Summary
Autophagy is a process in which cytosol and organelles are sequestered within double-membrane vesicles that deliver the contents to the lysosome/vacuole for degradation and recycling of the resulting macromolecules. It plays an important role in the cellular response to stress, is involved in various developmental pathways and functions in tumor suppression, resistance to pathogens and extension of lifespan. Conversely, autophagy may be associated with certain myopathies and neurodegenerative conditions. Substantial progress has been made in identifying the proteins required for autophagy and in understanding its molecular basis; however, many questions remain. For example, Tor is one of the key regulatory proteins at the induction step that controls the function of a complex including Atg1 kinase, but the target of Atg1 is not known. Although autophagy is generally considered to be nonspecific, there are specific types of autophagy that utilize receptor and adaptor proteins such as Atg11; however, the means by which Atg11 connects the cargo with the sequestering vesicle, the autophagosome, is not understood. Formation of the autophagosome is a complex process and neither the mechanism of vesicle formation nor the donor membrane origin is known. The final breakdown of the sequestered cargo relies on well-characterized lysosomal/vacuolar proteases; the roles of lipases, by contrast, have not been elucidated, and we do not know how the integrity of the lysosome/vacuole membrane is maintained during degradation.
Keywords: Lysosome, Pexophagy, Protein targeting, Vacuole, Yeast
Introduction
Autophagy is a ubiquitous process, occurring in all eukaryotic cells (Klionsky, 2004; Reggiori and Klionsky, 2002). There are three primary forms of autophagy: chaperone-mediated autophagy, microautophagy and macroautophagy. Chaperone-mediated autophagy is a secondary response to starvation and, unlike the other two processes, involves direct translocation of the targeted proteins across the lysosomal membrane (Massey et al., 2004). Currently, there is no known equivalent of this process in yeast. Microautophagy is the least-characterized process but is used to sequester cytoplasm by invagination and/or septation of the lysosomal/vacuolar membrane (Wang and Klionsky, 2004). By contrast, the most prevalent form, macroautophagy, involves the formation of cytosolic double-membrane vesicles that sequester portions of the cytoplasm (Fig. 1) (Klionsky and Ohsumi, 1999). During macroautophagy, the sequestering vesicles, termed autophagosomes, are not derived from the lysosome/vacuole membrane. Fusion of the completed autophagosome with the lysosome or vacuole results in the delivery of an inner vesicle (autophagic body) into the lumen of the degradative compartment. Subsequent breakdown of the vesicle membrane allows the degradation of its cargo and eventual recycling of the amino acids, etc., generated.
Fig. 1.
A schematic overview of autophagy and the Cvt pathway in yeast. The Cvt pathway is an autophagy-related process that operates under vegetative conditions and plays a biosynthetic role, delivering resident hydrolases such as aminopeptidase I (Ape1) to the yeast vacuole. The Cvt vesicle is approximately 150 nm in diameter and appears to exclude cytosol. In yeast, autophagy is induced by starvation, and the autophagosome, which is 300-900 nm in diameter, sequesters cytoplasm, including organelles; this pathway is also used for specific transport of prApe1. Both types of vesicle are thought to originate from the pre-autophagosomal structure (PAS). Upon completion, the vesicles fuse with the lysosome-like vacuole, releasing the inner vesicle, termed a Cvt body or autophagic body. These subvacuolar vesicles are broken down, allowing maturation of prApe1 and degradation of cytoplasm, with recycling of the resulting macromolecules.
Autophagy serves as a response to stress such as nutrient limitation and this is one of its primary roles in unicellular organisms such as yeast. However, autophagy also has homeostatic and biosynthetic functions. For example, under conditions in which they are no longer needed, peroxisomes are degraded through a specific type of autophagy termed pexophagy (Hutchins et al., 1999; Kim and Klionsky, 2000), which may occur through either a micro- or macroautophagic process. By contrast, the cytoplasm-to-vacuole targeting (Cvt) pathway (Fig. 1) is a biosynthetic pathway used for delivery of the resident vacuolar hydrolases aminopeptidase I (Ape1) and α-mannosidase (Ams1). The Cvt pathway uses most of the components needed for the degradative autophagy pathway (Harding et al., 1996; Scott et al., 1996). Here, I focus on macroautophagy, which hereafter I refer to simply as autophagy.
In addition to its role in degradation, autophagy may promote a type of programmed cell death that is separate from apoptosis, termed type II programmed cell death (Bursch et al., 2004). Autophagy also plays a role in a range of normal developmental processes, not all of which involve programmed cell death (Levine and Klionsky, 2004). For example, it is needed for sporulation in yeast, for entry into the dauer phase of the Caenorhabditis elegans life cycle, for pupa formation in Drosophila melanogaster and for development of the Dictyostelium discoideum fruiting body. Autophagy is also associated with the extension of lifespan that correlates with caloric restriction (Bergamini et al., 2003; Longo and Finch, 2003; Melendez et al., 2003; Vellai et al., 2003). The pathway might act as a means of defense against invasion by various bacteria and viruses; however, autophagy might also be subverted by pathogens to establish a replicative niche within the host cell (Dorn et al., 2002; Kirkegaard et al., 2004; Nakagawa et al., 2004). Finally, autophagy has been linked to various diseases in humans (reviewed by Shintani and Klionsky, 2004a), including cancer (Gozuacik and Kimchi, 2004; Qu et al., 2003; Yue et al., 2003), cardiomyopathy (Ueno et al., 2004) and neurodegenerative disorders such as Alzheimer’s, Parkinson’s and Huntington’s diseases, amyotrophic lateral sclerosis and prion diseases (Yuan et al., 2003).
The morphology of autophagy was first characterized in studies of mammalian cells (reviewed by Klionsky, 2004). With a few exceptions, however, the molecular components of autophagy were initially elucidated in yeast. Recent studies in various eukaryotic systems have revealed a conservation of the autophagic machinery (Reggiori and Klionsky, 2002; Wang and Klionsky, 2003; Levine and Klionsky, 2004). Here, I focus on our current understanding of the mechanism of autophagy and point out some of the unresolved questions in the field.
Multiple steps are required for the sequestration of cytoplasm through autophagy
Although autophagy and autophagy-related processes are dynamic, they can be broken down into several discrete steps for the purpose of discussion: (1) induction; (2) cargo selection and packaging; (3) nucleation of vesicle formation; (4) vesicle expansion and completion; (5) retrieval; (6) targeting, docking and fusion of the completed vesicle with the lysosome/vacuole; and (7) breakdown of the intralumenal vesicle (either Cvt body or autophagic body; Fig. 1) and its cargo and recycling of the macromolecular constituents. Many of the protein components required for each of these steps have been identified in Saccharomyces cerevisiae, Hansenula polymorpha and Pichia pastoris, and their roles in autophagy have been covered in other recent reviews (Abeliovich and Klionsky, 2001; Kiel et al., 2003; Klionsky, 2004; Ohsumi, 2001; Stromhaug and Klionsky, 2001). The nomenclature in this field has been unified, and the genes and gene products are now referred to as ATG and Atg, respectively (Klionsky et al., 2003).
