Abstract
Eukaryotic cells have evolved strategies to respond to stress conditions. For example, autophagy in yeast is primarily a response to the stress of nutrient limitation. Autophagy is a catabolic process for the degradation and recycling of cytosolic, long lived, or aggregated proteins and excess or defective organelles. In this study, we demonstrate a new pathway for the induction of autophagy. In the endoplasmic reticulum (ER), accumulation of misfolded proteins causes stress and activates the unfolded protein response to induce the expression of chaperones and proteins involved in the recovery process. ER stress stimulated the assembly of the pre-autophagosomal structure. In addition, autophagosome formation and transport to the vacuole were stimulated in an Atg protein-dependent manner. Finally, Atg1 kinase activity reflects both the nutritional status and autophagic state of the cell; starvation-induced autophagy results in increased Atg1 kinase activity. We found that Atg1 had high kinase activity during ER stress-induced autophagy. Together, these results indicate that ER stress can induce an autophagic response.
Autophagy is a cellular degradation process for long-lived proteins and unnecessary or damaged organelles (1, 2). During autophagy, double membrane vesicles termed autophagosomes are formed, which sequester the cytosolic proteins and/or organelles as cargoes and then are fused with the vacuole or lysosome. After fusion, the inner membrane vesicles (subsequently termed autophagic bodies) are released into the vacuole/lysosome lumen, and the contents are degraded by resident hydrolases (2, 3). This process is ubiquitous in eukaryotes from yeast to mammals and is essential for normal cellular development and differentiation (1, 4). Autophagy occurs at a basal level and can be significantly induced, depending on the cell type, when necessary. In yeast, for example, autophagy is induced in response to nutrient starvation; following breakdown of the cargo, the resulting macromolecules are presumably released from the vacuole lumen for reuse in the cytosol and supply the source for continued biosynthesis. Similarly, in higher eukaryotes, autophagy plays an important role in survival during starvation. In addition, mounting evidence shows that autophagy is associated with various pathophysiological conditions. For example, autophagy may remove aggregateprone proteins, such as mutant huntingtin and α-synuclein, which cause neurodegenerative disorders (5-7). Autophagy also functions in tumor suppression, possibly by removing damaged organelles to reduce the production of reactive oxygen species. In addition, autophagy is involved in the host immune response to invasion by certain bacterial and viral pathogens (8, 9).
Genetic analyses reveal that degradative autophagy shares mechanistic components with the biosynthetic cytoplasm to vacuole targeting (Cvt)2 pathway in yeast (10, 11). The Cvt pathway is highly selective and specifically transports at least two hydrolases, Ape1 (aminopeptidase I) and Ams1 (α-mannosidase), to the vacuole after sequestration within double-membrane Cvt vesicles (12-14). The protein components that function in these autophagy-related pathways are named Atg (15). Most of the Atg proteins localize to a perivacuolar site called the preautophagosomal structure (PAS), where the autophagosome and Cvt vesicle are thought to form (16, 17).
In eukaryotic cells, most proteins are either synthesized on soluble ribosomes or on ribosomes attached to the ER. Misfolded cytosolic and nuclear proteins are typically tagged with ubiquitin and degraded via the proteasome (18, 19). The accumulation of misfolded proteins in the ER induces the unfolded protein response (UPR), which results in the expression of chaperones and other proteins that act as folding catalysts (20). Several studies have reported a linkage between autophagy and ER function. For example, the early secretory pathway is required for autophagy, possibly supplying membrane for autophagosome formation (21-23). Moreover, it was shown that fragmented ER membrane structures are transported to the vacuole within autophagosomes under starvation conditions; however, no relationship has been described between autophagy and ER stress (24). In this study, we show that ER stress in yeast cells induces an autophagic response.
