine-fructose-6-phosphate transaminase (isomerizing) | NS | −1.1 |
| lp_1083 | Transketolase | NS | −1.3 |
| lp_1386 | Cation efflux protein (putative) | NS | −1.1 |
| lp_1409 | Amino acid efflux protein | −0.8 | NS |
| lp_1466 | Ferrous iron transport protein B | −0.7 | NS |
| lp_1468 | ABC transporter, ATP-binding protein | −0.7 | NS |
| lp_1469 | ABC transporter component (putative) | −0.8 | NS |
| lp_1471 | NifU-like protein | −0.9 | NS |
| lp_1472 | ABC transporter component, iron regulated (putative) | −0.8 | NS |
| lp_1473 | Iron chelatin ABC transporter, substrate binding protein (putative) | −0.8 | NS |
| lp_1686 | Acyl-CoA thioester hydrolase (putative) | −1.1 | −1.3 |
| lp_1921 | Transport protein | −0.7 | NS |
| lp_2531 | N-Acetylglucosamine and glucose PTS, EIICBA | −0.8 | NS |
| lp_2649 | N-Acetylgalactosamine PTS, EIIC | −0.9 | −1.0 |
| lp_2650 | N-Acetylgalactosamine PTS, EIIB | −1.0 | −1.3 |
| lp_2710 | Purine transport protein | −0.8 | −2.1 |
| lp_2776 | d-Serine dehydratase | −0.9 | NS |
| lp_2789 | Transport protein | −0.7 | NS |
| lp_2982 | Branched-chain amino acid ABC transporter, ATP-binding protein | NS | −1.0 |
| lp_2984 | Branched-chain amino acid ABC transporter, permease protein | −0.7 | NS |
| lp_3491 | Fumarate reductase, flavoprotein subunit precursor | −0.9 | NS |
| lp_3533 | Sugar transport protein | NS | −1.3 |
| lp_3543 | Bifunctional protein: transcriptional regulator; PTS, EIIA | −1.7 | NS |
| lp_3665 | p-Coumaric acid decarboxylase | NS | −1.0 |
| Fatty acid and phospholipid metabolism | |||
| lp_1675 | 3-Oxoacyl-[acyl-carrier protein] synthase II | −0.9 | −2.1 |
| lp_1673 | [Acyl-carrier protein] S-malonyltransferase | −0.9 | −2.1 |
| lp_1674 | 3-Oxoacyl-[acyl-carrier protein] reductase | −0.9 | −2.1 |
| lp_1678 | Acetyl-CoA carboxylase, biotin carboxylase subunit | −0.9 | −2.1 |
| lp_1680 | Acetyl-CoA carboxylase, carboxyl transferase subunit α | −0.8 | −2.0 |
| lp_1672 | Acyl carrier protein | −0.9 | −2.0 |
| lp_1676 | Acetyl-CoA carboxylase, biotin carboxyl carrier protein | −1.0 | −2.0 |
| lp_1679 | Acetyl-CoA carboxylase, carboxyl transferase subunit β | −0.8 | −2.0 |
| lp_1671 | 3-Oxoacyl-[acyl-carrier protein] synthase III | −0.9 | −1.9 |
| lp_1681 | Enoyl-[acyl-carrier protein] reductase (NADH) | −0.8 | −1.9 |
| lp_1682 | Phosphopantetheinyltransferase | −0.8 | −1.8 |
| lp_1670 | (3R)-Hydroxymyristoyl-[acyl carrier protein] dehydratase | −0.8 | −1.7 |
| Regulatory functions | |||
| lp_0057 | Transcription regulator | −0.9 | −1.0 |
| lp_0172 | Transcription regulator | −1.1 | −1.9 |
| lp_0173 | Transcription regulator | NS | −1.0 |
| lp_0435 | Transcription regulator | −1.1 | −1.9 |
| lp_0788 | Central glycolytic gene regulator | −0.7 | NS |
| lp_0836 | Regulatory protein Spx | −0.8 | −1.2 |
| lp_1685 | Transcription regulator | −0.9 | −1.4 |
| lp_2651 | Transcription regulator | −0.9 | −1.3 |
| lp_3506 | Transcription regulator | NS | −1.1 |
| Hypothetical protein | |||
| lp_0052 | Unknown | −1.3 | NS |
| lp_0154 | Unknown | 0.8 | −1.1 |
| lp_0438 | Unknown | NS | −1.2 |
| lp_0580 | HD superfamily hydrolase | NS | NS |
