Abstract
AA (arachidonic acid) hydroxylation to 20-HETE (20-hydroxyeicosatetraenoic acid) influences renal vascular and tubular function. To identify the CYP (cytochrome P450) isoforms catalysing this reaction in the mouse kidney, we analysed the substrate specificity of Cyp4a10, 4a12a, 4a12b and 4a14 and determined sex- and strain-specific expressions. All recombinant enzymes showed high lauric acid hydroxylase activities. Cyp4a12a and Cyp4a12b efficiently hydroxylated AA to 20-HETE with Vmax values of approx. 10 nmol·nmol−1·min−1 and Km values of 20–40 μM. 20-Carboxyeicosatetraenoic acid occurred as a secondary metabolite. AA hydroxylase activities were approx. 25–75-fold lower with Cyp4a10 and not detectable with Cyp4a14. Cyp4a12a and Cyp4a12b also efficiently converted EPA (eicosapentaenoic acid) into 19/20-OH- and 17,18-epoxy-EPA. In male mice, renal microsomal AA hydroxylase activities ranged between approx. 100 (NMRI), 45–55 (FVB/N, 129 Sv/J and Balb/c) and 25 pmol·min−1·mg−1 (C57BL/6). The activities correlated with differences in Cyp4a12a protein and mRNA levels. Treatment with 5α-dihydrotestosterone induced both 20-HETE production and Cyp4a12a expression more than 4-fold in male C57BL/6 mice. All female mice showed low AA hydroxylase activities (15–25 pmol·min−1·mg−1) and very low Cyp4a12a mRNA and protein levels, but high Cyp4a10 and Cyp4a14 expression. Renal Cyp4a12b mRNA expression was almost undetectable in both sexes of all strains. Thus Cyp4a12a is the predominant 20-HETE synthase in the mouse kidney. Cyp4a12a expression determines the sex- and strain-specific differences in 20-HETE generation and may explain sex and strain differences in the susceptibility to hypertension and target organ damage.
Keywords: arachidonic acid, cytochrome P450, eicosapentaenoic acid, gender, hydroxyeicosatetraenoic acid (HETE), mouse kidney
Abbreviations: AA, arachidonic acid; Ang II, angiotensin II; BK channel, calcium-activated potassium channel; 20-COOH-AA, eicosatetraen-1,20-dioic acid; CYP, cytochrome P450; CPR, NADPH-CYP reductase; cytb5, cytochrome b5; DDDA, dodecan-1,12-dioic acid; DHT, dihydrotestosterone; DOCA, deoxycorticosterone acetate; DTT, dithiothreitol; EET, epoxyeicosatrienoic acid; EETeTr, epoxyeicosatetraenoic acid; EPA, eicosapentaenoic acid; HEPE, hydroxyeicosapentaenoic acid; HETE, hydroxyeicosatetraenoic acid; LA, lauric acid; NP-HPLC, normal-phase HPLC; OH-LA, hydroxy LA; PUFA, polyunsaturated fatty acid; RP-HPLC, reverse-phase HPLC; RT, reverse transcriptase
INTRODUCTION
CYP (cytochrome P450) enzymes of the CYP4A subfamily are found in the liver, kidney, intestine, lung, heart and brain of various mammals including rat, rabbit, mouse and human [1]. They conduct ω-hydroxylation of fatty acids and related compounds. This reaction facilitates the degradation of long-chain fatty acids by initiating their conversion into dicarboxylic acids, which are efficiently oxidized by peroxisomal β-oxidation [2]. With AA (arachidonic acid; 20:4 ω-6), CYP4A-catalysed ω-hydroxylation yields 20-HETE (20-hydroxyeicosatetraenoic acid). 20-HETE plays an important role in the regulation of renal vascular and tubular function. Altered renal CYP4A expression is associated with hypertension [3–5]. Urinary 20-HETE excretion is increased in patients with cirrhosis [4,6] and endothelial dysfunction [7]. Moreover, a polymorphic CYP4A11 variant is associated with human hypertension [8]. The mechanisms linking 20-HETE to blood pressure regulation are only partially understood. Both increased and decreased 20-HETE levels can be associated with high blood pressure, probably reflecting the dual role of 20-HETE in inducing vasoconstriction (pro-hypertensive) and in promoting natriuresis (anti-hypertensive) [5].