EXPERIMENTAL PROCEDURES
Strains, Media, and Reagents
The S. cerevisiae strains used in this study are: AHY001 (25), SEY6210 (26), WHY1 (14), and YTS178 (27). Strain JLY27 (SEY6210 atg12Δ::Kan), UNY102 (SEY6210 TAP-Atg1), and UNY104 (UNY102 atg13Δ::LEU2) were generated through standard molecular genetics, using a PCR-based procedure. Yeast strains were grown or incubated in media as described previously (28). For induction of ER stress, cells were incubated in SMD medium (synthetic medium containing 0.67% yeast nitrogen base with auxotrophic amino acids and 2% glucose) containing 3 mM dithiothreitol (DTT) or 2 μg/ml tunicamycin (TM) as described previously (29, 30). All chemical reagents were from Sigma unless otherwise indicated.
Immunoblotting
Yeast cells were grown in SMD medium at 30 °C to A600 = 0.5, and either TM or DTT was added to each culture to induce ER stress. For starvation conditions, cells were cultured in SD-N medium (starvation medium containing 0.17% yeast nitrogen base without amino acids and 2% glucose). At the indicated times, cells were collected, and proteins were precipitated by the addition of trichloroacetic acid. Protein extracts were subjected to SDS-PAGE, followed by immunoblotting with anti-Atg8 (31), anti-Pgk1 (a generous gift from Dr. Jeremy Thorner (University of California, Berkeley)), anti-Kar2 (a generous gift from Dr. Jeffrey L. Brodsky, University of Pittsburgh, PA), anti-Ape1 (13) or anti-Atg1 antiserum (33), or anti-GFP antibodies (Covance Research Products, Berkeley, CA).
Fluorescence Microscopy
Yeast cells expressing fluorescent protein-fused chimeras were grown for 4h in SMD in the presence or absence of DTT or TM, or in SD-N. Fluorescence microscopy was carried out as described previously (28).
Atg1 Kinase Assay
An in vitro phosphorylation assay using TAP-tagged Atg1 was performed as described previously (34).
Protein Incorporation Assay
Yeast cells were incubated with or without either TM, DTT, or CCCP (100 μM)at30 °C for 4 h. Cells were incubated at 30 °C in SMD medium with 1.0 μCi of [35S]methionine. At each time point, cells were collected, and proteins were precipitated with trichloroacetic acid, resuspended in 0.1 N NaOH, and incubated at 40 °C for 10 min. The radioactivity was measured in a liquid scintillation counter (Beckman Coulter LS6500, Fullerton, CA).
RESULTS
The PAS is the putative site where Atg components localize to compose the vesicle-forming machinery for autophagy and the Cvt pathway (16, 17). Atg8 is a structural component on these vesicles and also a marker protein for the PAS. In order to test the effect of ER stress on autophagy, we first examined PAS formation by fluorescence microscopy by monitoring GFP-Atg8. Cells were treated with two distinct ER stressors, DTT, an inhibitor of disulfide bond formation, and TM, an inhibitor of N-glycosylation, at concentrations that induce ER stress. In the absence of DTT and TM, wild-type cells expressing GFP-Atg8 showed a punctate dot at the PAS under both growing and starvation conditions, because the PAS is formed for both the Cvt pathway and autophagy (Fig. 1A). In addition, we detected increased staining of the vacuole lumen in starvation conditions, reflecting the up-regulation of Atg8 and increased autophagy. In the presence of DTT and TM in growing conditions, there was an increase in the number of cells displaying punctate structures; 50% of the cells treated with DTT and 46% of the cells with TM showed punctate dots versus 19% of the cells with no treatment (Fig. 1A).
FIGURE 1.
