Abstract
A protein with a molecular mass of 42 kDa (P42) from Mycoplasma mobile, one of several mycoplasmas that exhibit gliding motility, was shown to be a novel NTPase (nucleoside triphosphatase). Although the P42 protein lacks a common ATP-binding sequence motif (Walker A), the recombinant proteins expressed in Escherichia coli certainly hydrolysed some nucleoside triphosphates, including ATP. The results of photoaffinity labelling by an ATP analogue supported that the P42 protein contains a specific binding site for ATP (or another nucleoside triphosphate). In the M. mobile genome, the P42 gene is located downstream of gli123, gli349 and gli521 genes, and they have been reported to be polycis-tronically transcribed. As the huge proteins encoded by gli123, gli349 and gli521 play a role in gliding motility of M. mobile, P42 might also have some kind of function in the gliding motility. The gliding motility of M. mobile is driven directly by ATP hydrolysis, but the key ATPase has not been identified. Our results showed that, among these four proteins, only P42 exhibited ATPase activity. Biochemical characteristics – optimal conditions for activity, substrate specificities, and inhibiting effects by ATP analogues – of the recombinant P42 proteins were very similar to those of a putative ATPase speculated from a previous analysis with a gliding ‘ghost’ whose cell membrane was permeabilized by Triton X-100. These results support the hypothesis that the P42 protein is the key ATPase in the gliding motility of M. mobile.
Keywords: affinity labelling, ATPase, gliding motility, motor, Mycoplasma mobile, nucleoside triphosphatase (NTPase)
Abbreviations: ATP[S], adenosine 5′-[γ-thio]triphosphate; MBP, maltose-binding protein; NTPase, nucleoside triphosphatase; ORF, open reading frame; p[NH]ppA, adenosine 5′-[β,γ-imido]triphosphate
INTRODUCTION
Mycoplasmas are small-genome parasitic or commensal bacteria that lack a peptidoglycan layer [1]. Several mycoplasma species form membrane protrusions [2–6] and exhibit gliding motility, the movement of cells on solid surfaces, in the direction of their membrane protrusion [3,7,8]. Mycoplasma pneumoniae, a causative organism of bronchitis and primary atypical pneumonia in humans, is also one of the gliding species [1–9]. Although gliding motility is believed to be involved in the pathogenicity of mycoplasmas, this relation has not been clearly established [1,3,7,8]. Mycoplasmas have no flagella or pili on their cell surfaces [10]. Their genomes contain no known genes related to other bacterial motility, and no genes homologous with motor proteins involved in eukaryotic motility [11–19]. These observations imply that the gliding mechanism of mycoplasmas might be distinct from other known mechanisms of motility [10,20,21].
Mycoplasma mobile, a piscine mycoplasma, is the fastest gliding mycoplasma, and its gliding mechanism has been under intense study [22–33]. Three huge proteins, Gli123 (123 kDa), Gli349 (349 kDa) and Gli521 (521 kDa), are involved in M. mobile gliding [30,31,33]. Analyses using gliding-defective mutants and gliding-inhibiting antibodies have indicated that the Gli123, Gli349 and Gli521 proteins form a cluster at the base of the membrane protrusion called the ‘neck’ and are likely to be responsible for the positioning of other gliding proteins, binding to glass during gliding, and generating and/or transmitting force respectively [24,30,31,33]. Rapid-freeze and freeze–fracture rotary-shadowing electron microscopy has revealed that many spike-like structures stick out around the neck and bind to the solid surface with their distal ends [29]. Therefore it has been speculated that the spike is composed of the Gli349 protein, and indirect support for this idea was provided by the molecular structure of the Gli349 protein unveiled by rotary-shadowing electron microscopy [22]. Based on these observations, the following gliding mechanism has been proposed [3,30,31]. The spike including the Gli349 molecule functions as a leg in the gliding mechanism. The leg, driven by force exerted from or through the Gli521 molecule, propels the cell, repeatedly binding to and being released from the solid surface.
Recently, by using a ‘ghost’ cell whose cell membrane was per-meabilized by Triton X-100, it was shown that the gliding motility of M. mobile must be driven by the energy of ATP [32]. This Triton model provided the first evidence that bacterial motility could also be driven directly by ATP hydrolysis. However, the key ATPase for the gliding remains to be identified. To clarify the gliding mechanism, identification of the ATPase for gliding is one of the most important issues. It is reasonable to treat the known gliding-related proteins as potential candidates for the ATPase. The genes encoding Gli123, Gli349 and Gli521 are located in tandem on the M. mobile genome with another gene encoding a 42 kDa function-unknown protein (P42), and these four genes seem to be polycistronically transcribed based on the results of RT (reverse transcriptase)–PCR analysis [33]. This relation with the Gli proteins led us to conjecture that the P42 protein might have some kind of function in the gliding motility.
Here, we propose that the P42 protein can hydrolyse some NTPs as well as ATP regardless of the lack of a common ATP-binding motif. Characteristics of the NTPase activity imply that this P42 protein is a prime candidate for the key ATPase generating gliding force for M. mobile.
MATERIALS AND METHODS
Cells, plasmids and materials
Escherichia coli strain HMS174(DE3) was from Novagen. The plasmids pET-43.1a(+), pMAL-c and pCold IV were purchased from Novagen, New England Biolabs and TaKaRa Bio respectively. Solutions of 100 mM ATP, UTP, GTP, CTP, dATP, dTTP, dUTP, dGTP and dCTP were from Promega, and chemical reagents of TTP, ADP, AMP, ATP[S] (adenosine 5′-[γ-thio]triphosphate) and p[NH]ppA (adenosine 5′-[β,γ-imido]triphosphate; ‘AMP-PNP’) were from Sigma. 8N3ATP[γ]biotin-LC-PEO-amine (8-azidoadenosine 5′-triphosphate [γ]-biotinyl-3,6,9-trioxaundecanediamine), an ATP analogue for photoaffinity labelling, was from Affinity Labeling Technologies.