| lp_1860 | Oxidoreductase | −0.8 | −1.2 |
| lp_2276 | Unknown | −0.7 | NS |
| lp_3175 | Integral membrane protein | NS | −1.0 |
| lp_3256 | DegV family protein | NS | −1.1 |
| lp_3501 | Unknown | −0.7 | NS |
| Cellular processes | |||
| lp_2131 | ComE operon protein 1 | NS | −1.0 |
| Purines, pyrimidines, nucleosides and nucleotides | |||
| lp_1058 | Adenylate kinase | NS | −1.0 |
| lp_2585 | Nucleotide-disulphide oxidoreductase | −1.0 | NS |
| lp_2728 | Phosphoribosylaminoimidazole carboxylase, ATPase subunit | NS | −1.0 |
| lp_3334 | Adenine deaminase | NS | −1.7 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | |||
| lp_1470 | Cysteine desulfurase | −0.8 | NS |
| lp_1480 | Molybdopterin precursor synthase MoaA | 0.9 | −1.2 |
| lp_3492 | Thiamin biosynthesis lipoprotein ApbE | −1.0 | −1.1 |
| Protein synthesis | |||
| lp_0538 | Aminoacyl-tRNA hydrolase | −0.7 | −1.1 |
| lp_0578 | Nonribosomal peptide synthetase NpsA | −1.2 | −1.1 |
| lp_1033 | Ribosomal protein L3 | NS | −1.0 |
| lp_1035 | Ribosomal protein L23 | NS | −1.0 |
| lp_1036 | Ribosomal protein L2 | NS | −1.0 |
| lp_1038 | Ribosomal protein S19 | NS | −1.0 |
| lp_1040 | Ribosomal protein S3 | NS | −1.3 |
| lp_1041 | Ribosomal protein L16 | NS | −1.0 |
| lp_1043 | Ribosomal protein L29 | NS | −1.0 |
| lp_1044 | Ribosomal protein S17 | NS | −1.1 |
| lp_1045 | Ribosomal protein L14 | NS | −1.2 |
| lp_1047 | Ribosomal protein L5 | NS | −1.0 |
| lp_1051 | Ribosomal protein L6 | NS | −1.2 |
| lp_1055 | Ribosomal protein L15 | NS | −1.1 |
| lp_1639 | tRNA (guanine-N1-)-methyltransferase | NS | −1.1 |
| Amino acid biosynthesis | |||
| lp_0256 | Cysteine synthase | NS | −1.1 |
| lp_0529 | Glutamate N-acetyltransferase | −0.8 | NS |
| lp_1084 | Shikimate 5-dehydrogenase | NS | −1.3 |
| lp_1085 | Phospho-2-dehydro-3-deoxyheptonate aldolase/chorismate | NS | −1.2 |
| lp_1169 | Glutamate dehydrogenase (NAD(P)+) | 0.7 | NS |
| lp_2033 | Shikimate kinase | NS | −1.2 |
| lp_2034 | Prephenate dehydrogenase | NS | −1.4 |
| lp_2035 | 3-Phosphoshikimate 1-carboxyvinyltransferase | NS | −1.1 |
| lp_2037 | Chorismate synthase | NS | −1.0 |
| lp_2551 | Histidinol-phosphate aminotransferase | −0.8 | −1.1 |
| lp_2553 | Phosphoribosyl-AMP cyclohydrolase | −0.9 | NS |
| lp_2556 | Phosphoribosylformimino-5-aminoimidazole carboxamideribotide | −1.0 | −1.3 |
| lp_2557 | Imidazole glycerol phosphate synthase, amidotransferase subunit | −0.8 | −1.2 |
| lp_2559 | Histidinol dehydrogenase | −0.9 | NS |
| lp_2830 | Aspartate ammonia-lyase | −0.9 | −1.2 |
| lp_3493 | 3-Dehydroquinate dehydratase | −1.0 | −1.4 |
| lp_3494 | Shikimate 5-dehydrogenase | −0.9 | −1.2 |
| Cell envelope | |||
| lp_1181 | Acyltransferase/acetyltransferase | NS | −1.0 |
| lp_2393 | Lipoprotein precursor | −1.4 | NS |
| lp_2975 | Extracellular protein | NS | −1.2 |
| lp_3093 | Muramidase (putative) | −0.7 | NS |
| lp_3393 | Cell surface hydrolase, membrane bound (putative) | NS | −1.1 |
| Other categories | |||
| lp_1687 | GTPase | NS | −1.1 |
M FOD1, log2(intensity of signal with scFOS at an OD600 of 1/intensity of signal with glucose at an OD600 of 1).