Induction of autophagy
In yeast and mammalian cells, autophagy occurs at a basal level under vegetative growth conditions. Accordingly, there must be a mechanism for sensing the extracellular milieu and transducing an appropriate signal to regulatory elements that allow autophagy to be induced. One of the major regulatory components is the protein kinase Tor (for ‘Target of rapamycin’) (Carerra, 2004), which inhibits autophagy under basal or nutrient-rich conditions. Tor acts in two ways. First, it directly or indirectly causes hyper phosphorylation of the autophagy protein Atg13 (Funakoshi et al., 1997; Scott et al., 2000) (Fig. 2A). This form of Atg13 has a lower affinity for the kinase with which it interacts, Atg1, and the reduced interaction might inhibit autophagy (Kamada et al., 2000). Inhibition of Tor through starvation or treatment with rapamycin results in partial dephosphorylation of Atg13 and allows autophagic induction (Abeliovich, 2004; Noda and Ohsumi, 1998). Second, Tor acts in a signal transduction cascade that controls phosphorylation of several effectors (e.g. Tap42, Sit4, Ure2 and Gln3) that regulate transcription and translation of certain proteins, some of which are required for autophagy. Autophagy is also regulated through additional factors such as protein kinase A (Budovskaya et al., 2004), Gcn2 (Tallóczy et al., 2002) and Snf1 (Huang et al., 1996).
Fig. 2.

Regulation of induction and vesicle nucleation. The regulation of autophagy has been characterized in studies of yeast and mammalian cells. (A) In yeast, a class III PI 3-K is required for autophagic activity and may function at the pre-autophagosomal membrane. A putative complex consisting of Atg1 kinase and several other proteins characterized as being required primarily for autophagy (in purple) or the Cvt pathway (in green) may be a downstream effector of Tor kinase to regulate the type of pathway that operates, depending on the nutritional conditions or other signals. Autophagy in yeast is primarily a starvation response; Tor, along with other regulatory components not shown (including PKA), responds to nutrient levels. In nutrient-rich conditions, Atg1 and Atg13 are more highly phosphorylated and have a lower affinity for each other; during starvation the two proteins are partially dephosphorylated. The PI 3-K complex I, consisting of Vps15, Vps34, Atg6/Vps30 and Atg14, is required for both the Cvt pathway and autophagy. (B) In mammalian cells, a class I PI 3-K is stimulated in response to the binding of a ligand to a receptor such as the insulin receptor (InR). PtdIns(3,4)P2 and PtdIns(3,4,5)P3 generated at the plasma membrane allow the binding and activation of 3-phosphoinositide-dependent protein kinase 1 (PDK1) and Akt/PKB, whereas PTEN antagonizes this pathway through its 3′-phosphoinositide phosphatase activity. Akt inhibits the GTPase-activating protein complex TSC1-TSC2, resulting in the stabilization of RhebGTP, which activates Tor, resulting in the inhibition of autophagy. Both Tor and PDK1 stimulate p70S6 kinase (p70S6k). The downregulation of p70S6k activity in starvation conditions (when Tor is inhibited) might prevent excessive autophagy (Scott et al., 2004). It is also possible that p70S6k indirectly inhibits Tor by interfering with activation of the class I PI 3-K, as suggested by studies in mammalian cells (Um et al., 2004). In nutrient-rich conditions, activation of p70S6k should inhibit PI 3-K, allowing a low level of autophagy for homeostatic purposes, whereas in starvation conditions the eventual inactivation of p70S6k should allow activation of PI 3-K to prevent excessive autophagy. The class III PI 3-K serves a stimulatory role possibly similar to that of the yeast enzyme complex.
Yeast Atg1 kinase (Matsuura et al., 1997) is part of a putative complex that includes several additional proteins (Fig. 2A). One intriguing point about this complex is that all of the proteins aside from Atg1 are characterized as functioning primarily in either the Cvt pathway or autophagy, but not both (Kamada et al., 2000; Kim et al., 2001b; Nice et al., 2002; Scott et al., 2000). This finding has led to speculation that the Atg1 complex is involved in switching between these two pathways. At present, however, not enough is known about most of the proteins in this complex to assign a specific function or to list them as participating in only one of these pathways. There is currently no evidence for an equivalent of the Cvt pathway outside yeast; so higher eukaryotes might not require the same capacity for switching between the biosynthetic and degradative types of autophagy.
In mammalian cells, Tor is regulated through the action of a class I phosphoinositide 3-kinase (PI 3-K) that allows the membrane binding and subsequent activation of Akt/protein kinase B (PKB) and 3-phosphoinositide-dependent protein kinase 1 (PDK1), which cause inhibition of autophagy (Codogno and Meijer, 2004) (Fig. 2B). PTEN, a 3′ phosphoinositide phosphatase, antagonizes the Akt/PKB pathway and is a positive regulator of autophagy. The TSC1-TSC2 complex acts as a GTPase-activating protein (GAP) for the GTPase Rheb. Inhibition of TSC1-TSC2 by Akt stabilizes the GTP-bound form of Rheb, which stimulates Tor. The GDP-bound form of Rheb might inhibit Tor (Tabancay et al., 2003). Both Tor and PDK1 activate ribosomal subunit S6 kinase (p70S6k), which is needed for maximal autophagic activity (Scott et al., 2004). The downregulation of p70S6k following Tor inactivation might prevent excessive autophagy in the absence of Tor activity. Regulation of mammalian autophagy involves additional components including eIF2α, Ras and heterotrimeric G proteins (Codogno and Meijer, 2004; Furuta et al., 2004; Tallóczy et al., 2002).
Questions
There are many questions that remain to be answered with regard to the yeast Atg1 complex. For example, interactions among the protein components of the putative complex depicted in Fig. 2A have been demonstrated in pairwise combinations. Hence, it is not clear that a stable holo-complex ever exists. Furthermore, our knowledge of the composition of this putative complex is based on genetic suppression studies, two-hybrid data and affinity isolation or coimmunoprecipitation using overexpressed proteins. It is possible that some of these interactions are transient in nature or do not even occur when the proteins are expressed at physiological levels.
The functions of the proteins in the putative Atg1 complex are largely not known. For example, Atg11 is thought to have a role in cargo selection (see below) but its reason for interacting with Atg1 is not clear, nor is it known when the two proteins interact-that is, whether the interaction is nutrient dependent (as suggested for the interaction between Atg1 and Atg13). Furthermore, we do not know whether the two proteins interact during the process of cargo selection or after the delivery of cargo to the pre-autophagosomal structure (PAS; see below). Similarly, the proposed function in this complex of Atg20, Atg24 and Vac8 is simply speculation. Atg20 and Atg24 bind to phosphatidylinositol (3)-phosphate [PtdIns(3)P] and interact with Atg11 and/or Atg17 (Nice et al., 2002). Vac8 has been implicated in homotypic vacuole fusion (Wang et al., 2001) and is required for the Cvt pathway, but does not appear to play a role in the fusion of Cvt vesicles with the vacuole (Scott et al., 2000). Vac8 interacts with Atg13 but it is not known whether it has any effect on the location or function of this protein. Atg13 and Atg17 appear to modulate the kinase activity of Atg1 (Kamada et al., 2000), but do Atg11, Atg20, Atg24 or Vac8 play a similar role? Furthermore, the conclusions about Atg1 regulation are based on in vitro studies using artificial substrates, because its physiological target is unknown. Indeed, the role of Atg1 kinase activity is also unclear. One report suggests that an increase in Atg1 kinase activity is needed to induce autophagy (Kamada et al., 2000). A more recent study came to the opposite conclusion and instead suggests that Atg1 has a structural role in autophagy and that its kinase activity is primarily required for the Cvt pathway (Abeliovich et al., 2003), although Atg1 kinase activity is probably also needed for autophagy.