Plasmid constructions
The DNA duplex consisting of the oligomeric DNAs 5′-AATTCCACCATCATCACCACCATTAATAAA-3′ and 5′-AGCTTTTATTAATGGTGGTGATGATGGTGG-3′ was ligated to the EcoRI–HindIII site of pCold IV. The resultant plasmid pCold-6His was constructed for overproduction of a C-terminal 6His-tagged recombinant protein. Plasmid pCold-P42-6His for overproduction of the C-terminal 6His-tagged P42 protein (P42-6His) was constructed by ligating the DNA fragment, which contains the ORF (open reading frame) MMOB1050 encoding the P42 protein, to the NdeI–XhoI site of pCold-6His. The DNA fragment was amplified by PCR using M. mobile 163K (ATCC 43663) genomic DNA [a gift from Dr A. Uenoyama (Graduate School of Science, Osaka City University, Osaka, Japan)] as a template. The primer sequences were 5′-GGCGGCGGCCATATGACAAAAAATTGgAATTTCGAA-3′ for the 5′-primer and 5′-GCCGCCCTCGAG-TCTTAGAAGAATTTCTTCTGTCAA-3′ for the 3′-primer, where underlined bases show the positions of the NdeI (5′-primer) and XhoI (3′-primer) sites. Since a codon of TGA is used for a tryptophan residue in mycoplasmas, those for the fifth and 67th tryptophan residues in the P42-encoding gene were replaced by a codon of TGG, for overproduction in E. coli cells. Replacement at the fifth residue was introduced by PCR with the 5′-primer. The lower-case letter in the described sequence shows the mutated base. Replacement at the 67th residue was introduced by site-directed mutagenesis with PCR as described previously [34].
Plasmid pCold-MBP-P42-6His for overproduction of the P42–6His fused to the MBP (maltose-binding protein) was constructed by ligating the DNA fragment containing the malE gene to the NdeI site of pCold-P42-6His. The DNA fragment was amplified by PCR using pMAL-c as a template. The primer sequences were 5′-GGACCATAGCATATGAAAACTGAAGAAGGTAAACT-3′ for the 5′-primer and 5′-TTTACTGAAATTAATCCTACCCTCGATGGATCCCC-3′ for the 3′-primer, where underlined bases show the positions of the NdeI (5′-primer) and PshBI (3′-primer) sites.
Plasmid pCold-Nus-P42-6His for overproduction of the P42–6His fused to E. coli NusA protein was constructed by ligating the DNA fragment containing the nusA gene to the NdeI site of pCold-P42-6His. The DNA fragment was amplified by PCR using pET-43.1a(+) as a template. The primer sequences were 5′-AGGAGATATACATATGAACAAAGAAATTTT-3′ for the 5′-primer and 5′-CCGCCGCCATTAATGGTACCGAGCTCCGCTTCGTCACCGAACCAGCAAAT-3′ for the 3′-primer, where underlined bases show the positions of the NdeI (5′-primer) and PshBI (3′-primer) sites.
Overproduction and purification of MBP–P42-6His
For overproduction, E. coli HMS174(DE3) was transformed with the pCold-MBP-P42-6His and grown in 200 ml of Luria–Bertani medium containing 0.1% glucose and 50 μg/ml ampicillin at 37 °C. When the D600 of the culture reached approx. 0.5, IPTG (isopropyl β-D-thiogalactoside) was added to the culture medium (final concentration: 0.3 mM). After additional cultivation at 37 °C for 30 min, the temperature of the growth medium was shifted to 15 °C and overproduction of the protein was carried out at 15 °C for 18 h. Cells were harvested by centrifugation at 5000 g for 5 min. The following protein purification was carried out at 4 °C. Cells were suspended in 20 mM Tris/HCl (pH 8.0) containing 1 mM EDTA (TE buffer), disrupted by sonication with an ultrasonic disruptor UD-201 from TOMY, and centrifuged at 18000 g for 30 min. The supernatant was applied to a column (2 ml) of amylose resin (New England Biolabs) equilibrated with TE buffer containing 0.5 M NaCl. After washing with 20 mM Tris/HCl (pH 8.0) containing 0.5 M NaCl (TS buffer), the protein was eluted from the column by 10 mM maltose in TS buffer. The eluent was applied to a column (1 ml) of HisTrap HP (GE Healthcare) charged with an NiSO4 solution and equilibrated with TS buffer. The protein was eluted from the column by a linear gradient of imidazole from 0 to 200 mM in TS buffer. The protein fractions were combined, concentrated, dialysed against 20 mM Tris/HCl (pH 8.0) containing 1 mM EDTA and 150 mM NaCl, and used for further analyses.
The protein concentration was determined by measuring UV absorption using an A2800.1% value of 1.15, which was calculated from molar absorption coefficient (ϵ) values of 1576 M−1·cm−1 for tyrosine and 5225 M−1·cm−1 for tryptophan at 280 nm [35].
Fractionation with gel filtration
Gel filtration was performed on a column (1.6 cm×60 cm) of HiLoad 16/60 Superdex 200 pg (GE Healthcare) equilibrated with 20 mM Tris/HCl (pH 8.0) containing 1 mM EDTA and 150 mM NaCl. The flow rate was set at 1.0 ml/min and 1.0 ml fractions were collected.