The CYP4A subfamily members expressed in mouse kidney include Cyp4a10, Cyp4a12 and Cyp4a14 [9–13]. Two Cyp4a12 genes exist, Cyp4a12a and Cyp4a12b, that result from a tandem 100 kb duplication within the Cyp4abx cluster on chromosome 4 [14]. We showed earlier that renal 20-HETE production is decreased in DOCA (deoxycorticosterone acetate)-salt-hypertensive mice [12]. Bezafibrate restored 20-HETE production and improved renal haemodynamics. Similarly, fenofibrate induced 20-HETE and prevented Ang II (angiotensin II)-induced hypertension [15]. In both models, the 20-HETE-producing CYP isoform(s) down-regulated in hypertension and induced by fibrates was not unequivocally identified. Targeted Cyp4a14 gene disruption resulted in increased AA ω-hydroxylase activities and caused hypertension in male mice [11]. The mechanism seems to involve increased plasma androgen levels in the Cyp4a14 gene-disrupted mice followed by androgen-induced up-regulation of Cyp4a12 [11]. Cyp4a10 gene disruption results in salt-sensitive hypertension by affecting the regulation of the kidney epithelial sodium channel [16].
We cloned and expressed the Cyp4a10, Cyp4a12a, Cyp4a12b and Cyp4a14 cDNAs, determined the substrate specificity of the recombinant enzymes and analysed their strain- and gender-specific relationship to renal 20-HETE production. We tested whether or not the two Cyp4a12 gene copies differ in terms of renal expression and enzymatic properties. For substrates, we relied on the CYP4A test substrate LA (lauric acid) to prove the successful reconstitution of enzymatic activities, AA to analyse the 20-HETE production capacity of the isoforms and the fish oil ω-3 PUFA (polyunsaturated fatty acid), EPA (eicosapentaenoic acid; 20:5 ω-3), which can replace AA in CYP-dependent eicosanoid production. Secondary hypertension and susceptibility to target organ damage in mice are largely sex- and strain-dependent. For example, 129/Sv mice are more susceptible to DOCA-salt hypertension than C57BL/6 mice [17]. Females are more resistant to Ang II-induced hypertension than males [18]. Since 20-HETE is involved in the pathophysiology of these hypertension forms [12,15], we investigated renal CYP expression and AA metabolism in male and female mice of different mouse strains.
MATERIALS AND METHODS
Animal strains and treatment
In the present study, 12–15-week-old male and female mice of the following strains were used (n=6–8 per gender and strain): NMRI, FVB/N, C57BL/6 (all three from Charles River Laboratories), 129 Sv/J and Balb/c (from DIMED Schönewalde). The mice were allowed free access to standard chow (0.25% sodium; SNIFF Spezialitäten) and drinking water ad libitum. In an additional set of experiments, male NMRI and C57BL/6 mice (n=6 per group) were subcutaneously implanted for 10 days with release pellets containing 5α-DHT (5α-dihydrotestosterone; 21-day pellet, 5 mg of DHT/day; Innovative Research of America).
Preparation of RNA and TaqMan analysis
Total RNA was isolated from liver, kidney, and renal arterioles using Qiashredder and RNeasy spin columns including chromosomal DNase digestion (Qiagen). For isolating preglomerular microvessels, the kidneys were cut in half along the cortico-papillary axis. The arterioles were prepared with the use of micro-scissors and forceps under an operating microscope and the surrounding tissue was carefully removed. TaqMan analysis of Cyp4a10, Cyp4a12a, Cyp4a12b and Cyp4a14 mRNA expression was conducted using isoform-specific primers and probes as described in the Supplementary online data at http://www.BiochemJ.org/bj/403/bj4030109add.htm.
Preparation of renal microsomes
Renal microsomes were prepared using one and a half kidneys per animal. The renal capsule and the fatty tissues were removed. Kidneys were minced and homogenized in 5 vol. of ice-cold 50 mM Tris/HCl buffer (pH 7.4) containing 0.25 M sucrose, 150 mM potassium chloride, 2 mM EDTA, 2 mM DTT (dithiothreitol), 1 μM FAD and FMN, and 0.25 mM PMSF in a motor-driven Teflon/glass Potter–Elvehjem homogenizer. After differential centrifugation (10 min, 1000 g; 20 min, 10000 g; 90 min, 100000 g), the microsomes were suspended and homogenized in 50 mM Tris/HCl buffer (pH 7.7) containing 20% (v/v) glycerol, 5 mM EDTA and 1 mM DTT. Aliquots were snap-frozen in liquid nitrogen and stored at −80 °C. The protein content was determined by the standard Lowry method [18a].