Whether Tor directly phosphorylates Atg13 and/or acts on other Atg proteins is unclear. Most or all of the components of the Atg1 complex are phosphoproteins and none of them appears to be a target of Atg1 (Scott et al., 2000) (D.J.K., unpublished). The kinases and phosphatases that act upon these proteins are not known. In the case of autophagy, the downstream effector(s) of Tor is not even clear. Tap42 is activated by Tor and is a regulatory subunit of protein phosphatase 2A (PP2A)-like phosphatases; however, it does not appear to function in autophagy (Kamada et al., 2000). Furthermore, Tor is thought to respond primarily to nitrogen starvation, but various other types of stress, including carbon starvation, can induce autophagy. The regulatory components that act under these different stress conditions to regulate autophagic induction are not known. In addition, although Atg1 and Atg13 appear to be centrally involved in autophagy in yeast, there is currently no evidence for an Atg11 or an Atg17 ortholog in higher eukaryotes, or an Atg13 or Vac8 ortholog except in plants. In many organisms, it is also not clear whether there is a true ortholog of Atg1.
Finally, even though most models suggest that Atg1 has a role in autophagic induction, this has not been firmly established. Such a role is proposed because of the effect of rapamycin on Atg13 phosphorylation and the apparent pathway specificity of the putative Atg1 complex components. However, this view is probably misleading because individual mutations in the genes encoding some of these ‘specific’ components (such as ATG11 and VAC8) result in defects in both pathways. Furthermore, Atg1 interacts with the cargopackaging factor Atg11 and is implicated in a late stage of autophagy-retrieval of Atg9 from the forming autophagosome (see below). Thus, Atg1 appears to act at multiple steps of autophagy and its role in induction remains to be verified.
Cargo selection and packaging
Autophagy is generally considered to be a nonspecific process in which bulk cytoplasm is randomly sequestered into the cytosolic autophagosome; however, there are instances of specific autophagy. For example, a recent report showed that the cytosolic protein Ald6 is delivered to the vacuole through autophagy at a rate inconsistent with nonspecific uptake (Onodera and Ohsumi, 2004). Similarly, during pexophagy, virtually all of the peroxisomes, but not other organelles, are recognized for rapid sequestration and vacuolar delivery (Hutchins et al., 1999; Tuttle et al., 1993; Veenhuis et al., 1983). In the case of the Cvt pathway, the cargo is taken up for biosynthetic rather than degradative purposes. In this pathway, the temporal order of action of several components has been determined (Fig. 3). Precursor ApeI (prApe1) forms dodecamers and these assemble to form a higher-order oligomer (Kim et al., 1997) termed the Ape1 complex (Shintani et al., 2002). This step appears to be dependent only upon the conformation of prApe1. The receptor protein Atg19 binds the propeptide domain of prApe1 (Scott et al., 2001); the propeptide contains the vacuolar-targeting information and is cleaved from the remainder of the protein following delivery into the vacuole lumen, allowing activation of the enzyme. At least one other biosynthetic cargo protein, Ams1, also binds to the Atg19 receptor (Hutchins and Klionsky, 2001; Shintani et al., 2002) to form the Cvt complex. Atg11 subsequently binds to Atg19 (Shintani et al., 2002) and acts as a tether or adapter to bring the Cvt complex towards the PAS, a site that might nucleate Cvt vesicle and autophagosome formation (Kim et al., 2002; Suzuki et al., 2001). Finally, the phosphatidylethanolamine (PE)-conjugated Atg8 protein binds to Atg19 to ensure inclusion of the Cvt complex into the forming Cvt vesicle (Shintani et al., 2002).
Fig. 3.
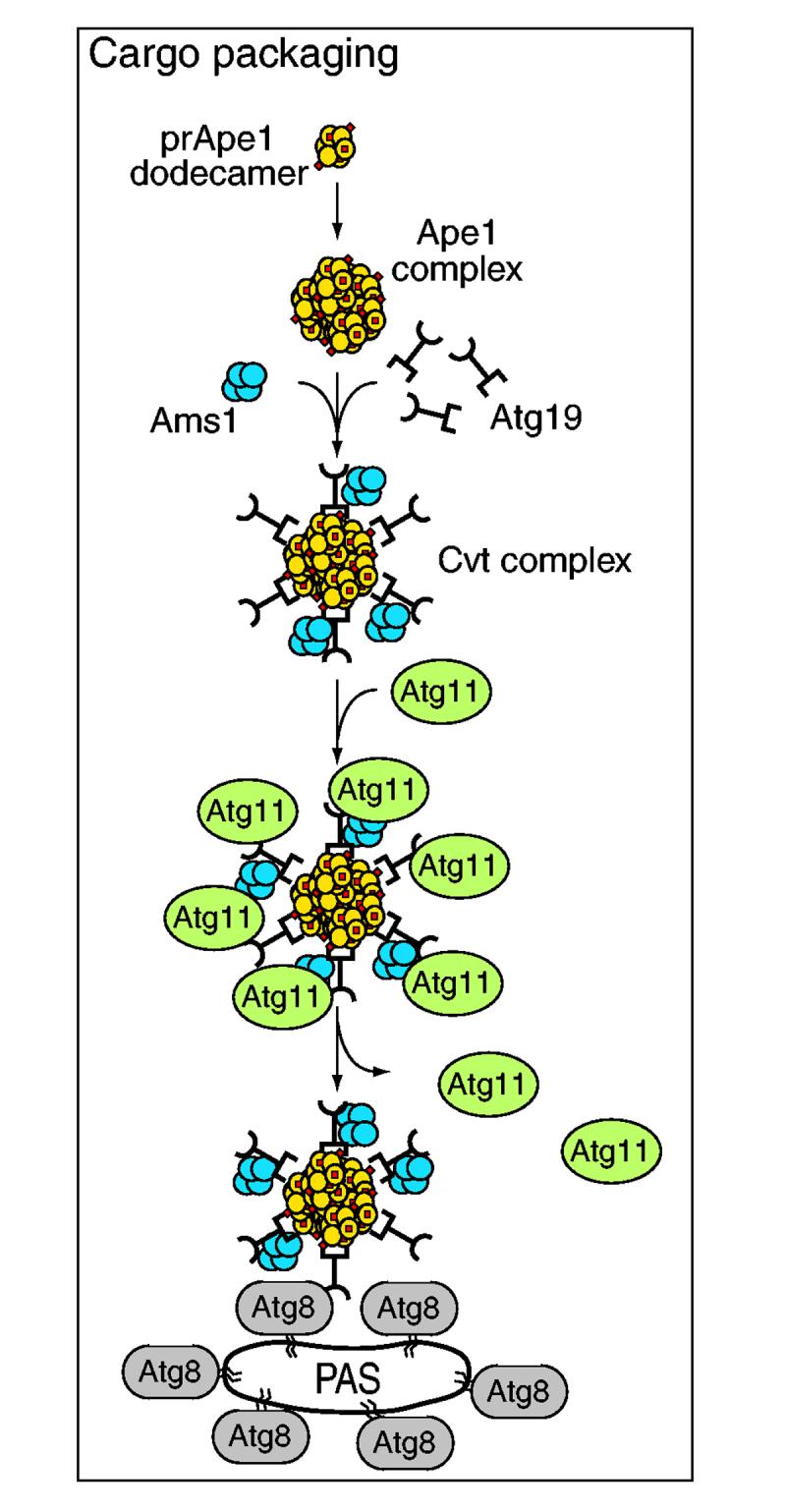
Temporal order of action of cargo-packaging components. The yeast Cvt pathway is an example of specific autophagy. The resident vacuolar hydrolases Ape1 and Ams1 assemble into oligomers in the cytosol. Precursor (pr)Ape1 dodecamers further collect into a large Ape1 complex. The receptor/adaptor Atg19 binds to both the Ape1 complex (through the prApe1 propeptide) and Ams1. Atg11, a component that is not essential for autophagy, interacts with this Cvt complex and is required to connect the complex with the pre-autophagosomal structure (PAS). Atg11 also interacts with Atg1 (not shown, see Fig. 2A), but the timing of this interaction relative to cargo packaging is not known. Atg11 is not part of the final Cvt vesicle or autophagosome and presumably detaches from Atg19 at some time during or after the arrival of the Cvt complex at the PAS. Atg19 subsequently binds to Atg8-phosphatidylethanolamine, which is localized to the PAS, and this event might trigger completion of the vesicle.