Overproduction and purification of Nus–P42–6His
Overproduction of the Nus–P42–6His protein and sonication lysis were carried out by a procedure similar to that described for the MBP–P42–6His, except that the pCold-Nus-P42-6His was used as a plasmid for overproduction. The supernatant obtained after sonication lysis was applied to a column (5 ml) of HiTrap Q HP (GE Healthcare) equilibrated with 20 mM Tris/HCl (pH 8.0). The flow-through fraction containing the protein was applied to a column (1 ml) of HisTrap HP charged with an NiSO4 solution and equilibrated with TS buffer. The protein was eluted from the column by a linear gradient of imidazole from 0 to 50 mM in TS buffer. The protein fractions were combined, concentrated, dialysed against 20 mM Tris/HCl (pH 8.0) containing 1 mM EDTA, 0.5 M NaCl and 3 mM imidazole, and used for further analyses.
The protein concentration was determined by measuring UV absorption using an A2800.1% value of 0.65, which was calculated as described above for MBP–P42–6His.
Measurement of nucleotide hydrolysis
Nucleotide hydrolysis was carried out at 37 °C for 30 min in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol, and in a final volume of 100 μl with a 96-well titre plate. The reaction was terminated by addition of 100 μl of Biomol Green solution (Biomol Research Laboratories) and its absorbance at 620 nm was measured using a model 680 microplate reader (Bio-Rad Laboratories) after incubation for 20 min at room temperature (20 °C). The standard curve was obtained with 0.8–25 μM phosphate. The effect of bivalent metal ions on the hydrolysis was analysed in the presence of 1 mM MnCl2, CoCl2, NiCl2, CuCl2, CaCl2 and ZnCl2 in place of MgCl2, or in the presence of 1 mM EDTA. The activity at different pH values was measured in a reaction buffer solution containing 10 mM Mes/NaOH (pH 5.6–7.1), 10 mM Hepes/NaOH (pH 6.8–8.3), 10 mM Tris/HCl (pH 7.1–8.8), or 10 mM glycine/NaOH (pH 8.3–10.0).
For the determination of kinetic parameters, the substrate concentration was varied from 0.06 to 1.0 mM. The hydrolysis of ATP, GTP, dATP or dGTP by the enzyme followed Michaelis–Menten kinetics, and the kinetic parameters were determined from a Lineweaver–Burk plot. For study of the inhibitory effect, the concentration of vanadate, ADP, AMP, ATP[S] or p[NH]ppA was varied from 0.001 to 0.5 mM. In the presence of these chemicals, ATPase activity was determined at ATP concentrations of 0.06–0.5 mM. Ki values were determined from a Dixon plot.
Affinity labelling with 8N3ATP[γ]biotin-LC-PEO-amine
For photoaffinity labelling, 10 μg of MBP–P42–6His was incubated on ice for 10 min with 0.5 mM 8N3ATP[γ]biotin-LC-PEO-amine in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol in a final volume of 50 μl, and then irradiated with a short-wave UV lamp (254 nm) at a distance of 1 cm on ice for 2 min. The reaction was stopped by the addition of 20 mM 2-mercaptoethanol, and free 8N3ATP[γ]biotin-LC-PEO-amine was removed by a Micro Bio-Spin chromatography gel-filtration column (Bio-Rad Laboratories). The labelled proteins were trapped with Tetralink™ tetrameric avidin resin (Promega). After washing five times with 20 mM Tris/HCl (pH 8.0) containing 0.2 M NaCl and 0.5% SDS, the resin was boiled at 100 °C for 10 min in SDS/PAGE sample buffer. The labelled proteins released to the sample buffer were separated by SDS/PAGE (10% gels), transferred on to a PVDF membrane, and detected by using ECL® (enhanced chemiluminescence) streptavidin–HRP (horseradish peroxidase) conjugate (GE Healthcare).
RESULTS
Overproduction of recombinant P42 proteins
To obtain the P42 protein in an amount sufficient for biochemical characterization, several overproducing E. coli strains were constructed. The C-terminal His-tagged proteins (P42–6His) were accumulated in a soluble form in E. coli cells, but they were so unstable that they became insoluble during purification. Similarly, the tag-free P42 proteins were also too unstable for purification (results not shown). On the other hand, the P42–6His proteins fused to the MBP (MBP–P42–6His) were purified easily and could be kept in a soluble form. In this construct, a cleavage site for Factor Xa was introduced between P42–6His and the MBP tag. However, cutting off the MBP tag by Factor Xa resulted in aggregation of the P42 proteins, probably because of the low solubility of the P42 proteins. Therefore the MBP–P42–6His protein without cleavage by Factor Xa was used for further analyses. As will be described below, the P42–6His fused to NusA protein (Nus–P42–6His) was also obtained in a soluble form.