M FOD1.5, log2(intensity of signal with scFOS at an OD600 of 1.5/intensity of signal with glucose at an OD600 of 1).
NS, nonsignificantly differentially expressed compared to expression with glucose at an OD600 of 1.
FIG. 4.
Structure of the locus up-regulated in L. plantarum WCFS1 during growth on scFOS, showing the sacK1 (fructokinase; lp_0184), pts1BCA (sucrose PTS EIIBCA; lp_0185), sacA (β-fructofuranosidase; lp_0187), sacR (sucrose operon repressor; lp_0188), and agl2 (α-glucosidase; lp_0189) operons. Black bar, conserved domain of the ROK family; gray bar, helix-turn-helix lactose operon repressor (smart00354); cross-hatched box, periplasmic binding proteins and sugar binding domain of the LacI family (pfam00532).
Genes for two other components of PTSs were activated, namely, the phosphocarrier protein Hpr (lp_1273) and a phosphoenolpyruvate-protein phosphotransferase (lp_1274) at an OD600 of 1. Generally, among the genes overexpressed, a large number were involved in carbohydrate metabolism, especially when comparing scFOS at an OD600 of 1 to glucose. Among these genes, those encoding pyruvate dehydrogenase (lp_2151-lp_2153) and citrate lyase (lp_1107-lp_1109) were two- to threefold up-regulated at OD600s of both 1 and 1.5 with scFOS. Numerous genes encoding hypothetical proteins with unknown function were found differentially expressed with scFOS, the majority being up-regulated. Genes involved in purine, pyrimidine, nucleoside, and nucleotide metabolism were in general highly up-regulated with scFOS at an OD600 of 1.5. This may be an effect of growth phase rather than carbohydrate, reflecting induction of nucleotide biosynthesis upon depletion of nucleotide precursors in the growth medium.
Among the down-regulated genes, another set of clustered genes (lp_0576-lp_0577) predicted to code for a mannose PTS was also distinguished, which was 15- to 50-fold differentially expressed. The mannose PTS is known to be a transporter for glucose in lactic acid bacteria (34). Other carbohydrate transporter systems were repressed such as the N-acetylglucosamine PTS, the cellobiose PTS, mannitol PTS, and N-acetylgalactosamine PTS. Genes which were also down-regulated included a large number involved in fatty acid and phospholipid metabolism and in amino acid and protein synthesis. This fact may be explained by a lower requirement to synthesize these materials, since the cells have a lower growth rate when growing on scFOS. Other differentially expressed genes were involved in transcription regulation, cellular processes, and cell envelope and prophage-related functions. Their connection with scFOS metabolism is unknown.
DISCUSSION
In this study, microarray techniques were used to determine genes differentially expressed when L. plantarum WCFS1 was grown in a medium where either scFOS or glucose was the only source of carbohydrate available.
L. plantarum is a facultative heterofermentative bacterium (21). Several studies have reported the utilization of glucose (17, 29). Little information is available, though, on the metabolism of scFOS in lactobacilli. It is known that some, but not all, lactobacilli are able to ferment this oligosaccharide (4, 22). A previous study (22) showed that L. plantarum strains tested were able to utilize scFOS, but the mechanisms were not elucidated. An operon involved in scFOS and FOS metabolism in L. acidophilus 1195 has thus far been described only once (4). This operon is composed of an active transporter (ABC transporter) and a β-fructofuranosidase. The strain was able to utilize both scFOS and FOS.