Cloning of the Cyp4a isoforms
The cDNAs for Cyp4a10, Cyp4a12a and Cyp4a14 were generated by RT (reverse transcriptase)–PCR (Ready-To-Go RT–PCR kit; Amersham Biosciences) using 300 ng of total renal RNA isolated from male NMRI mice as the template. The Cyp4a12b cDNA was amplified and selected starting from total hepatic mRNA isolated from male C57BL/6 mice. Reverse transcription was primed with oligo(dT) and PCR amplifications were done with the following primer pairs (5′–3′): TCGGAATTCGCAATGAGTGTCTCTGCTCTAA/CCGCTCGAGCGGTCAGTGGTGGTGGTGGTGGTGGAGCTTCTTGAGATGTAG (forward/reverse primer for Cyp4a10 with 5′-added EcoRI and XhoI sites respectively), ACGCGTAAGCTGTTGTATCATGAGTGC/GCTAGCCATTTGAGCTGTCTTGTTCTG (forward/reverse primer for Cyp4a12a and Cyp4a12b with 5′-added MluI and NheI sites respectively), and CGACTCGATCCAGAAACTAC/ACAGGACACATTGCAGAGAG (forward/reverse primer for Cyp4a14). The PCR products (∼1.6 kb) were cloned into the plasmid pCR2.1 (TOPOTA-Cloning kit; Invitrogen) and at least three different clones per isoform were sequenced (Invitek GmbH). For cloning of Cyp4a12b, the RT–PCR product obtained with the Cyp4a12a/Cyp4a12b-specific primers was first digested with AvrII, which selectively cleaves the Cyp4a12a cDNA and thus excludes it from cloning.
Expression of CYP enzymes
Co-expression of the individual CYP isoforms with the human CPR (NADPH-CYP reductase) was performed as described previously [19] using a baculovirus/Sf9 (Spodoptera frugiperda) insect cell system (for the details see Supplementary online data). The CYP concentrations were estimated by means of CO-difference spectra using a molar absorption coefficient of 91 mM−1·cm−1 [20]. CPR activities were assayed in 50 mM Tris/HCl buffer (pH 7.5) containing 0.1 mM EDTA, 0.05 mM cytochrome c, 0.1 mM NADPH and 2.2 mM KCN at 25 °C using a molar absorption coefficient of 21 mM−1·cm−1 at 550 nm. The microsomes used for kinetic analysis had molar CYP/CPR ratios in the range 1:0.9–1:1.2. For calculation, we assumed that 4.5 units (μmol of cytochrome c/min) correspond to 1 nmol of CPR based on the specific activity of the purified 79 kDa enzyme (60 units/mg).
Immunoblotting
Microsomal protein was separated on SDS/10% PAGE and transferred to Hybond ECL® nitrocellulose membranes (Amersham Biosciences). An antibody raised in goat against purified rat CYP4A1 (Daiichi Pure Chemicals) and a peptide-specific antibody raised in rabbits against mouse Cyp4a12a were used as primary antibodies. The antigenic peptide (NH3-SRRIQLQDEEELEKLKKKRR-CO2H) corresponded to amino acids 266–286 of the Cyp4a12a protein (peptide synthesis by BIOSYNTHAN GmbH and immunization by BioGenes GmbH). Anti-goat IgG and anti-rabbit IgG peroxidase conjugate (Sigma Chemical Co.) were used as secondary antibodies. Blots were developed with the chemiluminescence substrate from Boehringer–Mannheim and evaluated with the Image Reader LAS-1000 (Fuji Photo Film Co.). Ponceau staining was used for verifying equal protein loading.
Fatty acid metabolism
[1-14C]LA (2.11 GBq/mmol; Amersham Biosciences), [1−14C]AA (2.07 GBq/mmol; Amersham Biosciences) and [1−14C]EPA (2.0 GBq/mmol; PerkinElmer) were used as substrates. Standard reactions with the recombinant Cyp4a isoforms were performed in 100 μl of 100 mM potassium phosphate buffer (pH 7.2) containing 10 pmol of Cyp and the substrate at a concentration of 10 μM (AA and EPA) or 30 μM (LA). Reactions were started with NADPH (1 mM final concentration) after pre-incubating the microsomes with the substrates for 10 min at 37 °C. Reactions were terminated by adding citric acid (20 mM final concentration). Reaction products were extracted into ethyl acetate, evaporated under nitrogen, and resuspended in ethanol. Incubations with cytb5 (cytochrome b5; Calbiochem) were done in a molar Cyp/cytb5 ratio of 1:1. Microsomal enzymes and cytb5 were pre-incubated in a total volume of 15 μl for 10 min on ice. For kinetic analysis, standard reactions were performed with seven different substrate concentrations between 1.25 and 50 μM and the experiments were done in triplicate. Reactions were terminated after 1 min (LA) or 3 min (AA and EPA). For analysis of regio- and stereo-selectivities, standard reactions were scaled-up (4–10-fold; incubation times up to 10 min). With mouse renal microsomes, the standard reaction conditions described above were modified. The 100 mM potassium phosphate buffer (pH 7.2) was supplemented with 1 mM EDTA and 50 nM FAD and FMN and reactions were performed using 800 μg/ml renal microsomal protein with 40 μM AA for 30 min or with 200 μM LA for 3 min.