ATPase activity of MBP–P42–6His
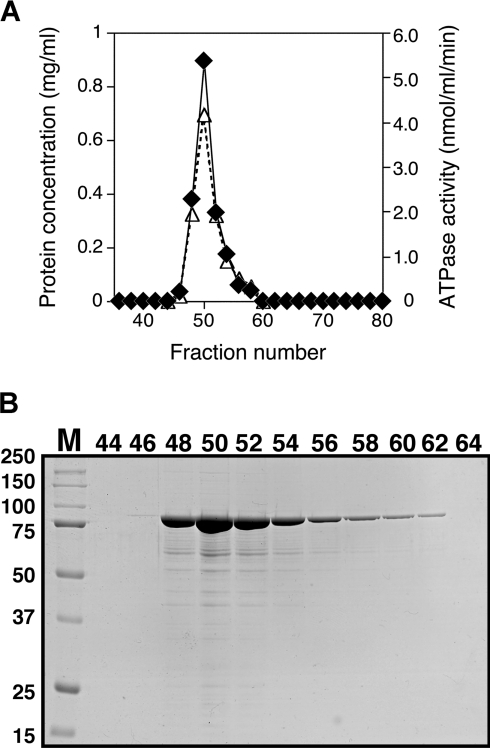
The recombinant MBP–P42–6His protein was purified with two kinds of column chromatography, amylose resin column chromatography for MBP tag and Ni2+-chelating column chromatography for His tag. When the purified protein was incubated with 1 mM ATP as described in the Materials and methods section, ATP hydrolysis was observed. To test for an association between MBP–P42–6His and the ATPase activity, the purified protein was further fractionated by gel-filtration column chromatography. As shown in Figure 1(A), the profile of ATPase activity showed a perfect match with the elution profile of the MBP–P42–6His protein, suggesting that ATPase activity was intrinsic to the MBP–P42–6His. As a control, a C-terminal His-tagged MBP (MBP–6His) was also purified and fractionated by gel-filtration column chromatography in the same manner as MBP–P42–6His, and its ATPase activity was examined under the same conditions as used for MBP–P42–6His. However, it exhibited no ATPase activity (results not shown), implying that the ATPase activity of MBP–P42–6His must be independent of the MBP tag.
Figure 1. Fractionation of the MBP–P42–6His preparation by gel-filtration column chromatography.
(A) Protein concentrations and ATPase activity of each elution fraction from the gel filtration column are shown by ◆ and △ respectively. Hydrolysis of ATP was carried out at 37 °C for 30 min in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol. A 5 μl portion from the elution fraction was used for each reaction in a final volume of 100 μl. (B) Coomassie Brilliant Blue-stained SDS/PAGE of each fraction. Numbers above the gel are the fraction numbers, and numbers along the gel represent the molecular masses of individual standard proteins. A small number of the degraded MBP–P42–6His proteins are also shown, which were confirmed by Western-blot analyses using P42 antiserum (a gift from Dr A. Uenoyama) and MBP antiserum (New England Biolabs; results not shown).
Characterization of ATPase activity
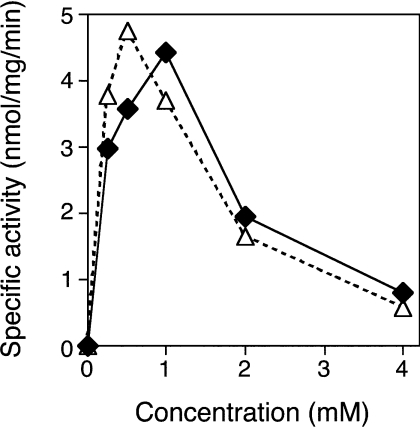
The assays for ATPase activity were carried out in the presence of 1 mM ATP under various conditions as described in the Materials and methods section. MBP–P42–6His exhibited the maximal ATPase activity of approx. 4.4 nmol·mg−1·min−1 at pH 8.0 in the presence of 1 mM MgCl2 and 10 mM NaCl. The enzyme exhibited activity only in the presence of Mg2+, and not in the presence of Mn2+, Co2+, Ni2+, Cu2+, Ca2+, Zn2+ or EDTA. The specific activity determined in the presence of 1 mM MgCl2 was approx. 4.2- or 2.4-fold higher than those in the presence of 0.1 or 10 mM respectively, and the activity at a pH of 8 was 3.5- or 1.3-fold higher than those at a pH of 6 or 10 respectively. Although the enzyme did not require NaCl or KCl for activity, NaCl or KCl at concentrations of <20 mM stimulated the reaction. The activities in the presence of NaCl were slightly (1.3-fold) higher than those in the presence of KCl. The effect of ATP concentration on the activity was also examined. As shown in Figure 2, ATP at concentrations of >1 mM dramatically inhibited the reaction. Because this inhibition was observed similarly in the presence of 5 mM MgCl2, the decline in hydrolysis might not be because of lack of Mg2+ ions. As described below, this enzyme also hydrolysed other nucleotides. In the case of dATP, a similar inhibition was observed (Figure 2). It seemed likely that the hydrolysis of nucleotide substrates by MBP–P42–6His was inhibited at substrate concentrations of >1 mM. Therefore, for the hydrolysis by the P42 protein, the total amount of nucleotides was determined to be limited to concentrations below 1 mM.
Figure 2. Effect of substrate concentrations on hydrolysis.
The specific activities of phosphatase were determined in the presence of ATP (◆) or dATP (△) at concentrations of 0.25–4 mM. Hydrolysis of the nucleotide was carried out at 37 °C for 30 min in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol. The specific activities shown are the average values for triplicate experiments and were reproducible within 20% of the mean values.