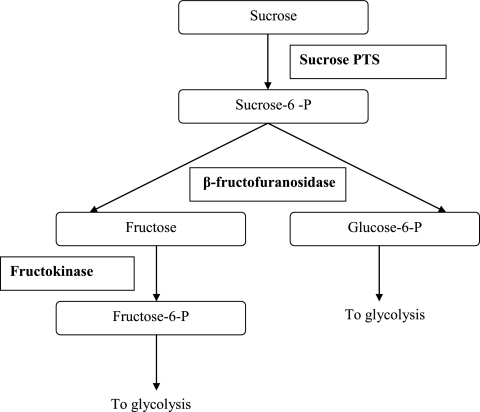
In this study, growth on scFOS and FOS was first evaluated in a chemically defined medium. L. plantarum WCFS1 was able to grow only with scFOS. This suggested that different enzymes were involved in degradation than those previously described for L. acidophilus 1195 (4). Using microarrays, a group of genes (lp_0184-lp_0189) were found to be likely involved in the transport and degradation of scFOS. The first stages in degradation of scFOS in L. plantarum probably reproduce the pathway for sucrose degradation (Fig. 5). Sucrose itself was unlikely to be responsible for up-regulation of these genes, as only traces of the compounds were detected at the beginning of the fermentation and at an OD600 of 1 in the medium with scFOS and was absent from the medium at an OD600 of 1.5. Three genes could participate in the degradation: a sucrose PTS, a β-fructofuranosidase, and a fructokinase. The sucrose PTS usually controls uptake of sucrose across the cytoplasmic membrane and its phosphorylation, but like some other PTS systems (2) its specificity may not be absolute and is likely to be able to phosphorylate both sucrose and scFOS, the latter consisting of a core sucrose to which one to three fructose units are linked. However, moieties with a higher DP of scFOS, such as of GF4, seemed to be hardly converted by L. plantarum, possibly because of limitation in this sucrose transporter. These results are in accordance with a previous study which concluded that an scFOS-fermenting Lactobacillus strain consumed rapidly GF2 and GF3 at approximately equal rates but that the GF4 moiety was not depleted (22). Moreover, in the growth medium, no accumulation of mono- or disaccharides was seen, which suggests that the fractions of scFOS used were totally degraded inside the cell. Once the fructooligosaccharide had entered the cells, the β-fructofuranosidase could cleave the β(2→1) bonds between the different glucose or fructose units. β-Fructofuranosidase from L. plantarum is predicted to be intracellular, as lp_0187 contained no known export signal such as a sortase signal (6). β-Fructofuranosidases, in other microorganisms, have been shown to exert the ability to degrade both sucrose and fructooligosaccharides (14, 37). The role of fructokinase could be to phosphorylate fructose units into fructose-6P to enter into the glycolysis pathway. Together, these different enzymes and transport systems could explain how scFOS can enter into the cells and be broken down. In the genome of L. plantarum WCFS1, two other genes code for a sucrose EII compound PTS (lp_3219 and lp_3522), but they were not differentially expressed in the presence of scFOS compared to glucose. At the protein level, the predicted product of lp_187 displayed only 35 and 37% degrees of identity with lp_3219 and lp_3522, respectively. The signal values for lp_3219 were very low, and this gene was likely unexpressed in any of the arrays. lp_3522 was possibly expressed, but not differentially. Recently, an operon with the same constitutive elements (sucrose transporter and a β-fructofuranosidase) was shown to be responsible for the metabolism of scFOS in Bifidobacterium breve (37). This strain was able to grow on a medium with scFOS but did not grow well with FOS derived from inulin. However, the β-fructofuranosidase activity of B. breve was relatively similar towards scFOS and FOS (84 and 69%, respectively). Limitation to FOS degradation might be due to the sucrose transporter, as the β-fructofuranosidase was predicted to be intracellular. Although the sucrose transporter and β-fructofuranosidase from B. breve and those from L. plantarum were not found to be similar (<25% identity), the growth of these two microorganisms on scFOS only could be explained by the limitation due to the sucrose transporter.
FIG. 5.
Enzymes involved in the uptake and degradation of sucrose in L. plantarum WCFS1 with a sucrose PTS, operating also for scFOS degradation.
The sucrose PTS and fructofuranosidase differentially expressed with scFOS seemed to differ in L. plantarum compared to other Lactobacillus strain but had a high similarity with enzymes from Pediococcus spp. strains. It has been suggested that a horizontal gene transfer had occurred for these genes between P. pentosaceus and L. plantarum (30).
