Abstract
The RNase E/G family of endoribonucleases plays the central role in numerous post-transcriptional mechanisms in Escherichia coli and, presumably, in other bacteria, including human pathogens. To learn more about specific properties of RNase E/G homologues from pathogenic Gram-positive bacteria, a polypeptide comprising the catalytic domain of Mycobacterium tuberculosis RNase E/G (MycRne) was purified and characterized in vitro. In the present study, we show that affinity-purified MycRne has a propensity to form dimers and tetramers in solution and possesses an endoribonucleolytic activity, which is dependent on the 5′-phosphorylation status of RNA. Our data also indicate that the cleavage specificities of the M. tuberculosis RNase E/G homologue and its E. coli counterpart are only moderately overlapping, and reveal a number of sequence determinants within MycRne cleavage sites that differentially affect the efficiency of cleavage. Finally, we demonstrate that, similar to E. coli RNase E, MycRne is able to cleave in an intercistronic region of the putative 9S precursor of 5S rRNA, thus suggesting a common function for RNase E/G homologues in rRNA processing.
Keywords: 5′-end-dependence, Escherichia coli, Mycobacterium tuberculosis, ribonuclease, RNA processing, RNase E/G
Abbreviations: BCG, Bacille Calmette–Guérin; DTT, dithiothreitol; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; MycRne, RNase E/G homologue from Mycobacterium tuberculosis; PNPase, polynucleotide phosphorylase
INTRODUCTION
The RNase E/G family of endoribonucleases is known for its essential role in RNA processing and decay in bacteria [1–4]. Our current knowledge about these enzymes mainly originates from studies of Escherichia coli RNase G and RNase E, site-specific ribonucleases that preferentially cleave in A/U-rich single-stranded regions of structured RNAs.
E. coli RNase E is encoded by the rne gene and is indispensable for cell viability [5,6]. Structural data obtained for an N-terminal polypeptide representing the evolutionarily conserved catalytic domain of this protein (Figure 1) have revealed the presence of discrete folds with putative functions in RNA recognition and cleavage [7]. Moreover, these data and others [8,9] indicate that the catalytic domain exists as a homotetramer, which has two non-equivalent subunit interfaces organizing the active site of the enzyme. In contrast with the N-terminal catalytic domain, the C-terminal part of E. coli RNase E shows very little or no similarity to the equivalent regions of other RNase E homologues [10]. This part of the E. coli enzyme contains an extra RNA-binding domain(s) [11,12] and multiple sites for interactions with the 3′ to 5′ phosphorolytic exonuclease PNPase (polynucleotide phosphorylase), the RNA helicase RhlB and the glycolytic enzyme enolase [10,13] to form the E. coli RNA degradosome [14].
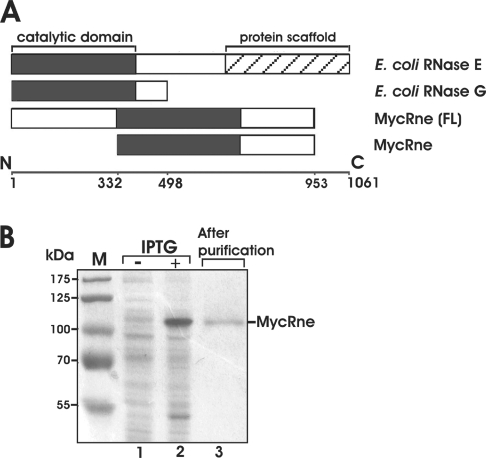
Figure 1. Purification of MycRne.
(A) Primary structures of E. coli and M. tuberculosis RNase E/G homologues. The evolutionarily conserved minimal catalytic domain [43] is shown by black boxes in the full-length E. coli RNase E, RNase G and M. tuberculosis RNase E/G [MycRne (FL)] polypeptides as well as in the N-terminally truncated form of M. tuberculosis RNase E/G (MycRne) used in the present study. The ‘protein scaffold’ region (hatched box) of E. coli RNase E (residues 688–1061) [13] is the location of the binding sites for the major components of the degradosome (enolase, RhlB RNA helicase and PNPase). (B) Purification of MycRne. Affinity-purified mycobacterial MycRne, as well as protein extracts prepared from BL21(DE3) cells harbouring the MycRne-coding plasmid before (−) and after (+) addition of IPTG respectively, were analysed in SDS/10% polyacrylamide gels followed by Coomassie Blue staining. Indicated are the positions of M. tuberculosis RNase E/G (MycRne) and the molecular-mass markers (in kDa; lane M).
E. coli RNase G [15,16] is encoded by the rng gene and was initially termed CafA owing to its effects on the formation of cytoplasmic axial filaments observed upon RNase G overproduction in vivo [17,18]. Further studies have revealed that this protein has an endoribonucleolytic activity [19] and is involved in maturation of 16S rRNA in vivo [15,16]. Although RNase G functionally overlaps with RNase E and shares 35% identity with and 50% similarity to the N-terminal catalytic part of RNase E [20], this endoribonuclease cannot fully compensate RNase E deficiency in rnets mutant strains [5,6].
Owing to their overlapping functions and apparently common origin, RNase E- and RNase G-like proteins are believed to belong to the same family of RNase E/G endoribonucleases that are predicted to exist in many bacteria, including pathogenic species [21–23]. Although their biochemical properties and biological functions have not yet been investigated in detail in human pathogens, such studies can offer important insights concerning the role of these endoribonucleases during infection and disease development. This consideration prompted us to purify and characterize the RNase E/G homologue (MycRne) from the intracellular pathogen Mycobacterium tuberculosis.
We have shown that, similar to E. coli RNase G [24] and the catalytic domain of E. coli RNase E [8,9], MycRne is a 5′-end-dependent endoribonuclease, which can exist in dimeric and tetrameric states in solution. Moreover, MycRne can cleave a putative 5S rRNA precursor in vitro, and the cleavage occurs very close to the 5′ end of mature 5S rRNA, thus suggesting a role for this endoribonuclease in rRNA processing in vivo. Although MycRne and E. coli RNase E share some common properties, we have shown in the present study that their substrate specificities are only partially overlapping. Taken together, our data expand the current knowledge about biochemical characteristics of RNase E/G enzymes and provide a basis for further analysis of their putative functions in mycobacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions
E. coli strains were routinely grown in LB (Luria–Bertani) medium supplemented with the appropriate antibiotic(s) and/or glucose at 37 °C. Mycobacterium bovis BCG (Bacille Calmette–Guérin) was grown in Middlebrook 7H9 broth (Difco) supplemented with 10% Middlebrook OADC (oleic acid/albumin/dextrose/catalase) enrichment (Difco) and 0.05% Tween 80 (Sigma).