Questions
If the yeast Atg11 and Atg19 proteins are not able to interact, because of mutations in the corresponding genes, the Cvt complex forms but does not localize to the PAS. However, it is not known how the function of Atg11 bridges the cargo with the vesicle-forming machinery. Along these lines, how does Atg11 target to the PAS as opposed to other locations within the cell? Does it interact with an unidentified component that acts as a targeting factor? Atg11 is not part of the final Cvt vesicle; so it must disengage from the receptor after delivery of the Cvt complex to the PAS. What triggers this event? Furthermore, Atg11 appears to interact with both Atg19 and Atg1. Does Atg11 bring Atg1 to the site of vesicle formation when cargo is present? As noted above, it is not known when Atg1 and Atg11 interact. Similarly, does binding of Atg8-PE to the Atg19 receptor signal to the PAS the presence of cargo? If so, how does this signaling occur? Ichimura et al. recently showed that Atg8 undergoes a conformational shift upon lipidation (Ichimura et al., 2004). Does a similar change occur when Atg8-PE binds to Atg19?
A recent study suggests that, in nutrient-rich conditions, the PAS does not form in the absence of prApe1, Atg19 or Atg11 (Shintani and Klionsky, 2004b). In the model proposed in this study, the cargo proteins induce or greatly facilitate the formation of the PAS. This makes sense in the biosynthetic Cvt pathway, in which the primary purpose is delivery of the Cvt complex. By contrast, formation of the PAS does not depend on the Cvt cargo or the cargo-recognition machinery during starvation conditions. In this situation, it is not known what factors determine the localization of the PAS components. Finally, it is possible that the peroxisome acts similarly to the Cvt complex to allow formation of the PAS or vesicle nucleation (the initiation of vesicle formation) during pexophagy. Atg19 is not needed for pexophagy; by contrast, Atg11 is required for this process. Is there another protein that substitutes for Atg19 during peroxisome degradation? Note that presently there is no evidence for the Cvt pathway existing outside S. cerevisiae; however, specific autophagy is likely to occur in all eukaryotes. For example, in addition to the uptake of peroxisomes, chloroplasts are also degraded by autophagy (Niwa et al., 2004), and mitochondria are probably targets for specific sequestration by autophagy under certain conditions (Xue et al., 2001). In these cases, less is known about the recognition mechanism. During pexophagy, Pex14 appears to play a role in peroxisome recognition (Bellu et al., 2001), but its binding partner has not been identified. Finally, there are also several studies indicating that pathogenic bacteria and viruses, as well as aberrant protein aggregates, can be targeted for degradation by autophagy (reviewed by Kirkegaard et al., 2004; Larsen and Sulzer, 2002). Again, we do not know what factors trigger autophagosome formation or cargo engulfment in these cases.
Vesicle nucleation
One of the least understood steps of the vesicle formation process is nucleation-the initial step that brings together the proteins and lipids that will constitute the autophagosome and Cvt vesicle, and the machinery that drives the process forward. There must be a nucleation step, a beginning point, in vesicle formation; however, it is difficult to define its hallmarks. In yeast, almost all of the autophagy proteins localize at least transiently to the PAS. Accordingly, arrival of Atg proteins at this site might mark the nucleation event.
In the case of most endomembrane traffic, a vesicle forms from a pre-existing organelle by budding. The resulting vesicle maintains the same cellular topology regarding the lumenal versus the cytosolic face as the organelle that serves as the membrane donor. In autophagy-related processes, the vesicle may form de novo. It is not tethered to an organelle and does not appear to form by extrusion from the surface of an organelle (Fig. 4). The result is that the autophagosome has a double membrane (in some cases it may be multi-lamellar) and the lumenal versus cytosolic topology of the cargo is reversed: the interior of the Cvt vesicle or autophagosome becomes equivalent to the lumen of an organelle or the extracellular space (Fig. 4). The specific components that act in vesicle nucleation are not completely clear, and some proteins may play a role in both nucleation and expansion. In yeast, one set of proteins that appear to function at the PAS is the PI 3-K complex I (Kametaka et al., 1998; Kihara et al., 2001) (Fig. 2A). There is only one PI 3-K in yeast, Vps34, but it is a component of two different complexes. Both complexes contain Vps34 and the presumed regulatory protein Vps15, along with Atg6 (also known as Vps30). In addition, complex I includes Atg14, whereas complex II includes Vps38. Complex I functions primarily, but not exclusively, at the PAS, whereas complex II functions at the endosome. The PtdIns(3)P resulting from complex I recruits proteins including Atg18, Atg20, Atg21, Atg24 and Atg27 to the PAS (Nice et al., 2002; Stromhaug et al., 2004; Wurmser and Emr, 2002); Atg20 and Atg24 are part of the putative Atg1 kinase complex (Fig. 2A).
Fig. 4.
Schematic model for formation of the autophagosome or Cvt vesicle. The origin of the membrane that forms the Cvt vesicle or autophagosome is not known, nor is the mechanism of vesicle formation. In one model (shown on the left), a membrane sheet from a pre-existing organelle such as the endoplasmic reticulum (ER) is induced to separate and undergo a deformation to form a spherical shape that eventually seals; no additional membrane is needed for expansion. It is not known when or how autophagy proteins such as Atg8 would be targeted to this membrane. In a second model (shown on the right), a portion of a membrane forms the nucleus of the autophagosome or Cvt vesicle; in yeast, this nucleus is the pre-autophagosomal structure (PAS). Additional membrane is added to allow expansion of the forming vesicle, but the origin of this membrane is not known. Most data support the second type of model. In either case, the original membrane is presumably equivalent on all surfaces in terms of protein content and phospholipid composition. During vesicle formation, the membrane may differentiate so that, upon vesicle completion, the two separate membranes have different properties; for example, Atg8-PE is located on both sides of the forming vesicle, but is removed from the outer membrane following cleavage by Atg4 (see Fig. 5). The outer vesicle membrane might contain SNARE elements needed for recognition and fusion with the vacuole (see Fig. 7), whereas the inner vesicle membrane might be more susceptible to degradation within the vacuole lumen. The double-membrane nature of the autophagosome or Cvt vesicle results in a transfer of the cargo from the cytosol into the lumenal space of the cell.