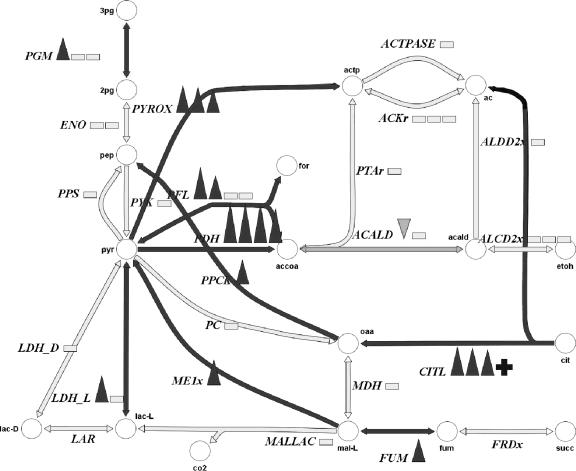
Another set of genes affected by scFOS metabolism encoded a pyruvate formate lyase (PFL) and a pyruvate dehydrogenase complex (PDH) (Fig. 6). Pyruvate, the product of glycolysis, is usually mainly reduced to lactate in L. plantarum but can also be converted into other end products, such as acetate (17, 28). Pyruvate in lactobacilli can be converted anaerobically into acetate via PFL or via a PDH (11). Although pdh genes have been identified in the genome sequence of L. plantarum WCFS1, no detectable activity under various growth conditions has been reported so far (12, 20, 24, 29, 42). Acetate was found to be the second major end product after lactate in the medium with scFOS.
FIG. 6.
Visualization using the Sympheny software (Genomatica, Inc.) of some of the enzymes involved in end product formation and differentially expressed by L. plantarum WCFS1 growth with scFOS at an OD of 1. Upward-pointing triangles, enzymes that are up-regulated; downward-pointing triangles, enzymes that are down-regulated; open bars, enzymes that are not differentially expressed. ACALD, acetaldehyde dehydrogenase; ACK4r, acetate kinase; ACTPASE, acetate phosphotransferase; ALCD2x, alcohol dehydrogenase; CITL, citrate lyase; ENO, enolase; LDH-L, l-lactate dehydrogenase; MALLAC, malolactic enzyme; MDH, malate dehydrogenase; ME1x, malic enzyme; PC, pyruvate carboxylase; PGM, phosphoglycerate mutase; PPCK, phoshoenolpyruvate carboxykinase; PPS, phosphoenal pyruvate synthase; PTAr, phosphate acetyl transferase; PYROX, pyruvate oxidase; PYK, pyruvate kinase; PFL, pyruvate formate lyase; PDH, pyruvate dehydrogenase; ac, acetate; accoa, acetyl coenzyme A; actp, acetyl phosphate; cit, citrate; co2, CO2; etoh, ethanol; for, formate; lac-D, d-lactate; lac-L, l-lactate; mal-L, l-malate; oaa, oxaloacetate; 3pg, 3-phosphoglycerate; 2pg, 2-phosphoglycerate; pep, phosphoenolpyruvate; pry, pyruvate; succ, succinate.
Employing microarrays, it was possible to not only determine the genes possibly responsible for breakdown of fructooligosaccharides but also to identify genes that were down-regulated, compared to growth on glucose. In this experiment, several PTSs and especially a mannose PTS were down-regulated. It is possible that during growth on scFOS, synthesis of enzymes involved in the metabolism of other sugars was negatively regulated, a phenomenon commonly termed carbon catabolite repression (CR). In gram-positive bacteria, catabolite repression has been attributed to a ROK family glucose kinase (25, 44). Here, a conserved domain ROK family (pfam00480) was found in the fructokinase sacK1 (lp_0184), which was up-regulated during growth on scFOS. An important role in overall regulation of sugar uptake and metabolism has also been assigned to the protein Hpr (36), which was up-regulated at an OD600 of 1 during growth on scFOS.
Measurement of gene expression using an alternative method, such as Northern blotting, has shown a good correlation with microarray results for L. plantarum WCFS1 (40). Recent comparisons between microarrays from Agilent platforms and quantitative PCR have also shown a good correlation for genome-wide studies (45).
Similar microarray experiments with a Lactobacillus acidophilus strain, on global transcription profiles determined on eight different carbohydrates, have shown that this type of experiment is very effective in identifying the genetic basis for transport and catabolism of carbohydrates (5). It was suggested that transcription of most carbohydrate transporters and hydrolases in this strain was specifically induced by their respective substrates.