Substrate specificity
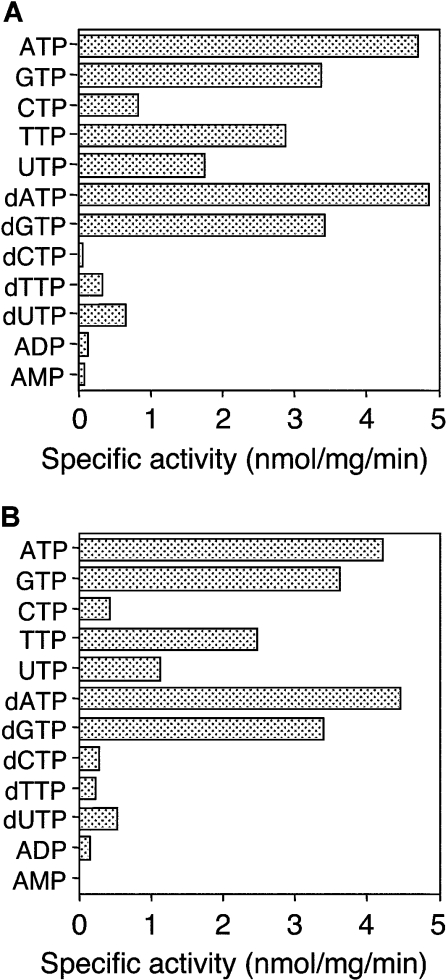
MBP–P42–6His hydrolysed not only ATP but also other nucleotides (Figure 3A). While the enzyme seemed to prefer purine NTPs such as ATP, GTP, dATP and dGTP, it weakly hydrolysed ADP or AMP. Therefore we recognized the P42 protein as an NTPase. Judging from the results of this assay and another general ATPase assay coupled with pyruvate kinase and lactate dehydrogenase [36], it was unlikely that the enzyme hydrolysed ATP[S] and p[NH]ppA (results not shown). The kinetic parameters for ATP, GTP, dATP and dGTP are shown in Table 1. The results suggested that the binding affinities to ATP or dATP were 3–5-fold higher than those to GTP or dGTP, but the catalytic efficiencies were almost the same among ATP, ATP, GTP and dGTP.
Figure 3. Substrate specificity of recombinant P42 proteins.
Specific activities of MBP–P42–6His (A) and Nus–P42–6His (B). Hydrolysis was carried out in the presence of 1 mM nucleotide at 37 °C for 30 min in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol. The specific activities shown are the average values for triplicate experiments and were reproducible within 20% of the mean values.
Table 1. Kinetic parameters of P42 proteins.
Hydrolysis of the nucleotide was carried out at 37 °C for 30 min in 10 mM Tris/HCl (pH 8.0) containing 1 mM MgCl2, 10 mM NaCl and 10% glycerol. The kinetic parameters were determined in triplicate experiments.
| MBP–P42–6His | Nus–P42–6His | |||
|---|---|---|---|---|
| Substrate | Km (mM) | Vmax (nmol·mg−1·min−1) | Km (mM) | Vmax (nmol·mg−1·min−1) |
| ATP | 0.13±0.01 | 4.51±0.20 | 0.18±0.02 | 5.68±0.33 |
| dATP | 0.16±0.01 | 4.70±0.71 | 0.12±0.02 | 6.44±0.02 |
| GTP | 0.64±0.07 | 3.94±0.08 | 0.78±0.06 | 10.90±0.89 |
| dGTP | 0.50±0.08 | 2.23±0.13 | 0.53±0.06 | 6.49±0.26 |
Inhibition of ATPase activity
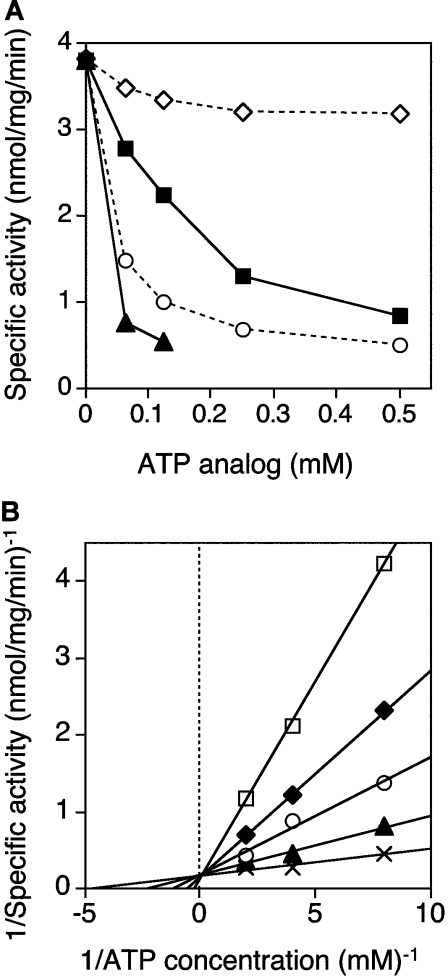
Unlike the activity of many ATPase enzymes, e.g. myosin [37], dynein [38], ABC transporter (ATP-binding-cassette transporter) [39] and MutS [40], the ATPase activity of MBP–P42–6His did not decrease in the presence of vanadate, an ATPase inhibitor acting as a phosphate analogue, at concentrations of 0.06–0.5 mM. On the other hand, ADP, ATP[S] and p[NH]ppA interfered with the ATP hydrolysis by MBP–P42–6His (Figure 4A). Lineweaver–Burk plots in the presence of different fixed concentrations of ADP showed that ADP inhibited the hydrolysis competitively with ATP (Figure 4B). Similar plots were obtained in the case of the other nucleotides (results not shown). The Ki values determined from Dixon plots (results not shown) were 0.033 mM for ADP, 0.0034 mM for ATP[S], and 0.0018 mM for p[NH]ppA. Although AMP also barely seemed to work on the hydrolysis as an inhibitor, its effect was too modest at concentrations of <0.5 mM to be detected clearly (Figure 4A). Because the nucleotides at concentrations of >1 mM non-competitively inhibited the ATP hydrolysis by the MBP–P42–6His (Figure 2), the Ki value for AMP could not be accurately determined at such high concentrations.
Figure 4. Inhibition of ATPase activity of MBP–P42–6His by ATP analogues.