Questions
Although the yeast PI 3-K complex I acts at the PAS, the role of PtdIns(3)P generated at this location is not entirely clear. PtdIns(3)P is needed to recruit the various proteins that bind to this lipid, but in most cases the functions of these proteins have not been elucidated. For example, Atg18 binds to PtdIns(3)P (Guan et al., 2001; Stromhaug et al., 2004) and is involved in the cycling of Atg9 (see below) (Reggiori et al., 2004a) but its function is not known. Atg18, Atg21 and Atg27 also bind to PtdIns(3)P (Stromhaug et al., 2004; Wurmser and Emr, 2002). In these cases, the nature of the lipid-binding domains is not known; these proteins do not contain PX or FYVE motifs.
The source of the membrane that forms the autophagosome or Cvt vesicle is another mystery, and whether this is the same for the Cvt pathway and autophagy is not clear. In yeast, one study suggests that membrane flow through the early secretory pathway is needed for autophagy but not the Cvt pathway (Hamasaki et al., 2003; Ishihara et al., 2001), whereas a more recent report indicates that the endoplasmic reticulum (ER) plays a role in both pathways (Reggiori et al., 2004b). This discrepancy might reflect differences in the kinetic parameters followed in each study. In mammalian cells, the most popular (but not the only) view is that the ER is the source membrane for autophagy. Studies of macrophages infected with Listeria monocytogenes or transfected COS-1 cells expressing poliovirus proteins also indicate a role for the ER (Rich et al., 2003; Suhy et al., 2000). However, even if this is the case, how is a portion of the ER caused to separate from the organelle and be delivered to the site of vesicle nucleation? It is also important to consider that the studies cited above reflect a cellular response to pathogens; it is not clear whether the process of vesicle formation is identical in non-infected cells, although evidence indicating a role for the ER is also seen in normal cell lines (Fengsrud et al., 2004).
There might be fundamental differences between the nucleation events in the yeast Cvt pathway and autophagy. For example, as mentioned above, Atg1 kinase activity might be required for nucleating only one type of vesicle. Similarly, there appear to be different requirements for Atg8 in the two processes: Cvt vesicles do not form in the absence of Atg8. By contrast, autophagosomes, albeit abnormally small ones, form without this protein (Abeliovich et al., 2000). These results suggest that Atg8 is needed for Cvt vesicle nucleation, but for autophagosome expansion not formation (see below). This result is supported by studies showing that the atg21Δ mutant has low levels of Atg8-PE and has a defective Cvt pathway but essentially normal autophagy (Stromhaug et al., 2004). Similarly, an Atg7 mutant (see below; Fig. 5) lacking the C-terminal thirteen residues has low levels of lipidated Atg8 and displays a defect only in the Cvt pathway (Yamazaki-Sato et al., 2003).
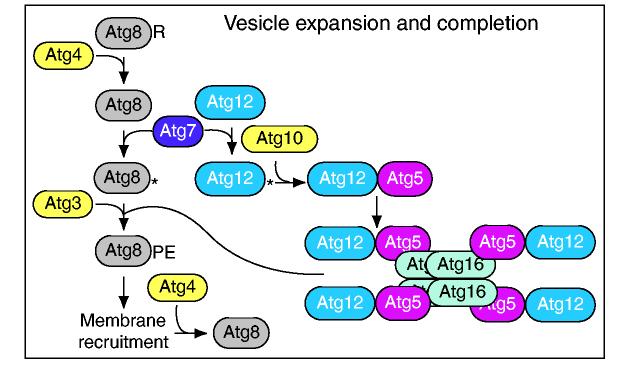
Fig. 5.
Vesicle expansion and completion. Two ubiquitin-like proteins participate in vesicle formation. The C-terminal arginine residue (R) of yeast Atg8 is removed by the Atg4 cysteine protease to reveal a glycine residue. Atg8 and Atg12 are ubiquitin-like proteins that are activated by the E1-like enzyme Atg7. Atg8 and Atg12 are then transferred to the E2-like enzymes Atg3 and Atg10 and are conjugated to phosphatidylethanolamine (PE) and Atg5, respectively. Atg8-PE becomes anchored in the membrane of the PAS and is a component of the forming and completed autophagosome or Cvt vesicle membrane. Atg12 and Atg5 bind Atg16 non-covalently and the self-interaction of Atg16 allows multimerization of the complex. A second cleavage by Atg4 releases Atg8 from the outer membrane of the completed vesicle. A similar set of reactions occurs in mammalian cells.
Vesicle expansion and completion
Most of the proteins that play a role in the Cvt and autophagy pathways act during vesicle formation, which includes the processes of expansion and completion. As noted above, the mechanism of vesicle formation is not known. If the autophagosome or Cvt vesicle forms by deformation of a pre-existing membrane (Fig. 4, left side), then there might be no expansion step; however, if formation proceeds by the sequential addition of membrane (Fig. 4, right side), the addition(s) that allows the membrane to grow constitutes the expansion phase. In either case, completion indicates the final step, at which the vesicle seals to separate the cargo from contact with the cytosol. The proteins that act in the expansion and completion steps include two sets of components involving ubiquitin-like (Ubl) proteins that participate in novel conjugation reactions (Ohsumi, 2001) (Fig. 5). Atg8 is a Ubl that undergoes proteolytic processing by the Atg4 protease to reveal a glycine residue that is then covalently attached to PE (Ichimura et al., 2000). This converts Atg8 from a soluble to a membrane-associated protein (Huang et al., 2000; Kirisako et al., 1999). Atg8-PE is the only known vesicle-associated protein that is thought to play a structural role. Membrane association of Atg8 must be important for some aspect of vesicle enlargement during autophagy because its absence results in the formation of aberrantly small autophagosomes (Abeliovich et al., 2000). By contrast, the absence of Atg8 during vegetative conditions prevents the formation of the Cvt vesicle, although the role of Atg8 or Atg8-PE in vesicle nucleation or expansion is not sufficiently understood to explain this difference. The conjugation reaction involves a ubiquitin-activating E1-like enzyme, Atg7 (Kim et al., 1999; Tanida et al., 1999; Yuan et al., 1999), which also plays a role in the second conjugation reaction: Atg12, another Ubl, is covalently attached to Atg5 (Kametaka et al., 1996) through the action of Atg7 and an E2-like enzyme, Atg10 (Mizushima et al., 1998; Shintani et al., 1999). A third protein, Atg16, multimerizes and links the Atg12-Atg5 conjugate to form a tetrameric complex (Kuma et al., 2002; Mizushima et al., 1999). The function of the Atg12-Atg5·Atg16 proteins is not known, but they might act as a transient coat.
Questions
There are three families of Atg8 proteins in mammalian cells: microtubule-associated protein 1 light chain 3 (MAP1LC3), Golgi-associated ATPase enhancer of 16 kDa (GATE-16) and γ-aminobutyric acid type A receptor-associated protein (GABARAP). Whether they all play a role in autophagy is not known, although a recent study suggests that all three proteins are localized to autophagosomal membranes (Kabeya et al., 2004). The rat MAP1LC3 ortholog appears to be conjugated to PE (Kabeya et al., 2004); however, one study suggests that one of the three human isoforms, MAP1LC3B, undergoes an unusual post-translational modification involving a C-terminal lysine rather than glycine residue (He et al., 2003). It has not yet been verified that the human MAP1LC3B ortholog is conjugated to PE, but, if lysine is the modified residue, activation probably does not involve Atg7 and Atg3. However, a recent study provides contradictory evidence and suggests that MAP1LC3B undergoes the same type of processing as the other isoforms (Tanida et al., 2004).