Soluble epoxide hydrolase assay
Soluble epoxide hydrolase activities were determined using the 10000 g supernatants obtained during the preparation of renal microsomes as described above. Reactions were performed at 37 °C for 10 min in 100 μl of 100 mM potassium phosphate buffer (pH 7.2) containing 50 μM [1−14C]14,15-EET (14,15-epoxyeicosatrienoic acid). [1−14C]14,15-EET was prepared by chemical oxidation of radiolabelled AA as described in [21]. The reactions were started by adding the 10000 g supernatant (5 μg of protein), and extracted as described above.
Analysis of metabolites
Metabolites were analysed using the HPLC system LC-10Avp from Shimadzu equipped with a radioactivity monitor (LB509, Berthold). Total metabolites were resolved by RP-HPLC (reverse-phase HPLC) on a Nucleosil 100-5C18 HD column (250 mm×4 mm; Macherey-Nagel). LA metabolites were esterified with diazomethane and resolved in RP-HPLC with a linear gradient of acetonitrile/water/acetic acid (29.5:70.5:0.1, by vol.) to acetonitrile/water/acetic acid (59.5:40.5:0.1, by vol.) over 30 min followed by 15 min acetonitrile/acetic acid (100:0.1, v/v) at a flow rate of 1 ml/min. AA and EPA metabolites were resolved with a linear gradient of acetonitrile/water/acetic acid (50:50:0.1, by vol.) to acetonitrile/acetic acid (100:0.1, v/v) over 40 min at a flow rate of 1 ml/min. 19- and 20-HETE as well as 19-HEPE (19-hydroxyeicosapentaenoic acid) and 20-HEPE were resolved by NP-HPLC (normal-phase HPLC) on a Nucleosil 100-5 column (250 mm×4 mm; Macherey-Nagel) utilizing a linear gradient from hexane/propan-2-ol/acetic acid (99:1:0.1, by vol.) to hexane/2-propanol/acetic acid (98.3:1.7:0.1, by vol.) over 40 min at a flow rate of 1 ml/min as described in [22]. 16-, 17-, and 18-HETE were resolved with hexane/2-propanol/acetic acid (100:0.4:0.1, by vol.) at a flow rate of 1.5 ml/min as described in [23]. 17,18-EETeTr (17,18-epoxyeicosatetraenoic acid) was treated with diazomethane and the methyl ester was then resolved into the (R,S) and (S,R) enantiomers on a Chiracel OB column (250 mm×4.6 mm; Daicell) using a linear gradient from hexane/2-propanol (99.7:0.3, v/v) to hexane/2-propanol (98:2, v/v) over 40 min at a flow rate of 1 ml/min [24]. Preparation of authentic LA [25], AA [26,27] and EPA [24,26,28] metabolites was performed as described in the Supplementary online data.
Statistical analysis
All data derived from the animal experiments are presented as means±S.E.M. and were analysed by ANOVA followed by the Tukey–Kramer multiple comparison test (InStat software; GraphPad Software). P<0.05 was considered statistically significant. Apparent Km and Vmax values of the recombinant Cyp4a enzymes were calculated using the Enzyme Kinetics Module of SigmaPlot 7 (SPSS).
RESULTS
Cloning of four different Cyp4a cDNAs
Cyp4a10
The Cyp4a10 cDNA cloned from NMRI showed 100% identity with the corresponding mRNA (GenBank® accession number AK002528) and genomic sequences (GenBank® accession number NT_039264) from C57BL/6 mice. However, it differed in two positions from the Cyp4a10 mRNA of FVB/N mice (GenBank® accession number NM_010011), which has been used as the provisional NCBI reference sequence for Cyp4a10: 190A>C and 207A>C (counted from the start ATG), resulting in Q64K and Q69H substitutions (NMRI and C57BL/6 versus FVB/N).
Cyp4a12a
The Cyp4a12-specific primer pair yielded cDNA clones from renal RNA of both NMRI and C57BL/6, whose sequences completely agreed with that of the MGC25972 gene (C57BL/6 contig NT_039264). The encoded Cyp isoform was termed ‘Cyp4a10-like’ or ‘similar to P450 4A8’ on contig NT_039264 but will be designated in the following as Cyp4a12a as described in [14]. cDNA sequences were so far only available for FVB/N mice (BC014721, NM_177406) and differ from that in NMRI and C57BL/6 by one non-synonymous (391A>C; M131L) and one silent nucleotide substitution (1233C>G).