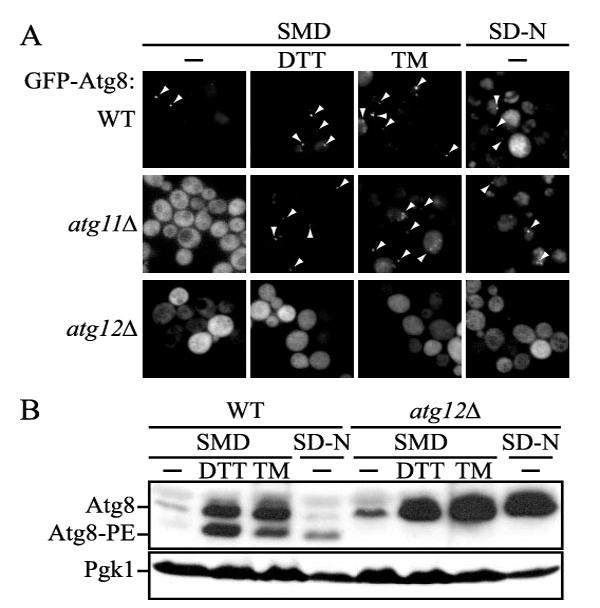
PAS formation was stimulated by ER stress. A, GFP-Atg8 localization under ER stress conditions. Wild-type (SEY6210), atg11Δ (AHY001), and atg12Δ (JLY27) cells expressing CEN plasmid-borne GFP-Atg8 from the endogenous promoter were grown in SMD to A600= 0.5. Cells were subsequently kept in SMD or shifted to SMD with 3 mM DTT or 2 μg/ml TM or to SD-N. After 4 h, cells were observed by fluorescence microscopy. Arrowheads mark the PAS. B, Atg8 lipidation was enhanced under ER stress conditions. Wild-type and atg12Δ cells were incubated for 4h in SMD with or without 3 mM DTT or 2 μg/ml TM or in SD-N as described in A. Atg8 and Atg8-PE were separated by 12% SDS-PAGE in the presence of 6 M urea followed by immunoblotting with anti-Atg8 antiserum or anti-Pgk1 as a loading control. WT, wild type.
In contrast to wild-type cells, GFP-Atg8 is not located at the PAS but is diffusely localized throughout the cytosol in vegetative conditions in the absence of the Cvt pathway-specific component Atg11, whereas PAS localization is restored, followed by transport to the vacuole, under starvation conditions reflecting an induction of autophagy (28) (Fig. 1A). We took advantage of the Cvt pathway-specific defect of the atg11Δ mutant to monitor the effect of DTT and TM. Treatment with DTT or TM in rich medium elicited localization of GFP-Atg8 at the PAS; essentially the same number of cells displayed punctate dots as seen with the wild-type strain (45% with DTT treatment and 46% with TM), suggesting that ER stress resulted in autophagic induction. Localization of GFP-Atg8 at the PAS was confirmed by the observation that TM- and DTT-induced GFP-Atg8 colocalized with RFP-Ape1, which is a PAS marker (data not shown). To check whether the PAS localization of GFP-Atg8 was dependent on the autophagic process under ER stress conditions, we examined GFP-Atg8 localization in atg12Δ cells. GFP-Atg8 did not show PAS localization in either growing or starvation conditions in the atg12Δ strain, as reported previously (17, 35). Similarly, in atg12Δ cells, treatment with TM or DTT was unable to cause localization of GFP-Atg8 to the PAS (Fig. 1A). These results suggested that PAS formation induced under ER stress was dependent on the normal autophagic machinery.
We extended our analysis by examining Atg8 conjugated to phosphatidylethanolamine (Atg8-PE) under ER stress conditions. When autophagy is induced in response to nutrient starvation, higher levels of Atg8/Atg8-PE are observed relative to growing conditions (31, 36). In nutrient-rich medium, the addition of either DTT or TM resulted in increased amounts of Atg8-PE as well as nonlipidated Atg8 at levels higher than those in starvation conditions (Fig. 1B). In contrast to the result in wild-type cells, Atg8-PE was not detected either in starvation conditions or in nutrient-rich conditions following treatment with DTT or TM in atg12Δ cells, although the expression level of Atg8 was increased significantly. This result was consistent with the microscopy data and suggested that ER stress induced autophagy in an Atg protein-dependent manner.