Purification of Rne498
E. coli BL21(DE3) strain (Novagen) carrying a pET16b-based plasmid that encodes the first 498 amino acids of E. coli RNase E (Rne498) [12] was used to overexpress His6-tagged Rne498, and the overexpressed protein was purified further using immobilized metal-affinity chromatography as described previously [12]. The eluate fractions, in which the purity of Rne498 was greater than 95%, were pooled, dialysed against SB buffer [50 mM Tris/HCl, pH 8.0, 0.1 mM EDTA, 0.5 M NaCl, 20% (v/v) glycerol and 1 mM DTT (dithiothreitol)] and stored in small (50 μl) aliquots at −80 °C.
Purification of M. tuberculosis RNase E/G
A 1.8 kb DNA fragment (gene MT2520) encoding the C-terminal portion of MycRne polypeptide (residues 332–953) was amplified from genomic DNA using primers Myc3 (5′-TAACATATGGTGGTGCGCGACC-3′) and Myc2 (5′-CGGAGA TCTGGTCAGTCTAGGCGG-3′). The NdeI/BglII-digested PCR product was ligated into NdeI/BamHI-treated p6HisF-11d (icl) [25] downstream of the T7 RNA polymerase promoter and in-frame with the preceding sequence of the N-terminal His6-tag. The resulting construct was named pMtRne, verified by sequencing and transformed into the E. coli strain BL21-CodonPlus (DE3)-RIL (Stratagene).
E. coli cells containing pMtRne plasmid were grown at 37 °C in LB medium supplemented with ampicillin (100 μg/ml), chloramphenicol (34 μg/ml) and glucose (0.75%). At an attenuance (D600) of ∼0.5, the cell culture was pre-cooled on ice for 45 min, and protein expression was induced by addition of IPTG (isopropyl β-D-thiogalactoside) to 1 mM and incubation for a further 16 h at 15 °C. Cells were harvested by centrifugation at 7000 g for 15 min, frozen at −20 °C, thawed and suspended in ice-cold binding buffer (0.5 M NaCl and 20 mM sodium phosphate, pH 7.4) supplemented with pepstatin (1 μg/ml) and protease inhibitor cocktail (one tablet per 3 g of wet cells) (Roche). The cells were disrupted by sonication in a Cell Disruptor W375 (Heat Systems Ultrasonics), the cell lysate was cleared further by centrifugation at 16000 g for 15 min at 4 °C, and the supernatant was applied to a HisTrap HP (Amersham Biosciences) column to purify His6-tagged MycRne according to the manufacturer's instructions. Column fractions containing MycRne were combined, supplemented with EDTA and DTT to final concentrations of 0.1 mM and 1 mM respectively, dialysed extensively against EB buffer (50 mM NaCl, 20 mM Hepes, pH 7.5, 1 mM DTT and 0.1 mM EDTA) and stored in small (50 μl) aliquots at −80 °C.
Gel filtration
Affinity-purified MycRne was dialysed against low-salt buffer (75 mM NaCl, 10 mM Hepes, pH 7.5, 1 mM DTT and 0.1 mM EDTA) and concentrated to 8.25 mg/ml employing a Centricon YM-30 device (Millipore). The concentration of MycRne was calculated based on its absorbance at 280 nm and using molar absorption coefficients estimated from its amino acid composition by ProtParam (ExPaSy server, http://www.expasy.org). Before loading an aliquot (∼150 μl) of the concentrated protein sample on a 25 ml Superose 12 HR10/30 column (Amersham Biosciences), the column was pre-equilibrated with low-salt buffer and calibrated using molecular-mass markers [aldolase (158 kDa), BSA (67 kDa) and chymotrypsinogen A (25 kDa)]. The column was run using the same buffer, and 0.5 ml fractions were subsequently collected and analysed by spectrophotometry and SDS/PAGE.
Analytical ultracentrifugation
An aliquot of affinity-purified MycRne was dialysed against 20 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM DTT and 0.1 mM EDTA and used further in sedimentation-equilibrium experiments, which were performed at 20 °C on a Beckman Optima XL-A ultracentrifuge with an AN60-Ti rotor. The same buffer was used to dilute samples and as a blank. Long runs were performed at six speeds (5000, 6000, 7000, 9500, 12000 and 45000 rev./min) using 150 μl samples at three concentrations (16, 8 and 1.6 μM) analysed at 292, 286 and 233 nm respectively, whereas short runs were processed at ten speeds (5000, 6000, 7000, 9500, 12000, 14000, 16000, 18000, 20000 and 45000 rev./min) using 40 μl samples at three concentrations (84, 42 and 16 μM) analysed at 297, 292 and 286 nm respectively. Absorbance analysis (220–320 nm) revealed that the equilibrium at 5000 rev./min was reached after 2 and 18 h for 40 and 150 μl samples respectively. Absorbance profiles were recorded five times with a step of 0.001 cm for every speed and fitted simultaneously on single species or self-association models. The molar absorption at 280 nm was computed from the protein sequence as ϵ280=33200 M−1·cm−1 [26] and extrapolated from protein spectra at other wavelengths (ϵ233=91721, ϵ292=12025 and ϵ297=2886 M−1·cm−1). Variances between experimental points and best fits were used to select the models. The MycRne specific partial volume v-bar=0.713 ml/g was computed from the protein sequence.
In vitro transcription and labelling of 9S RNA
A DNA fragment including the putative Mycobacterium bovis BCG 9S RNA sequence was PCR-amplified from genomic DNA using primers 9Sfor (5′-TGCTAACCGGCCGAAAACTTA-3′) and 9Srev (5′-AACATACAAAAACACCACCGT-3′), and cloned into the pGEM®-T Easy vector (Promega) at the T cloning site. The resulting plasmid (pMt9S) was verified further by sequencing, linearized by PstI (MBI Fermentas), blunted with Klenow enzyme (MBI Fermentas), purified in a 1% agarose gel and used to transcribe 9S RNA with a MEGAscript® T7 kit (Ambion). 9S RNA was purified further from a 6% polyacrylamide/urea gel, dephosphorylated with bacterial alkaline phosphatase (MBI Fermentas) and 5′-labelled using T4 polynucleotide phosphorylase (MBI Fermentas) and an excess of [γ-32P]ATP (Amersham Biosciences). 5′-End-labelled 9S RNA was purified further from a 6% (w/v) polyacrylamide sequencing gel as described in [27] and subsequently used for cleavage assays.