Cyp4a12b
The gene originally termed ‘Cyp4a12’ in the C57BL/6 genome (contig NT_039264) is located approx. 112 kbp downstream of Cyp4a12a. It will be designated here as Cyp4a12b as described in [14]. The coding sequences of Cyp4a12a and Cyp4a12b differ at only 23 nucleotide positions, resulting in 11 amino acid substitutions. The primer pair used in the present study allowed the amplification of both the Cyp4a12a and Cyp4a12b sequences. To enrich the Cyp4a12b cDNA, the RT–PCR product was digested with AvrII, which selectively cleaves the Cyp4a12a cDNA. In this way, clones were generated from C57BL/6 liver mRNA, which showed 100% sequence identity with the predicted coding sequence of the Cyp4a12b gene (‘Cyp4a12’ on contig NT_039264). However, compared with the provisional NCBI reference sequence for the ‘Cyp4a12’ mRNA (NM_172306, strain not specified), there is a non-synonymous 494G>A substitution (V165H in contig NT_039264 and own cDNA sequence).
Cyp4a14
The Cyp4a14 cDNA cloned from NMRI mice showed with one exception (13T>G resulting in L5V) 100% identity with the coding region of the mRNA reference sequence NM_007822 (strain C57BL/6×CBA) and to that predicted from the C57BL/6 genome (contig NT_039264). This polymorphism has been also observed in other mouse strains (rs3022990).
Metabolism of LA by recombinant mouse Cyp4a isoforms
Co-expression of the individual Cyp4a isoforms with CPR in Sf9 insect cells yielded highly active microsomal enzyme systems that hydroxylated LA with Vmax values between 40 and 76 nmol·nmol−1·min−1 and apparent Km values between 2 and 16 μM (Table 1). Addition of cytb5 in a 1:1 molar ratio doubled the Vmax values of Cyp4a10, Cyp4a12a and Cyp4a12b. In contrast, the activity of Cyp4a14 was not significantly influenced by cytb5 (results not shown).
Table 1. Metabolism of LA by recombinant Cyp4a enzymes.
| Product distribution (%)* | ||||
|---|---|---|---|---|
| CYP | Km (μM) | Vmax (nmol·nmol−1·min−1) | 11-OH-LA | 12-OH-LA |
| 4a10 | 2±0.4 | 55±3 | 6 | 94 |
| 4a12a | 16±6 | 76±17 | 17 | 83 |
| 4a12b | 2±0.6 | 40±3 | 52 | 48 |
| 4a14 | 8±2 | 40±5 | 38 | 62 |
*The products were resolved and quantified by RP-HPLC as shown in Supplementary Figure S1. The values are averages from experiments with at least three different microsomal preparations. The standard errors were less than 10% of the mean. The product distribution was independent of the LA concentration in the range used for kinetic analysis (1–50 μM).
The primary products of Cyp4a-catalysed LA hydroxylation were 11-OH-LA (11-hydroxy LA) and 12-OH-LA. The identity of these metabolites was confirmed by co-migration with the authentic standard compounds (for representative HPLC chromatograms see Supplementary Figure S1 at http://www.BiochemJ.org/bj/403/bj4030109add.htm) and by GLC–MS analysis as described in the Supplementary online data. The 12-OH:11-OH ratio was isoform-specific (Table 1). The regioselectivity was very high with Cyp4a10 (94:6), moderate with Cyp4a12a (83:17) and Cyp4a14 (62:38) and almost not expressed with Cyp4a12b (48:52). Prolonged incubation or increased enzyme concentrations yielded substantial amounts of DDDA (dodecan-1,12-dioic acid) as a secondary product resulting from further oxidation of 12-OH-LA (results not shown).
Metabolism of AA by recombinant mouse Cyp4a isoforms
AA was efficiently metabolized only by Cyp4a12a and Cyp4a12b (Table 2; for representative HPLC chromatograms, see Supplementary Figure S2 at http://www.BiochemJ.org/bj/403/bj4030109add.htm). Major primary metabolites were 20-HETE and 19-HETE, which were produced by both Cyp4a12a and Cyp4a12b in a ratio of approx. 90:10. Cyp4a12b produced another primary metabolite that co-migrated with 18-HETE in NP-HPLC and represented approx. 12% of the total product. Secondary product formation analogous to the oxidation of LA to DDDA was also observed with AA. The secondary metabolite co-migrated in RP-HPLC with authentic 20-COOH-AA (eicosatetraen-1,20-dioic acid). The same metabolite was also directly produced from [1−14C]20-HETE by the Cyp4a12 isoforms (Supplementary Figures S2B and S2H).