Next, we confirmed that treatment with DTT and TM induced the UPR by monitoring the level of Kar2, an ER chaperone, which is induced by the UPR (37). As expected, Kar2 was induced following treatment with either DTT or TM (Fig. 2A). In contrast, Kar2 was not substantially induced in starvation conditions that induce autophagy in the absence of these ER stressors. These results indicate that the UPR was induced under our experimental conditions of ER stress. We then extended our analysis by examining the transport of autophagosomes into the vacuole under conditions of ER stress. As a marker protein, we monitored GFP-Atg8-PE (referred to hereafter as GFP-Atg8), which remains associated with the inner autophagosome membrane and is transported into the vacuole concomitantly with the contents of the autophagic body (36, 38). After lysis of the autophagic body by vacuolar hydrolases, the GFP moiety is proteolytically cleaved from GFP-Atg8 and is relatively stable in the vacuolar lumen, whereas Atg8 is degraded. Accordingly, detecting free GFP through immunoblot can follow the delivery of autophagosomes to the vacuole. In wild-type cells, free GFP was quickly generated by starvation-induced autophagy (SD-N; Fig. 2B). This processing was blocked in an autophagy-defective atg1Δ strain, verifying its dependence on the autophagic machinery. A greatly reduced level of processing was seen under growing conditions (SMD). Cells that were treated with DTT or TM for 6 h showed the appearance of free GFP even in nutrient-rich conditions; however, the generation of free GFP was considerably lower and delayed relative to starvation conditions. atg1Δ cells did not show processed GFP following treatment with DTT or TM (Fig. 2B), although analysis of Kar2 indicated that the UPR was induced in the atg1Δ strain at a level similar to that seen in the wild-type strain (data not shown). These results indicated that processing of GFP-Atg8 occurred in an autophagy-dependent manner under conditions of ER stress.
FIGURE 2.
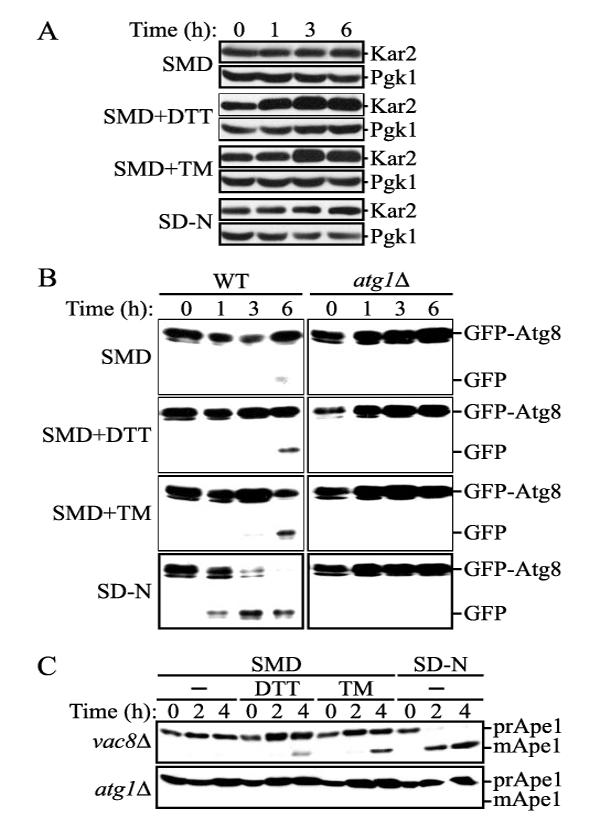
Autophagy was induced under ER stress. A, UPR induction. Wild-type (SEY6210) cells were incubated in SMD with or without 3 mM DTT or 2 μg/ml TM or in SD-N as described in the legend to Fig. 1A. At the indicated times, proteins were precipitated with trichloroacetic acid and resolved by SDS-PAGE followed by immunoblotting using anti-Kar2 serum. B, GFP-Atg8 processing under ER stress. Wild-type (SEY6210) and atg1Δ (WHY1) cells expressing CEN plasmid-borne GFP-Atg8 from the endogenous ATG8 promoter were grown and processed as above, followed by immunoblotting with anti-GFP antibodies. C, precursor Ape1 maturation occurred under ER stress. The vac8Δ (YTS178) and atg1Δ (WHY1) cells were grown and processed as above, followed by immunoblotting with anti-Ape1 antiserum. WT, wild type.