Primer extension
Total RNA from Mycobacterium bovis BCG was extracted using the hot phenol method [28]. The 5′-32P-labelled 5S rRNA-specific oligonucleotide 5SPEM (5′-CAGTATCATCGGCGCTGGC-3′) and total RNA (150 ng) were co-precipitated with ethanol, the pellet was air-dried and suspended in 5 μl of annealing buffer (75 mM KCl and 67 mM Tris/HCl, pH 8.3), heated to 70 °C, quenched on ice, mixed with 5 μl of RT buffet (75 mM Tris/HCl, pH 8.3, 20 mM MgCl2, 4 mM DTT and 0.8 mM each dNTP) containing 4 units of AMV (avian myeloblastosis virus) reverse transcriptase (Promega) and incubated for 30 min at 42 °C. The reaction was stopped by adding 10 μl of sequencing dye. The sample was denatured at 95 °C for 3 min and analysed further on an 8% sequencing gel along with a concurrently run sequencing ladder. The latter was prepared using PstI-digested pMt9S, the same 5′-32P-labelled primer and a Reader™ DNA sequencing kit (MBI Fermentas).
Synthetic oligonucleotide substrates
Ribo-oligonucleotides 5′-ACAGUAUUUG-3′ (BR10), 5′-ACAGAAUUUG-3′ (9SA), 5′-GAAGGAUUUAA-3′ (OmpC), 5′-AAAAAAAAAAAAAAAAAAAAAAAAAAA-3′ (A27), 5′-UUUUUUUUUUUUUUUUUUUUUUUUUUU-3′ (U27), 5′-UUUUUUUUUUUUUGUUUUUUUUUUUUU-3′ (U27G), 5′-UUUUUUUUUUUUUAUUUUUUUUUUUUU-3′ (U27A) and 5′-UUUUUUUUUUUUUCUUUUUUUUUUUUU-3′ (U27C), as well as oligonucleotides 5′-pGGGACAGUAUUUG-3′ (pBR13-Fluor) and 5′-HOGGGACAGUAUUUG-3′ (HO-BR13-Fluor) with a fluorescein tag at their 3′ ends, were purchased from VBC Genomics, whereas 5′-UUUUUUUUUUUUUXUUUUUUUUUUUUU-3′ (U27ab) containing an abasic residue (X) was purchased from Dharmacon.
RNase E/G cleavage of oligonucleotide substrates and 9S RNA
RNase E/G cleavage reactions were performed as described previously [27] using Rne498 (or MycRne) and 5′-labelled substrates. Ladders (1 nt) were prepared either by partial alkaline hydrolysis of each 5′-end-labelled substrate in 1 mM EDTA and 50 mM sodium carbonate, pH 9.2, at 85 °C for 20 min, or by partial digestion of 5′-end-labelled oligonucleotides with S1 nuclease in buffer provided by the manufacturer (MBI Fermentas). RNase T1 digests of 9S RNA were prepared by incubation of 5′-end-labelled 9S RNA in 10 mM Tris/HCl, pH 8.0, 100 mM NaCl and 5 mM MgCl2 with RNase T1 (MBI Fermentas) at 37 °C.
RESULTS
Cloning and purification of M. tuberculosis RNase E/G
To learn more about organization and biochemical properties of RNase E/G from Gram-positive bacteria, the M. tuberculosis RNase E/G homologue was overproduced in E. coli and purified by affinity chromatography (see the Materials and methods section). Since the full-length M. tuberculosis RNase E/G is difficult to purify in large amounts because of its tendency to form insoluble aggregates upon overexpression (A. Miczak, unpublished work), we cloned its truncated form of 621 amino acids, referred to as MycRne. The amino acid sequence of MycRne includes the entire catalytic domain and the adjacent C-terminal region of this protein (Figure 1A). The corresponding genomic fragment was amplified by PCR, ligated into an expression vector and verified by sequencing. The recombinant MycRne carrying a His6-tag was overexpressed in E. coli strain BL21-CodonPlus(DE3)-RIL and purified under native conditions. As seen in Figure 1(B), the 71 kDa MycRne protein exhibited an aberrant mobility, migrating as a 110 kDa polypeptide. Slower running of RNase E/G homologues in SDS/polyacrylamide gels has been reported previously and appears to be caused by their unusual amino acid composition [29]. MS analysis did not reveal any contamination of the MycRne preparation by E. coli RNase E/G polypeptides or other host ribonucleases (results not shown).
Quaternary structure of M. tuberculosis RNase E/G
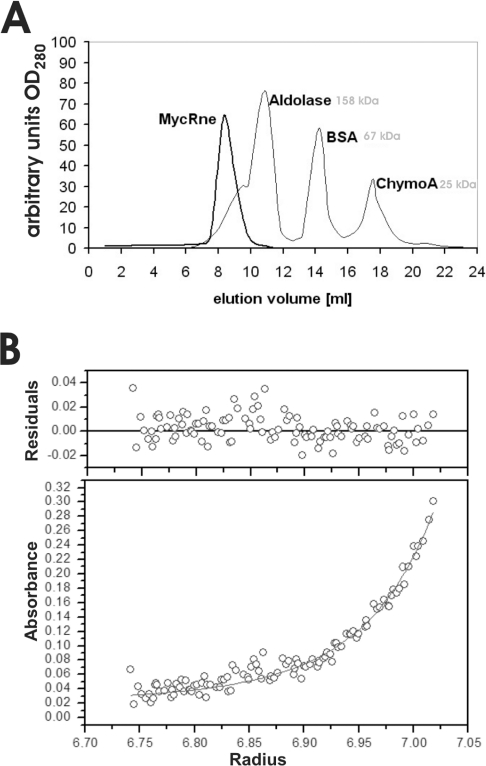
The putative catalytic domain of MycRne contains the highly conserved Zn-link motif known to be implicated in dimerization and tetramerization of E. coli RNase E/G [8,9]. To assess whether the ability of RNase E/G homologues to form dimeric and tetrameric forms is evolutionarily conserved, we analysed the oligomerization state of affinity-purified MycRne. According to the calibration profile obtained with aldolase, BSA and chymotrypsinogen A, MycRne was eluted as a complex with a hydrodynamic radius corresponding to a spherical protein with an apparent molecular mass of approx. 325 kDa (Figure 2A), suggesting that, similar to E. coli homologues, MycRne may exist in a tetrameric form.
Figure 2. Quaternary structure of RNase E/G.