One of the major questions concerning Cvt vesicle/autophagosome formation is what supplies the driving force for deformation/curvature of the membrane. In most vesicle-budding processes, coat proteins play a central role. At present, there is no evidence from either electron microscopy or vesicle purification for a coat on the completed Cvt vesicle or autophagosome. In yeast, there is clear evidence that Atg8-PE lines both sides of the membrane during autophagosome formation (Kirisako et al., 1999), although its role in this process in not known. It might interact with the Atg12-Atg5 system (see below), and the Atg12-Atg5·Atg16 complex, which can form a tetramer (Kuma et al., 2002), is the best candidate for a transient coat. Studies in mammalian cells provide additional evidence that these proteins act as a coat; time-lapse images show changes in Atg5-GFP fluorescence that seem to correspond to vesicle expansion and uncoating (Mizushima et al., 2001). One question is whether these proteins have the capacity to form higher-order assemblies. Another issue concerns the driving force for uncoating of the completed vesicle, a step that would be necessary prior to fusion with the lysosome/vacuole. Uncoating is usually a timed process that is regulated through the action of an ATPase, but such a protein has not been identified in the case of autophagy. Finally, there is a major difference in the size, and hence the curvature, of Cvt vesicles and autophagosomes (Baba et al., 1997). The Atg12-Atg5·Atg16 complex is required for formation of both types of vesicle, but it is not clear how conformational changes in these proteins could accommodate the two different degrees of curvature.
It is possible that vesicle formation occurs in the absence of coat proteins as in phagocytosis, where the driving force for engulfment is actin polymerization; however, the trigger for phagocytosis is binding of a target such as a bacterium that may act as a scaffold, making it possible for the membrane to enwrap the cargo. Whereas the Cvt complex could provide such a scaffold in the yeast Cvt pathway, it would not be sufficient to direct engulfment by the much larger autophagosome. Microfilaments are known to act during formation of the mammalian autophagosome (Kim and Klionsky, 2000), but we do not know whether they are involved in driving membrane deformation or whether they play a similar role in yeast. During phagocytosis, however, actin causes protrusion of the plasma membrane. Thus, this model may not be applicable to autophagy because the autophagosome is not thought to form by budding or protrusion from a pre-existing organelle (Fig. 4).
Several proteins control the modification, stability and localization of Atg8. For example, in the absence of Atg12-Atg5, Atg8-PE is not localized to the PAS and is unstable (Kim et al., 2001a; Suzuki et al., 2001). Evidence also exists for crosstalk between the two conjugation systems. For example, in mammalian cells, Atg10 binds to MAP1LC3 (Nemoto et al., 2003), and Atg3 facilitates formation of the Atg12-Atg5 conjugate (Tanida et al., 2002b). In addition, an increase in Atg10 levels facilitates MAP1LC3 processing (Nemoto et al., 2003). These results might reflect kinetic effects resulting from the release of Atg7 that can then function in the alternative pathway. However, similar studies revealed an increase in MAP1LC3 processing in the presence of excess Atg12 but a decrease in the presence of excess Atg12-Atg5 (Tanida et al., 2002a), although the significance of these observations is not clear. It is not known whether a similar type of crosstalk between the two different conjugation systems occurs in yeast or other systems.
Another major question concerns the function of Atg8. This protein might have different roles in the Cvt and autophagy pathways, being required for Cvt vesicle formation but for autophagosome expansion (Abeliovich et al., 2000). The mechanism(s) of autophagosome formation and expansion are not known (Fig. 4). Does a portion of the ER dedicate itself to forming the autophagosome and simply undergo a deformation process that forms a vesicular structure? Alternatively, does the autophagosome nucleate at the PAS and then expand by the addition of membrane from vesicles? If so, where do these vesicle come from and how are they targeted? The time-lapse studies in mammalian cells suggest that the autophagic vesicle expands over time (Mizushima et al., 2001). Some evidence for intermediate vesicular structures is also seen in yeast (George et al., 2000). In macrophages infected with L. monocytogenes, there is evidence for sequestration of bacteria by vesicles that fuse to form larger structures (Rich et al., 2003); however, it has not been shown that these vesicles contain autophagy markers. Also, it is not known what SNARE proteins would be involved in vesicle fusion. At present, there is no evidence for a role for SNAREs in completion of the Cvt vesicle/autophagosome and it is possible that fusion to seal the vesicle is SNARE independent.
Retrieval
In most targeting pathways, there is a process for retrieving certain components so that they can be reused. For example, many receptors dissociate from their cargo and recycle back to their original compartment for an additional round of loading. In other cases, it is critical to retrieve components that determine the identity or function of a compartment. This includes proteins such as v-SNAREs that need to be sent back to their donor membrane. Of the >20 proteins thought to have direct roles in yeast autophagy or the Cvt pathway, only two are known to remain associated with the autophagosomes or Cvt vesicles -Atg8-PE and Atg19. Most of the Atg proteins are soluble and can cycle off the vesicle during or after completion; however, Atg9 is a transmembrane protein that does not appear to be part of the completed autophagosome (Noda et al., 2000). Accordingly, there must be some mechanism for recycling this protein from the PAS, i.e. a retrieval pathway. Atg9 is unusual in that it displays a localization pattern different from that of most of the Atg proteins: in addition to localizing at the PAS, it is seen in multiple other punctate dots. One other protein, Atg23, shows a similar distribution (Tucker et al., 2003). Retrieval of Atg9, but not Atg23, from the PAS requires Atg18, Atg2 and PtdIns(3)P (Reggiori et al., 2004a) (Fig. 6). Retrieval of both Atg9 and Atg23 requires Atg1, but only in the case of Atg23 is Atg1 kinase activity necessary (Reggiori et al., 2004a). This latter result supports the idea that high levels of Atg1 kinase activity are crucial for the Cvt pathway rather than autophagy, because Atg23 is essential only for the former pathway.
Fig. 6.

Protein retrieval from the PAS. Only two proteins are known to remain associated with completed yeast Cvt vesicles or autophagosomes, the specific receptor Atg19 and Atg8-PE; other proteins that are involved in vesicle formation presumably recycle from the PAS or the autophagosome/Cvt vesicle during formation. Most Atg proteins are soluble or peripheral membrane proteins that can easily be released from the membrane surface, although the mechanism of targeting and release is not known. As shown here, retrieval of Atg9 from the PAS requires PtdIns(3)P, the PtdIns(3)P-binding protein Atg18, and Atg2, Atg13 and Atg1.
Questions
One of the major questions in the field is the source of the autophagosome or Cvt vesicle membrane. A related issue is how the membrane is targeted to the site of vesicle formation. In most cellular transport processes, some proteins serve as recognition markers to ensure proper membrane or vesicle delivery; these proteins must be retrieved back to the donor compartment for another round of delivery. Do Atg9 and/or Atg23 mark the origin of the donor membrane? If so, where are these proteins located within the cell? Although this is not generally postulated, multiple sources might contribute membrane. What components are needed for the targeting and delivery of Atg9 and Atg23 to the PAS? What are the functions of Atg1, Atg2 and Atg18 in the retrieval process? It is possible that these proteins function as part of a novel coat or adaptor for vesicle-mediated delivery or some novel delivery mechanism.