(A) ATPase activity in the presence of ADP (■), AMP (◇), ATP[S] (○) or p[NH]ppA (▲) at various concentrations. For hydrolysis, 0.5 mM ATP was added as described in the Materials and methods section. The specific activities shown are the average values for triplicate experiments and were reproducible within 20% of the mean values. (B) Lineweaver–Burk plots for ATP hydrolysis by MBP–P42–6His in the presence of ADP at various concentrations. Values in the presence of ADP at the concentrations of 0 (×), 0.06 (▲), 0.13 (○), 0.25 (◆) and 0.5 (□) mM.
NTPase activity of Nus–P42–6His
ATPase activity of another P42 construct, the Nus–P42–6His protein, was also examined as described for MBP–P42–6His. As shown in Figure 3(B), the substrate specificities of Nus–P42–6His were analogous to those of MBP–P42–6His. The kinetic parameters for Nus–P42–6His (Table 1) were also similar to those for MBP–P42–6His, although the Vmax value for GTP of Nus–P42–6His might be slightly higher than that of MBP–P42–6His. The similarity in the characteristics of NTPase activity between the two P42 constructs suggested that the NTPase activities of these constructs are independent of MBP tag or Nus tag and intrinsic to the P42 protein.
Photoaffinity labelling by ATP analogue
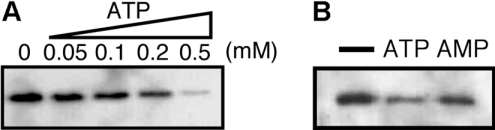
To confirm the site-specific binding of ATP, photoaffinity labelling by an ATP analogue was also examined. Since 8N3ATP[γ]biotin-LC-PEO-amine was used as a photoactivatable ATP analogue, the labelled proteins were recovered and detected by means of the affinity of biotin to avidin. Figure 5(A) shows the results of labelling of the MBP–P42–6His protein. A positive signal corresponding to MBP–P42–6His was detected. The signal was never observed without UV irradiation or 8N3ATP[γ]biotin-LC-PEO-amine (results not shown), suggesting that MBP–P42–6His was photoaffinity-labelled by 8N3ATP[γ]biotin-LC-PEO-amine. Next, the labelled MBP–P42–6His was exposed to Factor Xa and checked by Western-blot analyses using MBP antiserum. The result indicated that only the P42 region of MBP–P42–6His was labelled but the MBP tag was not (results not shown). As shown in Figure 5(A), this labelling was inhibited in the presence of ATP. Although the presence of AMP also seemed to inhibit the labelling slightly, the inhibition efficiency was lower than that in the presence of ATP (Figure 5B). The difference in the inhibiting efficiency between ATP and AMP seemed to agree with the results shown in Figure 4(A), which implies that P42 contains a specific binding site(s) for ATP (or another NTP).
Figure 5. Photoaffinity labelling of MBP–P42–6His by using 8N3ATP[γ]-biotin-LC-PEO-amine, an ATP analogue.
(A) Photoaffinity labelling in the presence of ATP at various concentrations. (B) The MBP–P42–6His protein labelled in the absence or in the presence of 0.5 mM ATP or AMP is shown.
DISCUSSION
P42 is a novel NTPase
It was shown that recombinant P42 proteins exhibited NTPase activity in in vitro assay. The similarity of biochemical characteristics between two different recombinant proteins, MBP–P42–6His and Nus–P42–6His, suggested that the NTPase activity depended on the P42 protein. The P42 proteins exhibited a specific ATPase activity of approx. 4.4 nmol·mg−1·min−1 (Figure 3). The specific activities of some secretion-associated ATPases/NTPases were as modest as those of the P42 proteins. For example, Aquifex aeolicus PilT [41], Legionella pneumophila DotB [42], Actinobacillus actinomycetemcomitans TadA [43], R64 PilQ [44] and R388 TrwD [45] have similar activities of between 1 and 15 nmol·mg−1·min−1. Although the specific activity of the P42 protein in vitro might be modest, it would be sufficient for the cellular function in vivo. Alternatively, some factors would be required to promote ATP hydrolysis by the P42 protein. The addition of phospholipids has often been reported to promote ATP hydrolysis by a membrane-associaed secretion NTPase [43,45,46]. The activity of the P42 protein might also be stimulated in the presence of phospholipids. It is because the P42 protein has the potential to work co-operatively with other membrane proteins (Gli123, Gli349 and Gli521) that it may be involved in M. mobile gliding as discussed below.
The lack of proteins homologous with P42, as well as the fact that P42 has no functional motif and no signal sequence, had made it difficult to predict its function. Only a product of ORF MYPU-2170 from Mycoplasma pulmonis, another gliding mycoplasma species, shows 28% amino acid sequence identities to the P42 protein, but its function is unknown. Although many ATPase enzymes contain a functional motif for ATP binding known as Walker A or P-loop (GXXXXGKT/S) [47], the P42 protein has no Walker A motif. Therefore the P42 protein was an unexpected novel NTPase.
Comparison with a putative ATPase for gliding motility
A previous analysis using a gliding ghost implied that a putative ATPase (NTPase) for gliding motility prefers reaction conditions of alkaline pH values and low salt concentrations, and is inhibited by ADP, AMP, ATP[S] and p[NH]ppA but not by vanadate [32]. These characteristics were also true for the ATPase activity of the P42 protein (Figure 4A). The putative ATPase is hypothesized to prefer to hydrolyse ATP, GTP, dATP and ATP[S] rather than ADP, AMP, p[NH]ppA and CTP [32]. The P42 protein also preferred to hydrolyse purine NTPs such as ATP, GTP, dATP and dGTP (Figure 3), although hydrolysis of ATP[S] was not detected in the in vitro ATPase assay. These similarities imply that the P42 protein is a potential candidate for a key ATPase in the generation of gliding force for M. mobile.