To further analyze the autophagic process under ER stress conditions, we monitored the processing of precursor Ape1 (prApe1). Ape1 is a vacuolar resident hydrolase and is synthesized as a precursor form in the cytosol. Precursor Ape1 is enwrapped within autophagosomes or Cvt vesicles, depending on the nutrient conditions, and is transported to the vacuole as a specific cargo via the Cvt pathway and autophagy. After transport to the vacuole, prApe1 is activated by removal of its N-terminal propeptide. In order to eliminate the constitutive maturation of prApe1 that occurs via the Cvt pathway, we used the vac8Δ mutant. Without Vac8, prApe1 maturation is blocked in growing conditions, whereas maturation is normal in starvation conditions (39). When vac8Δ cells were treated with DTT or TM, prApe1 maturation was observed, although it was again less efficient than that observed in starvation conditions (Fig. 2C). In contrast, neither starvation nor ER stress caused prApe1 processing in atg1Δ cells. Again, we observed induction of the UPR in both vac8Δ and atg1Δ cells based on analysis of Kar2 under these conditions (data not shown). This result was consistent with that observed with GFP-Atg8 processing. We confirmed that atg1Δ cells grew as well as the wild-type cells after the drug treatment, indicating that the defect of atg1Δ cells for GFP-Atg8 processing and prApe1 maturation was not due to loss of viability under ER stress conditions (data not shown). Taken together, we conclude that autophagy was induced in response to ER stress.
To gain a further mechanistic understanding of the induction process, we examined Atg1 kinase activity. In nutrient-rich conditions or in the absence of Atg13, Atg1 kinase has lower activity in vitro (40). Starvation or treatment with the autophagy inducer rapamycin results in increased Atg1 kinase activity. When purified from wild-type cells treated with rapamycin, TAP-tagged Atg1 was able to highly phosphorylate the myelin basic protein substrate compared with TAP-Atg1 isolated in the absence of rapamycin (Fig. 3A). In contrast, the phosphorylation of myelin basic protein was extremely reduced when incubated with TAP-Atg1 from atg13Δ cells, even when the cells had been treated with rapamycin, as reported previously (40). When cells were treated with DTT or TM, TAP-Atg1 displayed a high level of kinase activity based on phosphorylation of myelin basic protein, similar to the result seen with rapamycin treatment. The increased kinase activity of Atg1 further suggested that autophagy was induced during ER stress.
FIGURE 3.
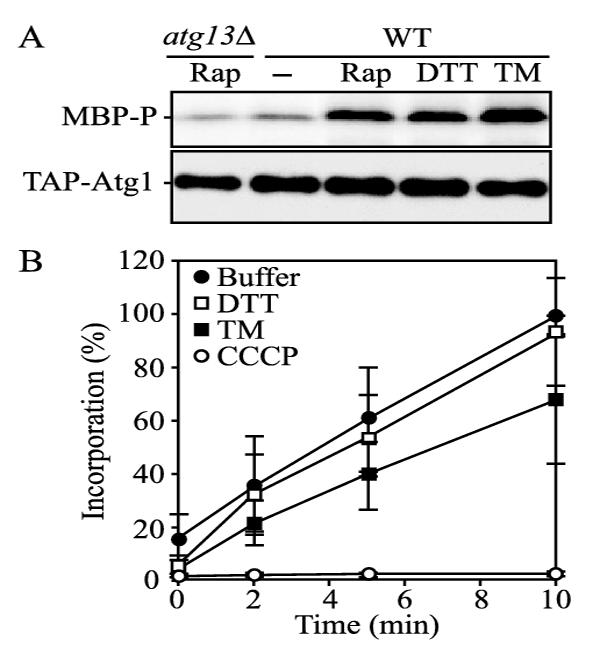
A, Atg1 kinase activity was increased during ER stress. Wild-type cells expressing TAP-Atg1 (UNY102) were grown in YPD with or without either 0.2 μg/ml rapamycin (Rap), 3mM DTT, or 2 μg/ml TM for 2 h. As a control, atg13Δ cells expressing TAP-Atg1 (UNY104) were also grown and treated with rapamycin. TAP-Atg1 was detected by immunoblotting with anti-Atg1 antibody. TAP-Atg1 was immunoprecipitated by IgG-Sepharose; a substrate protein, myelin basic protein, was mixed with the resulting immunocomplexes, and then Atg1 kinase activity was assayed as described under “Experimental Procedures.” B, cells treated with ER stressors were able to import and incorporate radioactive amino acids. Wild-type cells were incubated in SMD with no drug (closed circles), 3mM DTT (open squares), 2 μg/ml TM (closed squares), or 100 μMCCCP (open circles) for 4 h. After incubation, radiolabeled methionine was added into cell suspensions, and its uptake was measured at the indicated times as described under “Experimental Procedures.” Error bars indicate the S.D. of three independent experiments. WT, wild-type.