To conclude, the main finding of this study was that an association of a sucrose PTS and a β-fructofuranosidase could explain scFOS degradation in this strain of L. plantarum, which to our knowledge has not hitherto been reported for this species. Microarrays were a powerful tool to increase understanding of scFOS metabolism, and the same approach could be exploited with other prebiotics/carbohydrates for microorganisms whose genomes have already been sequenced. The results could warrant interest for further characterization of the biological properties of the enzymes strongly up-regulated with scFOS in L. plantarum.
Acknowledgments
This work has been carried out with financial support from the Commission of European Communities, specifically the RTD program “Quality of Life and Management of Living Resources” (QLRT-2001-00135, “Functional Assessment of Interactions Between the Human Gut Microbiota and the Host”). This work does not necessarily reflect the views and in no way anticipates the Commission's future policy in this area. We are members of The European Nutrigenomics Organization (http://www.nugo.org), which supported the transcriptomics work. The European Nutrigenomics Organization: linking genomics, nutrition and health research (NuGO, CT-2004-505944), is a Network of Excellence funded by the European Commission's Research Directorate General under Priority Thematic Area 5 Food Quality and Safety Priority of the Sixth Framework Programme for Research and Technological Development. We acknowledge this organization for their financial support.
We thank also Zeger Kruiswijk and Athanasios Goulas for technical assistance.
Footnotes
Published ahead of print on 19 January 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson, L. T. 1993. Lactic acid bacteria: classification and physiology, p. 456. In S. Salminen and A. Von Wright (ed.), Lactic acid bacteria. Marcel Dekker, Inc., New York, N.Y.
- 3.Bach Knudsen, K. E., and I. B. Hessov. 1995. Recovery of inulin from Jerusalem artichoke (Helianthus tuberosus L.) in the small intestine of man. Br. J. Nutr. 74:101-113. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA. 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA. 103:3816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boekhorst, J., M. W. de Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 187:4928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouhnik, Y., K. Vahedi, L. Achour, A. Attar, J. Salfati, P. Pochart, P. Marteau, B. Flourie, F. Bornet, and J. C. Rambaud. 1999. Short-chain fructo-oligosaccharide administration dose-dependently increases fecal bifidobacteria in healthy humans. J. Nutr. 129:113-116. [DOI] [PubMed] [Google Scholar]
- 8.Bringel, F., P. Quenee, and P. Tailliez. 2001. Polyphasic investigation of the diversity within Lactobacillus plantarum related strains revealed two L. plantarum subgroups. Syst. Appl. Microbiol. 24:561-571. [DOI] [PubMed] [Google Scholar]
- 9.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaplin, M. F., and J. F. Kennedy. 1994. Reductive cleavage of O-glycosidic linkages, p. 189. In M. F. Chaplin and J. F. Kennedy (ed.), Carbohydrate analysis: a practical approach, 2nd ed. Oxford University Press, Oxford, England.
- 11.Condon, S. 1987. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 46:269-280. [Google Scholar]
- 12.Dirar, H., and E. B. Collins. 1973. Aerobic utilization of low concentrations of galactose by Lactobacillus plantarum. J. Gen. Microbiol. 78:211-215. [DOI] [PubMed] [Google Scholar]
- 13.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Determination of sugars and related substances. Anal. Chem. 28:350-355. [Google Scholar]
- 14.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM10140T and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 15.Ellegard, L., H. Andersson, and I. Bosaeus. 1997. Inulin and oligofructose do not influence the absorption of cholesterol, or the excretion of cholesterol, Ca, Mg, Zn, Fe, or bile acids but increases energy excretion in ileostomy subjects. Eur. J. Clin. Nutr. 51:1-5. [DOI] [PubMed] [Google Scholar]
- 16.Enan, G., A. A. ElEssawy, M. Uyttendaele, and J. Debevere. 1996. Antibacterial activity of Lactobacillus plantarum UG1 isolated from dry sausage: characterization, production and bactericidal action of plantaricin UG1. Int. J. Food Microbiol. 30:189-215. [DOI] [PubMed] [Google Scholar]