The Ki values for ATP analogues of the putative ATPase (0.41 mM for ADP, 0.017 mM for ATP[S] and 0.36 mM for p[NH]ppA) [32] were higher than those of the P42 protein. The difference in Ki values might depend on a difference of evaluative criteria, gliding speed or phosphate generation from ATP. The gliding motility is likely to occur through a complicated mechanism involving various proteins and factors. These proteins or factors might play a role in the inhibiting effect of ATP analogues. On the other hand, the Km values of the P42 proteins shown in Table 1 were similar to those of 0.17 mM for ATP, 0.8 mM for GTP and 0.2 mM for dATP of the putative ATPase [32].
Is P42 the motor for gliding motility?
The genes encoding the P42 and other Gli proteins are located in tandem on the M. mobile genome, and seem to work as an operon [33]. Therefore it is reasonable to suppose involvement of the P42 in gliding. Neither Gli349 nor Gli521 native proteins purified from M. mobile cells exhibited detectable ATPase activity, and the recombinant Gli123 protein, which was expressed in E. coli and purified, also showed no ATPase activity (N. Ohtani, unpublished work). Among the proteins encoded in the operon, only P42 exhibited ATPase activity. In addition, the biochemical characteristics supported the idea that the P42 protein is the ATPase for gliding motility of M. mobile.
Is the P42 protein indeed a motor for gliding motility of M. mobile? To solve the problem, the effect of antibody against the P42 protein should be examined in the gliding ghost of M. mobile. Alternatively, disruption of the P42 gene in M. mobile is another way. Because of technical difficulties, however, these approaches remain to be examined. The P42 protein has no putative signal peptide and no putative transmembrane segment, suggesting that the P42 is a cytoplasmic protein. If the P42 protein is involved in gliding motility, it probably interacts with Gli proteins in order to get closer to the membrane and play its role in gliding. Analyses of the interactions between the P42 and other Gli proteins might provide some clue for establishing that the P42 protein is a key ATPase for the gliding motility of M. mobile.
Acknowledgments
This research was partially supported by a Grant-in-Aid for Young Scientists (B) (No. 17770115) from the Ministry of Education, Culture, Sports, Science and Technology of Japan at 2005–2006 (to N.O.), and by a Grant-in-Aid for Scientific Research on the Priority Areas ‘applied genomics’ and ‘structures of biological macromolecular assemblies’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to M.M.) and by a grant from the Institution for Fermentation Osaka (to M.M.).
References
- 1.Razin S., Yogev D., Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause D. C., Balish M. F. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 2004;51:917–924. doi: 10.1046/j.1365-2958.2003.03899.x. [DOI] [PubMed] [Google Scholar]
- 3.Miyata M. Gliding motility of mycoplasmas – the mechanism cannot be explained by current biology. In: Blanchard A., Browning G., editors. Mycoplasmas: Molecular Biology, Pathogenicity, and Strategies for Control. Norfolk, U.K.: Horizon Bioscience; 2005. pp. 137–163. [Google Scholar]
- 4.Seto S., Layh-Schmitt G., Kenri T., Miyata M. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 2001;183:1621–1630. doi: 10.1128/JB.183.5.1621-1630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seto S., Miyata M. Attachment organelle formation represented by localization of cytadherence protein and formation of an electron-dense core in wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 2003;185:1082–1091. doi: 10.1128/JB.185.3.1082-1091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimizu T., Miyata M. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 2002;44:431–434. doi: 10.1007/s00284-001-0014-8. [DOI] [PubMed] [Google Scholar]
- 7.Bredt W. Motility. In: Barile M. F., Razin S., Tully J. G., Whitcomb R. F., editors. The Mycoplasmas, Vol. 1. New York: Academic Press; 1979. pp. 141–145. [Google Scholar]
- 8.Kirchhoff H. Motility. In: Maniloff J., McElhaney R. N., Finch L. R., Baseman J. B., editors. Mycoplasmas-Molecular Biology and Pathogenesis. Washington DC: American Society for Microbiology; 1992. pp. 289–306. [Google Scholar]
- 9.Razin S., Jacobs E. Mycoplasma adhesion. J. Gen. Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 10.McBride M. J. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Chambaud I., Heilig R., Ferris S., Barbe V., Samson D., Galisson F., Moszer I., Dybvig K., Wroblewski H., Viari A., et al. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 2001;29:2145–2153. doi: 10.1093/nar/29.10.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser C. M., Gocayne J. D., White O., Adams M. D., Clayton R. A., Fleischmann R. D., Bult C. J., Kerlavage A. R., Sutton G., Kelley J. M., et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 13.Himmelreich R., Hilbert H., Plagens H., Pirkl E., Li B.-C., Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaffe J. D., Stange-Thomann N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M. G., Hafez N., et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minion F. C., Lefkowitz E. J., Madsen M. L., Cleary B. J., Swartzell S. M., Mahairas G. G. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J. Bacteriol. 2004;186:7123–7133. doi: 10.1128/JB.186.21.7123-7133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papazisi L., Gorton T. S., Kutish G., Markham P. F., Browning G. F., Nguyen D. K., Swartzell S., Madan A., Mahairas G., Geary S. J. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low) Microbiology. 2003;149:2307–2316. doi: 10.1099/mic.0.26427-0. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki Y., Ishikawa J., Yamashita A., Oshima K., Kenri T., Furuya K., Yoshino C., Horino A., Shiba T., Sasaki T., Hattori M. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 2002;30:5293–5300. doi: 10.1093/nar/gkf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vasconcelos A. T., Ferreira H. B., Bizarro C. V., Bonatto S. L., Carvalho M. O., Pinto P. M., Almeida D. F., Almeida L. G., Almeida R., Alves-Filho L., et al. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J. Bacteriol. 2005;187:5568–5577. doi: 10.1128/JB.187.16.5568-5577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Westberg J., Persson A., Holmberg A., Goesmann A., Lundeberg J., Johansson K. E., Pettersson B., Uhlen M. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP) Genome Res. 2004;14:221–227. doi: 10.1101/gr.1673304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geeves M. A., Fedorov R., Manstein D. J. Molecular mechanism of actomyosin-based motility. Cell. Mol. Life Sci. 2005;62:1462–1477. doi: 10.1007/s00018-005-5015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kull F. J. Motor proteins of the kinesin superfamily: structure and mechanism. Essays Biochem. 2000;35:61–73. doi: 10.1042/bse0350061. [DOI] [PubMed] [Google Scholar]