Atg20 and Atg24 are PX domain proteins that bind PtdIns(3)P and are involved in retrieval of material from endosomes (Hettema et al., 2003). Do these proteins play a similar role in retrieval of Atg9 and/or Atg23 from the PAS?
Vesicle targeting, docking and fusion
The timing of vesicle fusion with the lysosome/vacuole must be regulated; if the fusion process begins prior to completion of the double-membrane vesicle, the cargo will remain in the cytosol. The putative coat consisting of Atg12-Atg5·Atg16 might prevent premature fusion. Atg8-PE located on the outer surface of the autophagosome or Cvt vesicle might serve a similar purpose. At least in yeast, it is removed prior to fusion, as a result of a second cleavage by Atg4, although the timing of this processing event is not known. The expression of Atg8ΔR, a mutated form of Atg8 lacking the ultimate arginine residue, bypasses the need for the initial Atg4-dependent cleavage step (Fig. 5); however, inability to carry out the second cleavage event that releases Atg8 from PE results in a partial defect in autophagy and the Cvt pathway (Kirisako et al., 2000). This suggests that the liberated Atg8 is re-used, that Atg8-PE on the outer membrane interferes with a late stage of these pathways or that Atg4 has an additional function in the cell. An in vitro reaction that reconstitutes fusion of Cvt vesicles or autophagosomes with the vacuole has not been developed. However, molecular genetic studies have indicated that the machinery required for homotypic vacuole fusion is also required for the fusion of autophagosomes and Cvt vesicles with the vacuole (Fig. 7). This machinery includes: the SNARE proteins Vam3, Vam7, Vti1 and Ykt6; the NSF, SNAP and GDI homologs Sec17, Sec18 and Sec19; the Rab protein Ypt7; members of the class C Vps/HOPS complex; and two proteins recently shown to play a role in the fusion process, Ccz1 and Mon1 (Darsow et al., 1997; Fischer von Mollard and Stevens, 1999; Harding et al., 1995; Kim et al., 1999; Sato et al., 1998; Sato et al., 2000; Wang et al., 2003; Wang et al., 2002).
Fig. 7.

Docking and fusion. Vam3, Vam7, Vti1 and Ykt6 are members of the SNARE family, proteins that function in membrane fusion in a variety of cellular contexts. Ypt7 is a member of the Rab small GTPase family, whose members are again implicated in many instances of membrane fusion. Also shown is the class C Vps/HOPS complex and two proteins recently shown to play a role in the fusion process, Ccz1 and Mon1.
Questions
Analyses of the role of fusion components in yeast have been based on whether the corresponding mutants block maturation of prApe1 and accumulate a protease-resistant form; such mutants retain prApe1 within completed cytosolic vesicles, where it is resistant to exogenous protease following lysis of the spheroplast plasma membrane. These studies have generally been carried out under vegetative (Cvt pathway) conditions. It remains to be determined whether different SNARE and/or other fusion components operate during autophagy. Moreover, no one has demonstrated the presence of any fusion components on purified Cvt vesicles or autophagosomes. This has resulted in various uncertainties. For example, Vti1 is required for entry of prApe1 into the vacuole; however, we do not know whether it is on the autophagosome or Cvt vesicle, nor do we know whether it is needed for formation of the completed vesicle or fusion of this with the vacuole.
Vesicle breakdown
The main purpose of autophagy in yeast is to degrade cytoplasm and recycle the resulting macromolecules for use in the synthesis of essential components during nutrient stress. Accordingly, it must break down the single-membrane subvacuolar vesicles that result from fusion of the autophagosome/Cvt vesicle with the vacuole (Fig. 1). The vesicle lysis step depends on the acidic pH of the vacuole lumen and proteinase B (Prb1) (Nakamura et al., 1997; Takeshige et al., 1992); however, the function of Prb1 might be to activate vacuolar zymogens that play a direct role in the breakdown process. Two other proteins have also been implicated at this last step, Atg15 and Atg22 (Epple et al., 2001; Suriapranata et al., 2000; Teter et al., 2001). Atg15 is delivered to the vacuole through the multivesicular body pathway (Epple et al., 2003). This protein has sequence similarity to a family of lipases and seems likely to function directly in vesicle breakdown. Atg22 is an integral membrane protein located in the limiting membrane of the vacuole. In contrast to Atg15, Atg22 is needed only for the degradation of autophagic bodies.
Questions
Atg15 has a lipase motif, and mutations in the corresponding active site eliminate its function. However, this protein belongs to the triacylglycerol family of lipases; so its substrate is not clear. Furthermore, the site of action of Atg15-before or within the vacuole-has not been established. It will be difficult to address the latter issue until the origin of the autophagosome or Cvt vesicle membrane is known. Atg22 has sequence similarity to some permeases, but the role of this protein in autophagic body breakdown is unknown.
Another interesting issue concerns the differentiation and recognition and/or modification of the autophagosome or Cvt vesicle membranes. During the initial stage of vesicle formation, the two sides of the forming vesicle are presumably equivalent (Fig. 4). At some point, the two sides are likely to differentiate. For example, in yeast, Atg8-PE initially lines both sides of the forming autophagosome and Cvt vesicle (Figs 4 and 5). Following vesicle completion, Atg8-PE remains on the inside of the vesicle but Atg8 is removed from the outside membrane, whereas SNARE proteins may be restricted to the outer surface. Following fusion with the vacuole, the outer vesicle membrane is at least transiently continuous with the vacuole membrane. The cell cannot break down this membrane because it would be lethal to destroy the integrity of the vacuole. Presumably, this membrane is either modified so that it is protected similar to the rest of the vacuole membrane, or it is rapidly removed from the limiting membrane and recycled or degraded, possibly through microautophagy (Müller et al., 2000). By contrast, the inner vesicle membrane that gives rise to the subvacuolar vesicle must be degraded. Accordingly, this membrane-which started out equivalent to the outer membrane-cannot be inherently resistant to degradation by vacuolar hydrolases.
Conclusions
The identification of the first gene required specifically for autophagy was reported in 1997 (Matsuura et al., 1997; Straub et al., 1997). The subsequent seven years has seen the identification and characterization of 26 additional proteins that are involved in yeast autophagy, the Cvt pathway and/or pexophagy (Klionsky et al., 2003). Although additional proteins needed for these pathways may be discovered, it seems likely that the majority of proteins have now been identified. However, the work to date represents only the first step in understanding these complex processes that involve dynamic membrane rearrangements. A detailed molecular understanding will necessitate further molecular genetic and biochemical analyses coupled with in vitro and structural studies. Some steps of the yeast Atg8-PE conjugation system have been reconstituted in vitro (Ichimura et al., 2004; Kirisako et al., 2000), and a recent report describes the crystal structure of the mammalian ortholog MAP1LC3 (Sugawara et al., 2004). Thus, the next few years hold the promise of tremendous progress in our understanding of the mechanisms of autophagy and autophagy-related processes.
Footnotes
This work was supported by Public Health Service grant GM53396 from the National Institutes of Health to D.J.K.