- 17.Ferain, T., A. N. Schanck, and J. Delcour. 1996. C-13 nuclear magnetic resonance analysis of glucose and citrate end products in an ldhL-ldhD double-knockout strain of Lactobacillus plantarum. J. Bacteriol. 178:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, G. R., A. L. McCartney, and R. A. Rastall. 2005. Prebiotics and resistance to gastrointestinal infections. Br. J. Nutr. 93:S31-S34. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 20.Hickey, M. W., A. J. Hillier, and G. R. Jago. 1983. Metabolism of pyruvate and citrate in lactobacilli. Aust. J. Biol. Sci. 36:487-496. [DOI] [PubMed] [Google Scholar]
- 21.Kandler, O., and N. Weiss. 1986. Regular, nonsporing gram-positive rods, p. 1208-1260. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 22.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, H., and R. W. Hutkins. 2003. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl. Environ. Microbiol. 69:2217-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwakman, J., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Blay, G., C. Michel, H. M. Blottiere, and C. Cherbut. 1999. Prolonged intake of fructo-oligosaccharides induces a short-term elevation of lactic acid-producing bacteria and a persistent increase in cecal butyrate in rats. J. Nutr. 129:2231-2235. [DOI] [PubMed] [Google Scholar]
- 27.Molin, G. 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am. J. Clin. Nutr. 73:380S-385S. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, M. G., and S. Condon. 1984. Comparison of aerobic and anaerobic growth of Lactobacillus plantarum in a glucose medium. Arch. Microbiol. 138:49-53. [Google Scholar]
- 29.Murphy, M. G., and S. Condon. 1984. Correlation of oxygen utilization and hydrogen peroxide accumulation with oxygen induced enzymes in Lactobacillus plantarum cultures. Arch. Microbiol. 138:44-48. [DOI] [PubMed] [Google Scholar]
- 30.Naumov, D. G., and V. A. Livshits. 2001. Molecular structure of the locus for sucrose utilization by Lactobacillus plantarum: comparison with Pediococcus pentosaceus. Mol. Biol. 35:19-27. [PubMed] [Google Scholar]
- 31.Peyret, N., P. A. Seneviratne, H. T. Allawi, and J. SantaLucia. 1999. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A·A, C·C, G·G, and T·T mismatches. Biochemistry 38:3468-3477. [DOI] [PubMed] [Google Scholar]
- 32.Pieterse, B., R. H. Jellema, and M. J. Van der Werf. 2005. Quenching of microbial samples for increased reliability of microarray data. J. Microbiol. Methods 64:207-216. [DOI] [PubMed] [Google Scholar]
- 33.Poolman, B., and W. N. Konings. 1988. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 170:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postma, P. W., and J. W. Lengeler. 1985. Phosphoenolpyruvate carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 49:232-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rastall, R. A., G. R. Gibson, H. S. Gill, F. Guarner, T. R. Klaenhammer, B. Pot, G. Reid, I. R. Rowland, and M. E. Sanders. 2005. Modulation of the microbial ecology of the human colon by probiotics, prebiotics and synbiotics to enhance human health: an overview of enabling science and potential applications. FEMS Microbiol. Ecol. 52:145-152. [DOI] [PubMed] [Google Scholar]
- 36.Reizer, J. 1989. Regulation of sugar uptake and efflux in gram positive bacteria. FEMS Microbiol. Rev. 63:149-156. [DOI] [PubMed] [Google Scholar]
- 37.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3. http://www.bepress.com/sagmb/vol3/issu1/art3. [DOI] [PubMed]
- 39.Smyth, G. K., Y. H. Yang, and T. P. Speed. 2003. Statistical issues in microarray data analysis, p. 111-136. In M. J. Brownstein and A. B. Khodursky (ed.), Functional genomics: methods and protocols, vol. 224. Humana Press, Totowa, N.J. [Google Scholar]
- 40.Sturme, M. H., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 187:5224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teusink, B., and E. J. Smid. 2006. Modelling strategies for the industrial exploitation of lactic acid bacteria. Nat. Rev. Microbiol. 4:46-56. [DOI] [PubMed] [Google Scholar]
- 42.Tseng, C. P., and T. J. Montville. 1990. Enzyme activities affecting end product distribution by Lactobacillus plantarum in response to changes in pH and O2. Appl. Environ. Microbiol. 56:2761-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]
- 44.Wagner, E., S. Marcandier, N. O. Egeter, J. Deutscher, F. Gotz, and R. Bruckner. 1995. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J. Bacteriol. 177:6144-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y., C. Barbacioru, F. Hyland, W. Xiao, K. L. Hunkapiller, J. Blake, F. Chan, C. Gonzalez, L. Zhang, and R. R. Samaha. 2006. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]