- 22.Adan-Kubo J., Uenoyama A., Arata T., Miyata M. Morphology of isolated Gli349, a leg protein responsible for Mycoplasma mobile gliding via glass binding, revealed by rotary shadowing electron microscopy. J. Bacteriol. 2006;188:2821–2828. doi: 10.1128/JB.188.8.2821-2828.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe J. D., Miyata M., Berg H. C. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 2004;186:4254–4261. doi: 10.1128/JB.186.13.4254-4261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kusumoto A., Seto S., Jaffe J. D., Miyata M. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology. 2004;150:4001–4008. doi: 10.1099/mic.0.27436-0. [DOI] [PubMed] [Google Scholar]
- 25.Metsugi S., Uenoyama A., Adan-Kubo J., Miyata M., Yura K., Kono H., Go N. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics. 2005;1:33–43. doi: 10.2142/biophysics.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyata M., Yamamoto H., Shimizu T., Uenoyama A., Citti C., Rosengarten R. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology. 2000;146:1311–1320. doi: 10.1099/00221287-146-6-1311. [DOI] [PubMed] [Google Scholar]
- 27.Miyata M., Uenoyama A. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 2002;215:285–289. doi: 10.1111/j.1574-6968.2002.tb11404.x. [DOI] [PubMed] [Google Scholar]
- 28.Miyata M., Ryu W. S., Berg H. C. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 2002;184:1827–1831. doi: 10.1128/JB.184.7.1827-1831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyata M., Petersen J. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 2004;186:4382–4386. doi: 10.1128/JB.186.13.4382-4386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto S., Uenoyama A., Miyata M. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 2005;187:3502–3510. doi: 10.1128/JB.187.10.3502-3510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uenoyama A., Kusumoto A., Miyata M. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 2004;186:1537–1545. doi: 10.1128/JB.186.5.1537-1545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uenoyama A., Miyata M. Gliding ghosts of Mycoplasma mobile. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12754–12758. doi: 10.1073/pnas.0506114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uenoyama A., Miyata M. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 2005;187:5578–5584. doi: 10.1128/JB.187.16.5578-5584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanaya S., Oobatake M., Nakamura H., Ikehara M. pH-dependent thermostabilization of Escherichia coli ribonuclease HI by histidine to alanine substitutions. J. Biotechnol. 1993;28:117–136. doi: 10.1016/0168-1656(93)90129-b. [DOI] [PubMed] [Google Scholar]
- 35.Goodwin T. W., Morton R. A. The spectrophotometric determination of tyrosine and tryptophan in proteins. Biochem. J. 1946;40:628–632. doi: 10.1042/bj0400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugh B. F., Cox M. M. High salt activation of recA protein ATPase in the absence of DNA. J. Biol. Chem. 1988;263:76–83. [PubMed] [Google Scholar]
- 37.Goodno C. C. Inhibition of myosin ATPase by vanadate ion. Proc. Natl. Acad. Sci. U.S.A. 1979;76:2620–2624. doi: 10.1073/pnas.76.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibbons I. R., Cosson M. P., Evans J. A., Gibbons B. H., Houck B., Martinson K. H., Sale W. S., Tang W. J. Potent inhibition of dynein adenosinetriphosphatase and of the motility of cilia and sperm flagella by vanadate. Proc. Natl. Acad. Sci. U.S.A. 1978;75:2220–2224. doi: 10.1073/pnas.75.5.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson D. S., Adhikari P., Nowalk A. J., Chen C. Y., Mietzner T. A. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 2004;186:6220–6229. doi: 10.1128/JB.186.18.6220-6229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pezza R. J., Villarreal M. A., Montich G. G., Argarana C. E. Vanadate inhibits the ATPase activity and DNA binding capability of bacterial MutS. A structural model for the vanadate-MutS interaction at the Walker A motif. Nucleic Acids Res. 2002;30:4700–4708. doi: 10.1093/nar/gkf606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herdendorf T. J., McCaslin D. R., Forest K. T. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J. Bacteriol. 2002;184:6465–6471. doi: 10.1128/JB.184.23.6465-6471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sexton J. A., Pinkner J. S., Roth R., Heuser J. E., Hultgren S. J., Vogel J. P. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 2004;186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharjee M. K., Kachlany S. C., Fine D. H., Figurski D. H. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J. Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai D., Horiuchi T., Komano T. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J. Biol. Chem. 2001;276:17968–17975. doi: 10.1074/jbc.M010652200. [DOI] [PubMed] [Google Scholar]
- 45.Rivas S., Bolland S., Cabezon E., Goni F. M., de la Cruz F. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 46.Krause S., Pansegrau W., Lurz R., de la Cruz F., Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saraste M., Sibbald P. R., Wittinghofer A. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]