(A) Gel filtration of MycRne. Affinity-purified MycRne (thicker line) was analysed by size-exclusion chromatography using a 25 ml Superose® 12 HR 10/30 column (Amersham Biosciences) connected to an FPLC system. The column was calibrated with aldolase (158 kDa), BSA (67 kDa) and chymotrypsinogen A (25 kDa) (thinner line). Elution profiles were recorded online; the absorbance at 280 nm is indicated on the y-axis, whereas elution volumes are indicated on the x-axis. (B) Sedimentation equilibrium analysis. A representative radial concentration distribution of RNase E/G at 16 μM is shown after reaching equilibrium (18 h) at 12000 rev./min at 20 °C. The lower panel plots the absorbance at 292 nm against the radial position (cm). The absorbance offset was set to 0.022 absorbance unit. The best fit of the data set (six speeds, 1502 points) was consistent at this sample concentration with a monomer–dimer equilibrium (solid lines) with a Kd of 1.8 μM. Residual values between experimental data and a self-associating model of ideal species (molecular mass, 71100 Da) are shown in the upper panel.
In order to confirm the oligomerization state of MycRne by an independent method, we performed analytical ultracentrifugation (Figure 2B). To analyse the self-association state of MycRne, sedimentation equilibrium experiments were performed at different concentrations (1.6, 8, 16, 42 and 84 μM) and several speeds. The absorbance scans (fitted simultaneously on single species or self-association models) indicated an apparent molecular mass of 71100 Da for the monomer. The records at 16 μM and 42 μM fitted best on a monomer–dimer equilibrium (1502 points/concentration; variance, 0.000163) with a Kd of ∼1.8 μM, while the profiles at 84 μM could best be modelled as a dimer–tetramer equilibrium (862 points; variance, 0.000277) with a Kd of ∼95 μM. According to these data, the molecular species of MycRne appear mainly as monomers below concentrations of approx. 1 μM, predominantly as dimers up to approx. 100 μM and as tetramers in more concentrated solutions.
Cleavage of oligonucleotide substrates
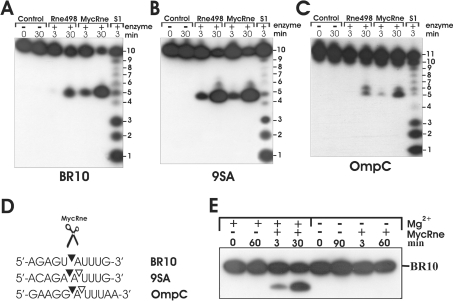
To test whether MycRne has an endoribonucleolytic activity and to compare its specificity with that of E. coli RNase E, we performed cleavage assays using affinity-purified MycRne and Rne498 (E. coli RNase E; residues 1–498), a C-terminally truncated form of E. coli RNase E representing its catalytic domain [12]. The assays were carried out with synthetic ribo-oligonucleotides BR10, 9SA and OmpC, all of which are well-characterized E. coli RNase E substrates [27,30,31]. As seen in Figure 3, MycRne could cut all three oligonucleotides yielding cleavage patterns that were largely indistinguishable from the analogous patterns generated by its E. coli counterpart. A minor difference was only observed in the cleavage pattern of OmpC. Although Rne498 cleaved this oligonucleotide at positions 5 and 6 with equal efficiencies, MycRne showed some preference for cleavage at position 5. We also found that, similar to its RNase E/G homologues from Aquifex aeolicus and E. coli, MycRne requires Mg2+ ions for its activity (Figure 3E).
Figure 3. Cleavage patterns of oligonucleotides BR10, 9SA and OmpC.
Each 5′-end-labelled oligonucleotide (A, B and C respectively) was incubated without enzyme (control), affinity-purified E. coli RNase E (Rne498) or M. tuberculosis RNase E/G (MycRne), and aliquots withdrawn at the times indicated above each lane were analysed on 20% polyacrylamide/urea gels. Lanes S1, 1 nt ladder generated by partial digestion of BR10, 9SA or OmpC with S1 nuclease respectively. The sequence of BR10, 9SA and OmpC is shown in (D). The internucleotide bonds that are sensitive to MycRne cleavage are indicated by triangles. Filled triangles point towards the bonds that have higher sensitivity to the endonucleolytic activity of MycRne. (E) Magnesium-dependence of MycRne cleavages. BR10 was incubated with MycRne in the absence (−) or presence (+) of Mg2+ ions, and aliquots withdrawn at times indicated above each lane were analysed on a 15% sequencing gel. Each assay was repeated at least twice, and one representative gel image is shown.
5′-end-dependence of M. tuberculosis RNase E/G cleavages
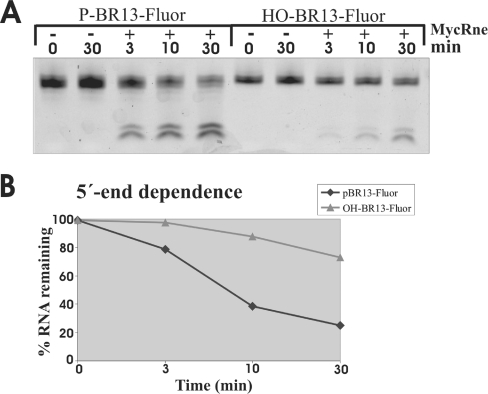
RNase E/G homologues from Gram-negative bacteria are known to preferentially cleave 5′-monophosphorylated substrates over non-phosphorylated [19,32,33] or 5′-triphosphorylated ones [34]. Employing 5′-monophosphorylated and non-phosphorylated BR13 (5′-GGGACAGUAUUUG-3′) tagged with a fluorescein group at their 3′ ends, we demonstrated that, similar to its counterparts, MycRng cleaves 5′-monophosphorylated substrates faster than the non-phosphorylated ones (Figures 4A and 4B). Thus our data suggest that the so-called 5′-end-dependence is apparently a common feature of RNase E/G homologues from both Gram-negative and Gram-positive bacteria.
Figure 4. 5′-end-dependence of MycRne cleavages.
(A) Relative efficiency of MycRne cleavage of fluorescently labelled 5′-phosphorylated and non-phosphorylated oligonucleotides (P-BR13-Fluor and HO-BR13-Fluor respectively). Each oligonucleotide (∼5 pmol) was incubated without (control) or with affinity-purified MycRne, and aliquots withdrawn at the times indicated above each lane were analysed on a 15% (w/v) sequencing gel. The signals corresponding to the fluorescently labelled oligonucleotides and their products of cleavage were visualized using a PhosphorImager (Typhoon 8600, Molecular Dynamics) and quantified further employing ImageQuant software. (B) Graphical representation of uncleaved RNA (%) plotted as a function of time (min) indicates that MycRne cleaves the 5′-phosphorylated substrate faster than the non-phosphorylated one. The graph represents the data from one single experiment, which was repeated twice with similar results.