DTT and TM interfere with protein biosynthesis in the ER. Accordingly, these treatments might induce a starvation response by blocking the production of plasma membrane nutrient transporters. To eliminate this possibility, we investigated the uptake and incorporation of amino acids in cells treated with ER stressors. We incubated cells with or without TM or DTT for 4 h and examined the cellular uptake and incorporation of radioactive methionine into nascent proteins (Fig. 3B). Wild-type cells subjected to mock treatment with buffer showed a linear increase in acid-precipitable radioisotope with increasing time. Cells treated with DTT showed a level of incorporation similar to those without treatment. TM-treated cells displayed a reduced level of incorporation, but it was significantly higher than that seen with cells treated with the proton ionophore CCCP. These results suggest that cells were able to import extracellular amino acids and that they were not experiencing starvation conditions due to a block in nutrient uptake.
DISCUSSION
Eukaryotic cells are able to respond to various types of stress caused by changes in the extracellular environment. Autophagy is one type of stress response. This process is normally maintained at low levels in nutrient-rich conditions, whereas it is highly activated in response to nutrient starvation (2, 4). In this study, we describe a new induction pathway of autophagy; DTT and TM, which are commonly used to cause ER stress (29), allowed cells to induce autophagy in vegetatively growing conditions. Treatment with either drug results in accumulation of misfolded/unfolded proteins in the ER lumen. Subsequently, the UPR is activated to stimulate the expression of proteins that can relieve the stress condition (20, 41). Although the early secretory pathway is known to be required for starvation-induced autophagy (21-23), no relationship has been noted between autophagy and ER stress. Previous microarray results have characterized up-regulation of the transcription of hundreds of genes by ER stress. Among them, the mRNA of genes related to autophagy, including ATG8, ATG14, and APE1 are also transcriptionally induced (41). In addition, it was very recently shown that in mammalian cells, polyglutamine- and drug-induced ER stress facilitated the conversion of LC3, a mammalian homolog of Atg8, from LC3-I to -II, which is an important step for autophagosome formation (42); however, it was still unclear whether functional autophagy actually occurred under these conditions of ER stress. Here we show that ER stress induced autophagy.
We took advantage of the defect in PAS assembly in atg11Δ cells and monitored induction of autophagy by following GFP-Atg8 localization in response to ER stress. Atg8 is conserved from yeast to mammalian cells and is commonly used as a marker for the PAS and the autophagosome (17). Unlike the situation in normal growing conditions, ER stress facilitated PAS formation in atg11Δ cells in an Atg protein-dependent manner (Fig. 1A). Next, we biochemically monitored the delivery of autophagosomes to the vacuole using two established marker proteins, GFP-Atg8 and Ape1. The delivery of autophagosomes, measured by the formation of free GFP or maturation of precursor Ape1, was observed in response to ER stress, although the level of the delivery was less efficient than that seen under starvation conditions (Fig. 2). Finally, Atg1 purified from cells under ER stress conditions showed essentially the same increase in kinase activity as that from cells treated with the autophagy inducer rapamycin (Fig. 3A).
It remains to be determined whether ER stress affects Tor kinase or whether it acts indirectly on a downstream component or even on a different kinase, such as protein kinase A, that might also be involved in the Atg1 signaling pathway (43). To eliminate the possibility that DTT or TM induced autophagy indirectly by generating a starvation response, we monitored amino acid uptake of cells treated with these ER stressors. DTT-treated cells displayed essentially the same level of uptake and incorporation of radioactive methionine as nontreated cells, suggesting that these cells did not experience starvation conditions due to inhibition of the biosynthesis of plasma membrane permeases. Similarly, there was apparently no significant interference with the cell’s translational machinery. The uptake and incorporation of methionine in TM-treated cells was less efficient, being about 70% of that seen with untreated or DTT-treated cells (Fig. 3B); however, this level was well above that seen with CCCP treatment.