Michaelis–Menten kinetics were observed determining the AA conversion rates at different substrate concentrations. The apparent Km values were 25 and 43 μM for Cyp4a12a and Cyp4a12b respectively. Both enzymes reached high Vmax values of approx. 10 nmol·min−1·nmol−1. The Vmax values were almost doubled in the presence of stoichiometric amounts of cytb5 (Table 2).
Table 2. Metabolism of AA by recombinant Cyp4a enzymes.
n.d., Not determined.
| Km (μM) | Vmax (nmol·nmol−1·min−1) | Product distribution (%)* | ||||||
|---|---|---|---|---|---|---|---|---|
| CYP | + cytb5 | + cytb5 | 18-HETE | 19-HETE | 20-HETE | 11,12-EET | ||
| 4a10 | n.d. | n.d. | 0.02±0.01† | 0.05±0.01† | – | 20‡ | 80‡ | – |
| 4a12a | 25±4 | 34±6 | 8±1 | 16±2 | – | 13 | 87 | – |
| 4a12b | 43±12 | 72±10 | 10±2 | 19±2 | 12 | 10 | 78 | – |
| 4a14 | n.d. | n.d. | 0.01±0.003† | 0.01±0.001† | – | – | – | 100 |
*The metabolites were resolved and quantified by RP- and NP-HPLC as shown in Supplementary Figure S2. The values are averages for experiments with at least three different microsomal preparations. The standard errors were less than 10% of the mean.
†Activities of Cyp4a10 and Cyp4a14 are given for a substrate concentration of 10 μM since a detailed kinetic analysis was not possible due to the very weak activities of these isoforms towards AA. For comparison, under the same conditions, the corresponding values for Cyp4a12a were 2.3 and 3.7 (+cytb5) nmol·nmol−1·min−1 and for Cyp4a12b 1.9 and 2.3 (+cytb5) nmol·nmol−1·min−1.
‡Roughly estimated from RP-HPLC analysis.
With Cyp4a10 and Cyp4a14, AA metabolites were clearly detectable only after increasing the enzyme concentration to 500 nM and prolonging the reaction time (Table 2 and Supplementary Figures S2A and S2D). The metabolites observed with Cyp4a10 co-migrated with 19/20-HETE. Cyp4a14 was almost completely inactive as an AA hydroxylase, but produced small amounts of 11,12-EET. At a substrate concentration of 10 μM, Cyp4a10 reached an activity of 0.05 nmol·nmol−1·min−1 (in the presence of cytb5) and was thus 70-fold less active than Cyp4a12a. At higher AA concentrations, Cyp4a10 showed specific activities of 0.14 (40 μM), 0.21 (100 μM) and 0.63 nmol·nmol−1·min−1 (200 μM), without reaching substrate saturation. Owing to these low conversion rates, a further detailed kinetic analysis was not possible.
Metabolism of EPA by recombinant mouse Cyp4a isoforms
EPA was converted by Cyp4a12a and Cyp4a12b with high catalytic efficiencies that even slightly exceeded those determined for AA (Table 3). In comparison, Cyp4a10 and Cyp4a14 showed only a weak capacity to convert EPA resembling their very low activities in AA conversion (Table 3; for representative HPLC chromatograms, see Supplementary Figure S3 at http://www.BiochemJ.org/bj/403/bj4030109add.htm).
Table 3. Metabolism of EPA by recombinant Cyp4a enzymes.
n.d., Not determined.
| Km (μM) | Vmax (nmol·nmol−1·min−1) | Product distribution (%)* | Stereoselectivity (%)* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYP | + cytb5 | + cytb5 | 19-HEPE | 20-HEPE | 17,18-EETeTr | EETeTr | 17(R),18(S)-EETeTr | 17(S),18(R)-EETeTr | ||
| 4a10 | n.d. | n.d. | 0.1±0.01† | 0.2±0.03† | 60‡ | 40‡ | – | n.d. | n.d. | |
| 4a12a | 29±4 | 33±2 | 14±1 | 29±1 | 12 | 32 | 56 | – | 73 | 27 |
| 4a12b | 41±5 | 62±20 | 15±1 | 30±7 | 11 | 21 | 68 | – | 51 | 49 |
| 4a14 | n.d. | n.d. | 0.02±0.01† | 0.03±0.01† | 11‡ | – | 89‡ | n.d. | n.d. | |
*The metabolites were resolved and quantified by RP-HPLC followed by NP-HPLC or chiral-phase HPLC as shown in Supplementary Figure S3. The values are averages for experiments with at least three different microsomal preparations. The standard errors were less than 10% of the mean.