Probing the substrate specificity of M. tuberculosis RNase E/G
In order to identify specific sequence determinants of MycRne cleavage sites, we used essentially the same oligonucleotide-based approach that was employed previously to characterize its E. coli counterpart [35]. By comparing the cleavage patterns of poly(A) and poly(U) oligonucleotides and their mutant variants carrying single-base substitutions of G, C, U or A respectively for adenosine (or uridine), this method allows us to determine the effect of each nucleotide at multiple positions to the point of cleavage [35].
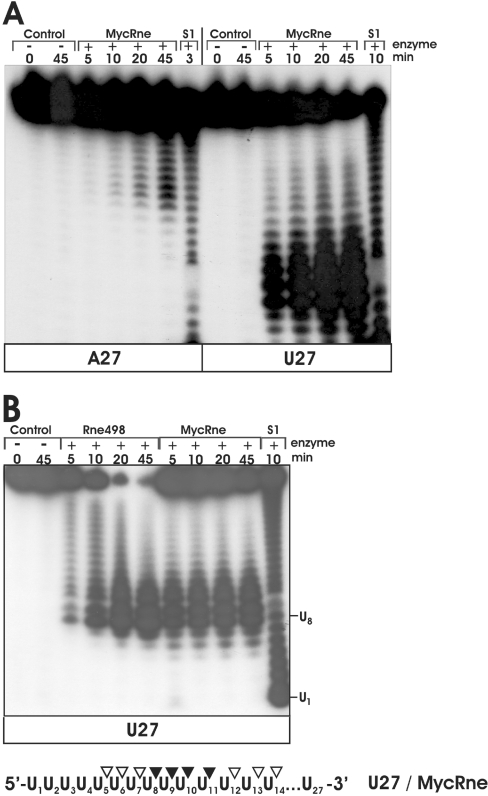
Unlike E. coli RNase E [36,37], MycRne cleaved A27 inefficiently. As seen in Figure 5(A), despite a 5-fold excess of MycRne (when compared with the cleavage conditions of U27), A27 cleavage products can be readily detected only on overexposed X-ray films (Figure 5A). In the following assays, we therefore decided to use only U27 (control) and its derivatives, U27A, U27G, U27C and U27ab oligonucleotides, that had a single-base substitution of A, G, C or abasic residue respectively for U at position 14.
Figure 5. Comparative cleavage of A27 and U27 by MycRne.
(A) 5′-End-labelled A27 (left) or U27 (right) were incubated without enzyme (control) or with 1.5–0.3 μg of native M. tuberculosis RNase E/G (MycRne), and aliquots withdrawn at the times indicated above each lane were analysed on 15% polyacrylamide/urea gels. The 1 nt ladders (lanes S1) were generated by partial digestion of A27 and U27 with S1 nuclease respectively. Each experiment was repeated at least twice, and one representative gel image is shown. (B) Cleavage patterns of U27 generated by of Rne498 and MycRne. 5′-End-labelled U27 was incubated without enzyme (control), with E. coli RNase E (Rne498) or with M. tuberculosis RNase E/G (MycRne), and aliquots withdrawn at the times indicated above each lane were analysed on a 15% polyacrylamide/urea gel. Lane S1, a 1 nt ladder prepared by partial digestion of U27 with S1 nuclease. The co-ordinates of U1 and U8 within U27 are indicated. Internucleotide bonds with a moderate (open triangles) or high (closed triangles) sensitivity to MycRne cleavage are indicated in a schematic view of U27 shown at the bottom. Each experiment was repeated at least twice, and one representative gel image is shown.
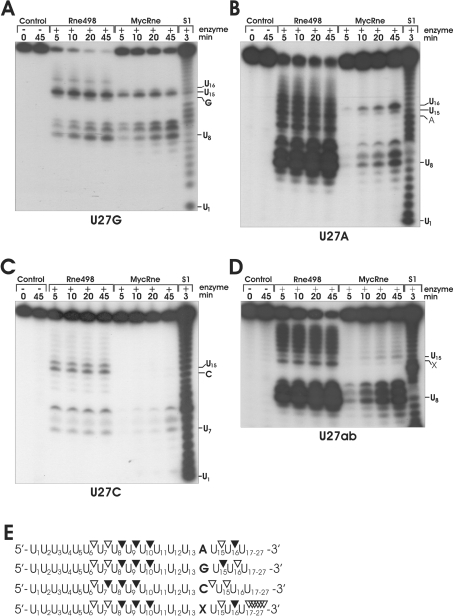
Similar to cleavage of U27 (Figure 5B), incubation of U27G with MycRne and Rne498 resulted in nearly identical patterns (Figure 6A). The major cleavages of U27G at positions U8, U9, U10 and U15 were observed for both enzymes, whereas Rne498 (but not MycRne) could additionally cleave this substrate at position U18. In the case of E. coli RNase E, it has been reported previously that a G nucleotide, which is 5′ and in close vicinity to the scissile bond, is an important determinant of cleavage [31,33,35]. Likewise, we found that M. tuberculosis RNase E/G cleavage is more efficient if there is a G nucleotide located two nucleotides upstream of the scissile bond (Figure 6A). These data are fully consistent with the cleavage patterns of BR10, 9SA and OmpC oligonucleotides, showing that MycRne cleavage frequently occurs two nucleotides downstream of a G nucleotide (Figure 3).
Figure 6. Cleavage patterns of U27A, U27G, U27C and U27ab generated by Rne498 and MycRne.
Each 5′-end-labelled substrate derived from U27 by replacement of the U at position 14 by a G (U27G) (A), A (U27A) (B), C (U27C) (C) or abasic (U27ab) (D) residue was incubated without enzyme (control), with E. coli RNase E (Rne498) or with M. tuberculosis RNase E/G (MycRne), and aliquots withdrawn at the times indicated above each lane were analysed on 15% polyacrylamide/urea gels. Indicated are the positions of several nucleotides including the substituted base [A, G, C or the abasic residue (X) respectively] as well as site(s) sensitive to MycRne cleavage. Lanes S1, 1 nt ladders generated by partial digestion of each oligonucleotide with S1. Similar patterns were obtained for each substrate in three independent experiments. (E) Schematic views of the substrates. The internucleotide bonds with a moderate and high susceptibility to MycRne cleavage are depicted by open and closed triangles respectively.
In contrast with nearly identical cleavage patterns that were generated by MycRne and Rne498 using U27G (Figure 6A), the cleavage patterns of U27A, U27C and U27ab showed relatively low resemblance (Figures 6B, 6C and 6D respectively). Although Rne498 can cut at multiple positions within U27A (Figure 6B), MycRne cleavage of the same substrate was not observed at many of these locations, especially downstream and upstream of U16. Similarly, the presence of a cytosine at position 14 in U27C (Figure 6C) strongly inhibits MycRne (and to a lesser degree Rne498) cleavage at nearly all locations, suggesting the overall inhibiting effect of this nucleotide. Finally, we found that substitution of an abasic residue for the U at position 14 in U27ab (Figure 6D) decreased the efficiency of MycRne cleavage at multiple locations. This finding strongly suggests that the presence of uridines at many locations close to the scissile bond promotes MycRne cleavage.