Ire1 is an ER membrane protein that senses accumulation of unfolded proteins in the ER lumen. Active Ire1 splices HAC1 mRNA, and the spliced mRNA, which encodes a transcription factor, is translated into a functional protein. The active Hac1 protein induces the expression of genes encoding proteins involved in the UPR (44). We observed that under ER stress conditions, depletion of either Ire1 or Hac1 blocked autophagy based on GFP-Atg8 processing and prApe1 maturation; however, both ire1Δ and hac1Δ cells also showed low viability under ER stress conditions.3 Thus, it was not possible to determine whether the Ire1-Hac1 signaling pathway is involved in induction of autophagy under ER stress conditions.
The ER-associated degradation (ERAD) pathway is the primary degradation mechanism for handling misfolded proteins that cause ER stress (19). During ERAD, unfolded proteins that accumulate in the ER lumen are exported (dislocated) into the cytosol through the translocon channel and then degraded by the proteasome. We speculate that autophagy functions as backup to ERAD if degradative substrates overwhelm the ERAD capacity (30, 32). It has been known that there is a regulatory link between ERAD and the UPR (30). One of the next questions to address is how autophagy is associated with these two pathways under starvation and other stress conditions.
In summary, our current report shows that autophagy is induced by ER stress. Previous studies have shown that stress caused by accumulation of cytosolic protein aggregates can mediate autophagy in mammalian cells (6, 7). To our knowledge, this is the first report of ER stress-induced autophagy. Further study will be needed to gain additional details about the regulatory association of this type of autophagy with the UPR and ERAD.
Acknowledgments
Acknowledgments— We thank Dr. Jeffrey L. Brodsky for anti-Kar2 antiserum, Dr. Jeremy Thorner for anti-Pgk1 antiserum, and members of the Klionsky laboratory for helpful discussions and suggestions.
Footnotes
This work was supported by National Institutes of Health Public Health Service Grant GM53396 (to D. J. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
- Cvt
- cytoplasm to vacuole targeting
- Atg8-PE
- Atg8 conjugated to phosphatidylethanolamine
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- PAS
- pre-autophagosomal structure
- TM
- tunicamycin
- UPR
- unfolded protein response
- prApe1
- precursor Ape1
- GFP
- green fluorescent protein.
T. Yorimitsu, U. Nair, Z. Yang, and D. J. Klionsky, unpublished data.
REFERENCES
- 1.Levine B, Klionsky DJ. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Yorimitsu T, Klionsky DJ. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ. J. Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N. Cell Death Differ. 2005;12:1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 5.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 6.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 8.Levine B. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Shintani T, Klionsky DJ. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 11.Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchins MU, Klionsky DJ. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klionsky DJ, Cueva R, Yaver DS. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostova Z, Wolf DH. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meusser B, Hirsch C, Jarosch E, Sommer T. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 20.Patil C, Walter P. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 21.Hamasaki M, Noda T, Ohsumi Y. Cell Struct. Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- 22.Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Mol. Biol. Cell. 2004;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamasaki M, Noda T, Baba M, Ohsumi Y. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Mol. Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yorimitsu T, Klionsky DJ. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. Methods. 2005;35:395–416. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. Nat. Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 31.Huang W-P, Scott SV, Kim J, Klionsky DJ. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 32.Spear ED, Ng DT. Mol. Biol. Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abeliovich H, Zhang C, Dunn WA, Jr., Shokat KM, Klionsky DJ. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada Y, Jung US, Piotrowski J, Levin DE. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 35.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Normington K, Kohno K, Kozutsumi Y, Gething MJ, Sambrook J. Cell. 1989;57:1223–1236. doi: 10.1016/0092-8674(89)90059-7. [DOI] [PubMed] [Google Scholar]
- 38.Shintani T, Klionsky DJ. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 40.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 42.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. Cell Death Differ. 2006 doi: 10.1038/sj.cdd.4401984. in press. [DOI] [PubMed] [Google Scholar]
- 43.Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13933–13938. doi: 10.1073/pnas.0501046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox JS, Walter P. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]