†Activities of Cyp4a10 and Cyp4a14 are given for a substrate concentration of 10 μM since a detailed kinetic analysis was not possible due to the very weak activities of these isoforms towards EPA. For comparison, under the same conditions, the corresponding values for Cyp4a12a were 3.6 and 6.7 (+cytb5) nmol·nmol−1·min−1 and for Cyp4a12b 3.0 and 4.2 (+cytb5) nmol·nmol−1·min−1.
‡Estimation from RP-HPLC only.
Whereas Cyp4a12a and Cyp4a12b functioned solely as hydroxylases when converting AA, they catalysed both a hydroxylation and an epoxidation reaction with EPA. The ratio of hydroxylase to epoxygenase activities was 1:1.3 with Cyp4a12a and 1:2 with Cyp4a12b. Accordingly, the epoxidation product represented the major EPA metabolite with both isoforms. The epoxidation product co-migrated in RP-HPLC with authentic 17,18-EETeTr. Chiral-phase HPLC resolved the 17,18-EETeTr peak collected from RP-HPLC into the respective (R,S)- and (S,R)-enatiomers. Cyp4a12a produced preferentially the (R,S)-enantiomer, whereas Cyp4a12b displayed no stereoselectivity (Table 3). The hydroxylase product consisted of 19- and 20-HEPE and both enzymes showed a regioselectivity in favour of producing the 20-OH metabolite (Table 3).
Gender- and strain-specific renal expression of mouse Cyp4a isoform
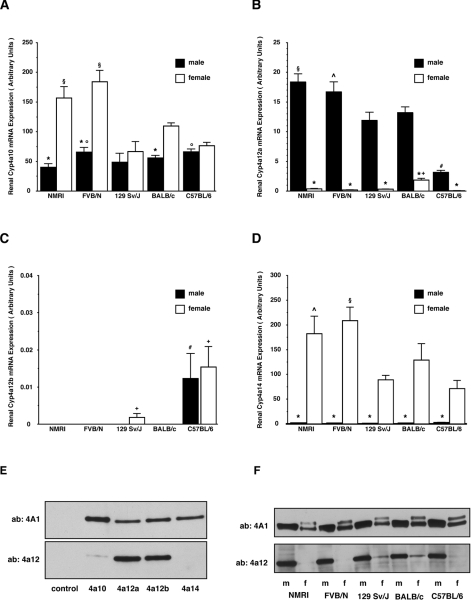
TaqMan-PCR analysis demonstrated that Cyp4a10 is highly expressed in the kidneys of both genders, whereas Cyp4a12a is predominantly a male- and Cyp4a14 a female-specific isoform (Figures 1A–1D).
Figure 1. Gender- and strain-specific renal expression of Cyp4a-isoforms.
(A–D) Renal mRNA expression of Cyp4a10, Cyp4a12a, Cyp4a12b and Cyp4a14 in male and female NMRI, FVB/N, 129 Sv/J, Balb/c and C57BL/6 mice. Results shown are means±S.E.M. (n=5 in each group). Significant differences (P<0.05) between the groups are marked as follows: *, compared with other gender of the same strain; #, compared with other males; +, compared with other females; §, compared with 129 Sv/J, Balb/c and C57BL/6; ^, compared with 129 Sv/J and C57BL/6; °, compared with NMRI. (E) Specificity of the antibodies used for Western-blot analysis. Control: microsomal protein (8 μg) isolated from Sf9 cells not expressing any of the Cyp isoforms; other lanes: individual recombinant Cyp4a isoforms (0.2 pmol each); ‘ab: 4A1’: antibody raised against rat CYP4A1; ‘ab: 4a12’: antibody raised against a Cyp4a12a-specific peptide. (F) Western-blot analysis of renal Cyp4a protein expression using the primary 4A1 and 4a12 antibodies; 7.5 μg of total microsomal protein was loaded per lane. m, male; f, female.
Among the isoforms efficiently producing 20-HETE, only Cyp4a12a but not Cyp4a12b showed significant renal expression. Cyp4a12a levels in the males largely exceeded that in the females (45-fold for NMRI, 88-fold for FVB/N, 40-fold for 129 Sv/J, 7-fold for Balb/c and 48-fold for C57BL/6; Figure 1B). Moreover, we observed marked differences in the renal Cyp4a12a mRNA levels between the five different male strains (Figure 1B). The highest expression was observed in NMRI followed by FVB/N, Balb/c and 129 Sv/J. The renal Cyp4a12a expression level in male C57BL/6 reached only approx. 17% of male NMRI and was statistically significantly lower compared with all other male strains.