On the basis of the above experimental data, we determined and summarized the effects of each nucleotide within MycRne cleavage sites (Figure 6E). Interestingly, according to these data, the C at position +2 in 9SA should confer higher resistance of this oligonucleotide (in comparison with BR10 lacking a cytosine at the equivalent position) to MycRne. In contrast, we did not observe any significant differences in the efficiency of cleavage (Figure 3, and M.-E. Zeller and V. R. Kaberdin, unpublished work). This may imply that the expected inhibiting effect of the cytosine was efficiently counteracted by other nucleotides, for example by the A and G nucleotides at positions −3 and −2 respectively. In other words, strong enhancing effects of nucleotides at certain positions of different cleavage sites can potentially neutralize or override negative effects of other sequence determinants, resulting in approximately equal efficiency of cleavage.
As in the case of E. coli RNase E [35,37], the efficiency of MycRne cleavage of U27 and its mutant variants decreases with the number of the remaining nucleotides that are 5′ to the scissile bond (the ‘end-proximity effect’), thereby leading to accumulation of short oligomers that are further resistant to the nucleolytic activity of this enzyme.
9S RNA processing by M. tuberculosis RNase E/G
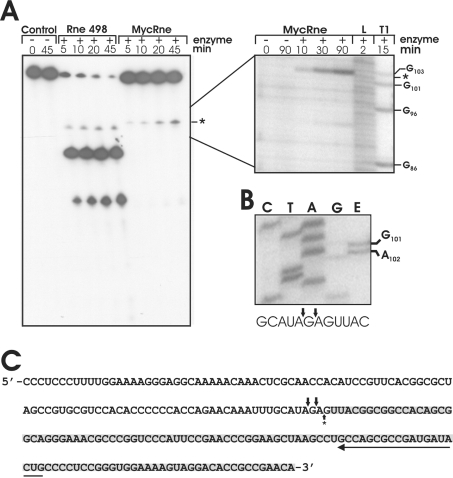
While playing an essential role in maturation of stable RNA in E. coli, RNase E is known to process the 9S RNA precursor of the 5S rRNA [38]. To find out whether MycRne might act likewise in the processing of rRNA in M. tuberculosis, we examined the cleavage pattern obtained after incubation of in vitro transcribed M. tuberculosis 9S RNA with Rne498 and MycRne. Although both enzymes could cleave this RNA, the cleavages occurred at different locations (Figure 7A). This finding is consistent with our data (Figure 6) suggesting some differences in the substrate specificities of E. coli and M. tuberculosis RNase E/G homologues. Further analysis (Figure 7B) revealed that MycRne cleaved 9S RNA at the position, which nearly coincides with the position of the 5′ end of mature 5S rRNA in vivo, which was mapped by primer extension of total RNA (Figures 7B and 7C).
Figure 7. 9S RNA processing in vitro and in vivo.
(A) 5′-End-labelled mycobacterial 9S RNA was incubated without enzyme (control), with E. coli RNase E (Rne498) or with M. tuberculosis RNase E/G (MycRne). Aliquots were withdrawn at the times indicated above each lane and analysed on 10% polyacrylamide/urea gels. The position of the M. tuberculosis RNase E/G cleavage site (*) was determined employing concomitantly run partial alkaline (L) and RNase T1 (T1) digests of the same transcript shown on the right. The experiment was repeated three times with similar results. (B) Primer-extension analysis of total mycobacterial RNA (lane E) was performed using a 5′-32P-labelled primer specific for 5S rRNA. The 5′-end of the in vivo processed 5S rRNA was determined by employing a concomitantly run sequencing ladder (lanes C, T, A and G) generated with the same primer (for details, see the Materials and methods section). The experiment was repeated twice with similar results. (C) The sequence of M. tuberculosis 5S rRNA (highlighted with grey shading) and its 5′-end-flanking region. A long horizontal arrow shows the direction and position of the primer used for primer extension. The position of in vitro (A) and in vivo (B) MycRne cleavage sites that were mapped close to the 5′ end of 5S rRNA are indicated by a single upward and two downward arrows respectively.
DISCUSSION
Mycobacteria are Gram-positive bacteria that are known to enter a state of metabolic dormancy when they encounter anoxic or nutritionally deprived environments [39]. This mechanism allows pathogenic species such as M. tuberculosis to survive within their host cells and involves numerous changes in gene expression to convey the required physiological adjustments. Adaptive processes in the RNA metabolism of latent mycobacteria differentially affect the steady-state levels of numerous transcripts [40], ribosome biosynthesis [41] as well as RNase E/G levels [42], suggesting an important regulatory role of this enzyme during disease development. To learn more about the function and biochemical properties of the RNase E/G homologue from M. tuberculosis (MycRne), we cloned, purified and characterized a polypeptide including the centrally located catalytic domain as well as the flanking C-terminal part of this protein (Figure 1).
We found that, similar to its homologues, E. coli RNase E (residues 1–530) [7–9] and RNase G [24] polypeptides, purified MycRne can exist in solution in dimeric or tetrameric forms. Previous work has shown that a pair of cysteine residues in the conserved CPXCXGXG motif, which co-ordinates a Zn2+ ion (so-called Zn-link motif), is critical for assembly of the E. coli RNase E homotetramers [7]. Given that the Zn-link motif exists in MycRne and other members of the RNase E/G family, our results suggest that the protein–protein interfaces, which are involved in intermolecular association of MycRne (the present study) and its E. coli counterparts [7–9,24], apparently resemble each other and that the association as dimer-of-dimers may be a common feature of RNase E/G homologues. Interestingly, although the tetrameric form of the catalytic domain of E. coli RNase E (amino acid residues 1–530) is more active in vitro than the monomeric one [9], a truncated E. coli RNase E polypeptide (amino acid residues 1–395), which retains an endoribonucleolytic activity (but has no potential to self-associate), can functionally substitute for the full-length E. coli RNase E in vivo [43]. Although this finding suggests that bacterial cells can survive with RNase E functioning as a monomer, a tetrameric quaternary structure of this enzyme may be still important to confer efficient cleavage of RNA in fast-growing cells.
By employing oligonucleotide substrates, we have also shown that MycRne has an endoribonucleolytic activity, which is dependent on the 5′-phosphorylation status of its substrates, thereby suggesting that MycRne is a 5′-end-dependent endoribonuclease. This property of MycRne (the present study) and other RNase E/G homologues [19,32,34,44,45] implies that these enzymes more efficiently cleave already damaged or partially degraded forms of cellular RNAs (i.e. RNA species that usually carry 5′-monophosphate groups) rather than primary transcripts, which are naturally triphosphorylated.