References
- Abeliovich H. Regulation of autophagy by the target of rapamycin (Tor) proteins. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 60–69. [Google Scholar]
- Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol. Mol. Biol. Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Zhang C, Dunn WA, Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellu AR, Komori M, van der Klei IJ, Kiel JAKW, Veenhuis M. Peroxisome biogenesis and selective degradation converge at Pex14p. J. Biol. Chem. 2001;276:44570–44574. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- Bergamini E, Cavallini G, Donati A, Gori Z. The anti-ageing effects of caloric restriction may involve stimulation of macroautophagy and lysosomal degradation, and can be intensified pharmacologically. Biomed. Pharmacother. 2003;57:203–208. doi: 10.1016/s0753-3322(03)00048-9. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursch W, Ellinger A, Gerner C, Schulte-Hermann R. Autophagocytosis and programmed cell death. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 287–303. [Google Scholar]
- Carrera AC. TOR signaling in mammals. J. Cell Sci. 2004;117:4615–4616. doi: 10.1242/jcs.01311. [DOI] [PubMed] [Google Scholar]
- Codogno P, Meijer AJ. Signaling pathways in mammalian autophagy. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 26–47. [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn BR, Dunn WA, Jr, Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell. Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- Epple UD, Suriapranata I, Eskelinen E-L, Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple UD, Eskelinen E-L, Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Fengsrud M, Sneve ML, øverbye A, Seglen PO. Structural aspects of mammalian autophagy. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 11–25. [Google Scholar]
- Fischer von Mollard G, Stevens TH. The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 1999;10:1719–1732. doi: 10.1091/mbc.10.6.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Furuta S, Hidaka E, Ogata A, Yokota S, Kamata T. Ras is involved in the negative control of autophagy through the class I PI3-kinase. Oncogene. 2004;23:3898–3904. doi: 10.1038/sj.onc.1207539. [DOI] [PubMed] [Google Scholar]
- George MD, Baba M, Scott SV, Mizushima N, Garrison BS, Ohsumi Y, Klionsky DJ. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004;23:2891–2906. doi: 10.1038/sj.onc.1207521. [DOI] [PubMed] [Google Scholar]
- Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr, Klionsky DJ. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol. Biol. Cell. 2001;12:3821–3838. doi: 10.1091/mbc.12.12.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct. Funct. 2003;28:49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 2003;278:29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HRB. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Farkas I, Roach PJ. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000;275:5845–5851. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 1999;112:4079–4087. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene. 1996;178:139–143. doi: 10.1016/0378-1119(96)00354-x. [DOI] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:22284–22291. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- Kiel JAKW, Komduur JA, van der Klei IJ, Veenhuis M. Macropexophagy in Hansenula polymorpha: facts and views. FEBS Lett. 2003;549:1–6. doi: 10.1016/s0014-5793(03)00794-4. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy. Landes Bioscience; Georgetown, TX: 2004. [Google Scholar]
- Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol. Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- Melendez A, Tallóczy Z, Seaman M, Eskelinen E-L, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O, Sattler T, Flotenmeyer M, Schwarz H, Plattner H, Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;106:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Matsuura A, Wada Y, Ohsumi Y. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- Nemoto T, Tanida I, Tanida-Miyake E, Minematsu-Ikeguchi N, Yokota M, Ohsumi M, Ueno T, Kominami E. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J. Biol. Chem. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Kato T, Tabata S, Seki M, Kobayashi M, Shinozaki K, Moriyasu Y. Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma. 2004;223:229–232. doi: 10.1007/s00709-004-0037-7. [DOI] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang W-P, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:16071–16076. doi: 10.1074/jbc.M312706200. [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen E-L, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol. Cell. Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 2000;275:25840–25849. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell. 2001;7:1131–1141. doi: 10.1016/s1097-2765(01)00263-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004a;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004b;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Mizushima N, Ogawa Y, Matsuura A, Noda T, Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J. Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug PE, Klionsky DJ. Approaching the molecular mechanism of autophagy. Traffic. 2001;2:524–531. doi: 10.1034/j.1600-0854.2001.20802.x. [DOI] [PubMed] [Google Scholar]
- Stromhaug PE, Reggiori F, Guan J, Wang C-W, Klionsky DJ. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004;9:611–618. doi: 10.1111/j.1356-9597.2004.00750.x. [DOI] [PubMed] [Google Scholar]
- Suhy DA, Giddings TH, Jr, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 2000;74:8953–8965. doi: 10.1128/jvi.74.19.8953-8965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suriapranata I, Epple UD, Bernreuther D, Bredschneider M, Sovarasteanu K, Thumm M. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J. Cell Sci. 2000;113:4025–4033. doi: 10.1242/jcs.113.22.4025. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabancay AP, Jr, Gau CL, Machado IM, Uhlmann EJ, Gutmann DH, Guo L, Tamanoi F. Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K. J. Biol. Chem. 2003;278:39921–39930. doi: 10.1074/jbc.M306553200. [DOI] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallóczy Z, Jiang W, Virgin HW, IV, Leib DA, Scheuner D, Kaufman RJ, Eskelinen E-L, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Nishitani T, Nemoto T, Ueno T, Kominami E. Mammalian Apg12p, but not the Apg12p·Apg5p conjugate, facilitates LC3 processing. Biochem. Biophys. Res. Commun. 2002a;296:1164–1170. doi: 10.1016/s0006-291x(02)02057-0. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002b;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 2004;279:47704–47710. doi: 10.1074/jbc.M407016200. [DOI] [PubMed] [Google Scholar]
- Teter SA, Eggerton KP, Scott SV, Kim J, Fischer AM, Klionsky DJ. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001;276:2083–2087. doi: 10.1074/jbc.C000739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KA, Reggiori F, Dunn WA, Jr, Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 2003;278:48445–48452. doi: 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 1993;60:283–290. [PubMed] [Google Scholar]
- Ueno T, Tanida I, Kominami E. Autophagy and neuromuscular disease. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 264–286. [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Douma A, Harder W, Osumi M. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch. Microbiol. 1983;134:193–203. doi: 10.1007/BF00407757. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wang C-W, Klionsky DJ. The molecular mechanism of autophagy. Mol. Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Klionsky DJ. Microautophagy. In: Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. pp. 107–114. [Google Scholar]
- Wang C-W, Stromhaug PE, Shima J, Klionsky DJ. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J. Biol. Chem. 2002;277:47917–47927. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1Mon1 complex during the tethering/docking stage. J. Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Kauffman EJ, Duex JE, Weisman LS. Fusion of docked membranes requires the armadillo repeat protein Vac8p. J. Biol. Chem. 2001;276:35133–35140. doi: 10.1074/jbc.M103937200. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Novel PtdIns(3)P-binding protein Etf1 functions as an effector of the Vps34 PtdIns 3-kinase in autophagy. J. Cell Biol. 2002;158:761–772. doi: 10.1083/jcb.200112050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Fletcher GC, Tolkovsky AM. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- Yamazaki-Sato H, Tanida I, Ueno T, Kominami E. The carboxyl terminal 17 amino acids within Apg7 are essential for Apg8 lipidation, but not for Apg12 conjugation. FEBS Lett. 2003;551:71–77. doi: 10.1016/s0014-5793(03)00899-8. [DOI] [PubMed] [Google Scholar]
- Yuan J, Lipinski M, Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40:401–413. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]
- Yuan W, Stromhaug PE, Dunn WA., Jr Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol. Biol. Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]