The enzyme can cleave short oligonucleotides with the specificity, which is similar to that observed previously for E. coli RNase E (Figure 3). However, despite their seemingly indistinguishable sequence specificities (Figure 3), further analysis using homo-oligomeric substrate U27 and its monosubstituted derivatives U27A, U27C, U27G and U27ab (Figure 6) revealed a number of important differences in the optimal location of G, C, U and A nucleotides within E. coli RNase E and MycRne cleavage sites. Both endoribonucleases cleave more efficiently in the presence of a G nucleotide upstream of and in close proximity to the point of cleavage. However, unlike E. coli RNase E sites that are usually rich in A/U nucleotides [35,46,47], MycRne apparently prefers to cleave sequences enriched mainly in U (but not A) nucleotides. Likewise, the C in U27C differentially affects cleavages by E. coli and M. tuberculosis RNase E/G homologues. When placed in certain locations close to the scissile bond, the cytosine not only inhibits but also enhances the efficiency of E. coli RNase E cleavages ([35], Figure 6C), whereas, in the case of M. tuberculosis RNase E/G, it only inhibits cleavage at nearly every position. The present study and others [19,32] demonstrate that the substrate specificity of RNase E/G endoribonucleases can significantly vary and cannot be easily inferred from previous data that were obtained for a particular RNase E/G homologue.
Our recent work demonstrates that the knowledge of specific sequence determinants within RNase E/G cleavage sites can be instructive to suggest mutations that increase/decrease the efficiency of cleavage within the site of interest [35] and to infer differences in the substrate specificities of various RNase E/G homologues (the present study). However, this information may itself not be sufficient to predict potential RNase E cleavage sites within a regular transcript. Additional factors such as RNA structure or interactions with RNA-binding proteins should be also taken into consideration.
Given that RNase E and RNase G are involved in maturation of ribosomal and transfer RNAs in E. coli [48,49], we anticipated that this function of RNase E/G-like proteins might be also conserved in mycobacteria. Indeed, we found that MycRne was able to cleave mycobacterial 9S RNA (Figure 7A), a putative precursor of its cognate 5S rRNA, and the cleavage occurred in close vicinity to the mature 5′-end of 5S rRNA (Figures 7B and 7C), thereby suggesting a role for this enzyme in rRNA processing. Although the internucleotide bonds at the 5′ end of 5S rRNA that are generated by MycRne in vitro (Figure 7A) and during rRNA processing in vivo (Figure 7B) were not exactly the same, this minor (one or two nucleotide) difference could be well explained by the action of ancilliary factors or associated ribosomal proteins that can affect MycRne cleavage in vivo and therefore slightly influence the selection of the scissile bond(s). Further efforts will be necessary to get a better understanding of diverse roles of MycRne in RNA processing and to reveal and characterize the MycRne-mediated post-transcriptional mechanisms that are involved in adaptive stress responses and the associated alterations in RNA metabolism in mycobacteria.
Acknowledgments
We thank Dr Denise Barlow for critically reading the manuscript. This work was supported by grant no. F1707 from the Austrian Science Foundation (V.R.K.), by CNRS (Centre National de la Recherche Scientifique) (T.R. and T.B.), and by OTKA (Országos Tudományos Kutatási Alapprogramok) 034820 from the Hungarian Scientific Research Fund (A.M.). A.Cs. was a recipient of the FEMS (Federation of European Microbiological Societies) and Hungarian State Eötvös fellowships.
References
- 1.Cohen S. N., McDowall K. J. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 1997;23:1099–1106. doi: 10.1111/j.1365-2958.1997.tb02593.x. [DOI] [PubMed] [Google Scholar]
- 2.Coburn G. A., Mackie G. A. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acids Res. Mol. Biol. 1999;62:55–108. doi: 10.1016/s0079-6603(08)60505-x. [DOI] [PubMed] [Google Scholar]
- 3.Régnier P., Arraiano C. M. Degradation of mRNA in bacteria: emergence of ubiquitous features. BioEssays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Kushner S. R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deana A., Belasco J. G. The function of RNase G in Escherichia coli is constrained by its amino and carboxyl termini. Mol. Microbiol. 2004;51:1205–1217. doi: 10.1046/j.1365-2958.2003.03905.x. [DOI] [PubMed] [Google Scholar]
- 6.Ow M. C., Perwez T., Kushner S. R. RNase G of Escherichia coli exhibits only limited functional overlap with its essential homologue, RNase E. Mol. Microbiol. 2003;49:607–622. doi: 10.1046/j.1365-2958.2003.03587.x. [DOI] [PubMed] [Google Scholar]
- 7.Callaghan A. J., Marcaida M. J., Stead J. A., McDowall K. J., Scott W. G., Luisi B. F. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 8.Callaghan A. J., Grossmann J. G., Redko Y. U., Ilag L. L., Moncrieffe M. C., Symmons M. F., Robinson C. V., McDowall K. J., Luisi B. F. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry. 2003;42:13848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]
- 9.Callaghan A. J., Redko Y., Murphy L. M., Grossmann J. G., Yates D., Garman E., Ilag L. L., Robinson C. V., Symmons M. F., McDowall K. J., Luisi B. F. “Zn-link”: a metal-sharing interface that organizes the quaternary structure and catalytic site of the endoribonuclease, RNase E. Biochemistry. 2005;44:4667–4675. doi: 10.1021/bi0478244. [DOI] [PubMed] [Google Scholar]
- 10.Kaberdin V. R., Miczak A., Jakobsen J. S., Lin-Chao S., McDowall K. J., von Gabain A. The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11637–11642. doi: 10.1073/pnas.95.20.11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leroy A., Vanzo N. F., Sousa S., Dreyfus M., Carpousis A. J. Function in Escherichia coli of the non-catalytic part of RNase E: role in the degradation of ribosome-free mRNA. Mol. Microbiol. 2002;45:1231–1243. doi: 10.1046/j.1365-2958.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- 12.McDowall K. J., Cohen S. N. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA-binding site. J. Mol. Biol. 1996;255:349–355. doi: 10.1006/jmbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 13.Vanzo N. F., Li Y. S., Py B., Blum E., Higgins C. F., Raynal L. C., Krisch H. M., Carpousis A. J. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcaida M. J., Depristo M. A., Chandran V., Carpousis A. J., Luisi B. F. The RNA degradosome: life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Wachi M., Umitsuki G., Shimizu M., Takada A., Nagai K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5′ end of 16S rRNA. Biochem. Biophys. Res. Commun. 1999;259:483–488. doi: 10.1006/bbrc.1999.0806. [DOI] [PubMed] [Google Scholar]
- 16.Li Z., Pandit S., Deutscher M. P. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada Y., Wachi M., Hirata A., Suzuki K., Nagai K., Matsuhashi M. Cytoplasmic axial filaments in Escherichia coli cells: possible function in the mechanism of chromosome segregation and cell division. J. Bacteriol. 1994;176:917–922. doi: 10.1128/jb.176.3.917-922.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y., Shibata T., Matsuhashi M. Possible function of the cytoplasmic axial filaments in chromosomal segregation and cellular division of Escherichia coli. Sci. Prog. 1993;77:253–264. [PubMed] [Google Scholar]
- 19.Tock M. R., Walsh A. P., Carroll G., McDowall K. J. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′- end-dependent ribonuclease that has context-dependent broad sequence specificity. J. Biol. Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 20.McDowall K. J., Hernandez R. G., Lin-Chao S., Cohen S. N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J. Bacteriol. 1993;175:4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpousis A. J. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- 22.Condon C., Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K., Cohen S. N. A Streptomyces coelicolor functional orthologue of Escherichia coli RNase E shows shuffling of catalytic and PNPase-binding domains. Mol. Microbiol. 2003;48:349–360. doi: 10.1046/j.1365-2958.2003.03435.x. [DOI] [PubMed] [Google Scholar]
- 24.Briant D. J., Hankins J. S., Cook M. A., Mackie G. A. The quaternary structure of RNase G from Escherichia coli. Mol. Microbiol. 2003;50:1381–1390. doi: 10.1046/j.1365-2958.2003.03775.x. [DOI] [PubMed] [Google Scholar]
- 25.Honer Zu Bentrup K., Miczak A., Swenson D. L., Russell D. G. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 1999;181:7161–7167. doi: 10.1128/jb.181.23.7161-7167.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDowall K. J., Kaberdin V. R., Wu S. W., Cohen S. N., Lin-Chao S. Site-specific RNase E cleavage of oligonucleotides and inhibition by stem–loops. Nature. 1995;374:287–290. doi: 10.1038/374287a0. [DOI] [PubMed] [Google Scholar]
- 28.Lin-Chao S., Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol. Gen. Genet. 1986;203:143–149. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- 29.Casaregola S., Jacq A., Laoudj D., McGurk G., Margarson S., Tempete M., Norris V., Holland I. B. Cloning and analysis of the entire Escherichia coli ams gene. ams is identical to hmp1 and encodes a 114 kDa protein that migrates as a 180 kDa protein. J. Mol. Biol. 1992;228:30–40. doi: 10.1016/0022-2836(92)90489-7. [DOI] [PubMed] [Google Scholar]
- 30.Afonyushkin T., Moll I., Bläsi U., Kaberdin V. R. Temperature-dependent stability and translation of Escherichia coli ompA mRNA. Biochem. Biophys. Res. Commun. 2003;311:604–609. doi: 10.1016/j.bbrc.2003.10.040. [DOI] [PubMed] [Google Scholar]
- 31.Kaberdin V. R., Walsh A. P., Jakobsen T., McDowall K. J., von Gabain A. Enhanced cleavage of RNA mediated by an interaction between substrates and the arginine-rich domain of E. coli. ribonuclease E. J. Mol. Biol. 2000;301:257–264. doi: 10.1006/jmbi.2000.3962. [DOI] [PubMed] [Google Scholar]
- 32.Kaberdin V. R., Bizebard T. Characterization of Aquifex aeolicus RNase E/G. Biochem. Biophys. Res. Commun. 2005;327:382–392. doi: 10.1016/j.bbrc.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Redko Y., Tock M. R., Adams C. J., Kaberdin V. R., Grasby J. A., McDowall K. J. Determination of the catalytic parameters of the N-terminal half of. E. coli ribonuclease E and the identification of critical functional groups in RNA substrates. J. Biol. Chem. 2003;278:44001–44008. doi: 10.1074/jbc.M306760200. [DOI] [PubMed] [Google Scholar]
- 34.Mackie G. A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. [DOI] [PubMed] [Google Scholar]
- 35.Kaberdin V. R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 2003;31:4710–4716. doi: 10.1093/nar/gkg690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang H. J., Liao J., Cohen S. N. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature. 1998;391:99–102. doi: 10.1038/34219. [DOI] [PubMed] [Google Scholar]
- 37.Walsh A. P., Tock M. R., Mallen M. H., Kaberdin V. R., von Gabain A., McDowall K. J. Cleavage of poly(A) tails on the 3′-end of RNA by ribonuclease E of Escherichia coli. Nucleic Acids Res. 2001;29:1864–1871. doi: 10.1093/nar/29.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghora B. K., Apirion D. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 39.Cardona P. J., Ruiz-Manzano J. On the nature of Mycobacterium tuberculosis-latent bacilli. Eur. Respir. J. 2004;24:1044–1051. doi: 10.1183/09031936.04.00072604. [DOI] [PubMed] [Google Scholar]
- 40.Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beste D. J., Peters J., Hooper T., Avignone-Rossa C., Bushell M. E., McFadden J. Compiling a molecular inventory for Mycobacterium bovis BCG at two growth rates: evidence for growth rate-mediated regulation of ribosome biosynthesis and lipid metabolism. J. Bacteriol. 2005;187:1677–1684. doi: 10.1128/JB.187.5.1677-1684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Archuleta R. J., Hoppes P. Y., Primm T. P. Mycobacterium avium enters a state of metabolic dormancy in response to starvation. Tuberculosis. 2005;85:147–158. doi: 10.1016/j.tube.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Caruthers J. M., Feng Y., McKay D. B., Cohen S. N. Retention of core catalytic functions by a conserved minimal RNase E peptide that lacks the domain required for tetramer formation. J. Biol. Chem. 2006;281:27046–27051. doi: 10.1074/jbc.M602467200. [DOI] [PubMed] [Google Scholar]
- 44.Jiang X., Diwa A., Belasco J. G. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J. Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin-Chao S., Cohen S. N. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell. 1991;65:1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- 46.McDowall K. J., Lin-Chao S., Cohen S. N. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 47.Lin-Chao S., Wong T. T., McDowall K. J., Cohen S. N. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNAI encoded by the pBR322 plasmid. J. Biol. Chem. 1994;269:10797–10803. [PubMed] [Google Scholar]
- 48.Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J. Biol. Chem. 1979;254:11154–11159. [PubMed] [Google Scholar]
- 49.Li Z., Deutscher M. P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]