In contrast, Cyp4a12b mRNA was almost undetectable in the kidneys of both genders of all strains tested. Reliable TaqMan-PCR amplification curves were only obtained with renal samples from female 129 Sv/J, and male and female C57BL/6 (Figure 1C). Renal samples from all other strains yielded low unspecific signals not different from the non-template controls. However, Cyp4a12b mRNA was easily detectable under the same experimental conditions using lung instead of renal samples (results not shown) indicating that Cyp4a12b shows a different tissue specificity of expression.
We also analysed the gender and strain differences in Cyp4a isoform expression at the protein level. The antibody raised against a Cyp4a12a-derived peptide efficiently recognized recombinant Cyp4a12a and Cyp4a12b, showed a very weak cross-reactivity with Cyp4a10 and did not react with Cyp4a14 (Figure 1E). In contrast, the CYP4A1-antibody detected all four mouse Cyp4a isoforms (Figure 1E). Western-blot analysis of mouse renal microsomes confirmed the gender differences in Cyp4a12 isoform expression (Figure 1F). The Cyp4a12-specific antibody yielded a single strong band with male and almost no reaction with female renal microsomes with the exception of a very weak but reproducible Cyp4a12 protein expression in female 129 Sv/J and Balb/c (Figure 1F).
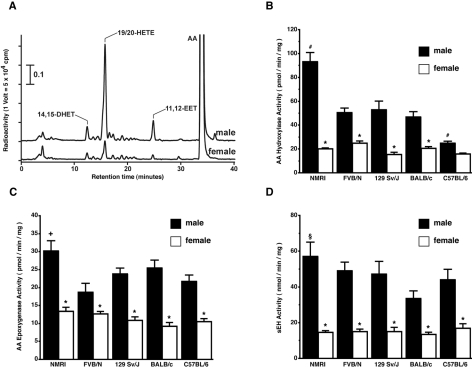
Gender- and strain-specific renal microsomal AA hydroxylase activities
Mouse renal microsomes of all strains tested showed both AA hydroxylase and AA epoxygenase activities (Figure 2A). Renal microsomes of female mice had low AA hydroxylase activities ranging from 15 to 25 pmol·min−1·mg−1. Males showed 3–5 times higher activities than the respective female strain (Figure 2B). The gender difference was most pronounced in NMRI, where males reached a renal AA hydroxylase activity of 93±4 pmol·min−1·mg−1 compared with 20±0.5 pmol·min−1·mg−1 in females. No significant gender-related difference was observed in renal microsomal 20-HETE production of C57BL/6 (25±1 versus 16±1 pmol·min−1·mg−1, male versus female).
Figure 2. Gender- and strain-specific expression of enzymatic activities involved in mouse renal AA metabolism.
(A) Representative RP-HPLC chromatogram of AA metabolites produced by renal microsomes isolated from male and female NMRI mice. (B–D) Renal AA hydroxylase, AA epoxygenase, and soluble epoxide hydrolase activities of male and female NMRI, FVB/N, 129 Sv/J, Balb/c and C57BL/6. Results shown are means±S.E.M. (n=6 in each group). Significant differences (P<0.05) between the groups are marked as follows: *, compared with other gender of the same strain; #, compared with other males; +, compared with FVB/N and C57BL/6; §, compared with Balb/c.
Clear gender differences were also found in renal microsomal AA epoxygenase and in soluble epoxide hydrolase activities (Figures 2C and 2D). Compared with the large differences in 20-HETE production among male strains (Figure 2B), strain differences in epoxygenase and hydrolase activities were less pronounced.
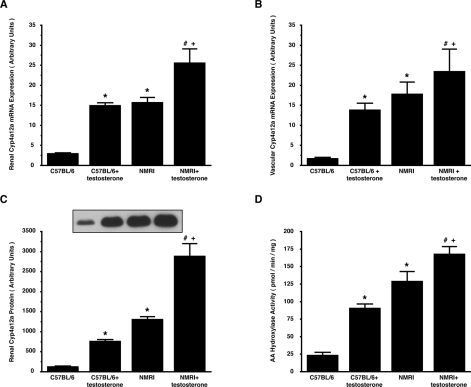
Induction of Cyp4a12a and of renal microsomal 20-HETE production by androgen treatment
Androgen treatment induced total renal and renal arteriole Cyp4a12a mRNA expression more than 4-fold in C57BL/6 and approx. 1.4-fold in NMRI (Figures 3A and 3B). Western-blot analysis showed that the Cyp4a12 protein level was significantly higher in male NMRI compared with male C57BL/6 (Figure 3C). 5α-DHT strongly increased the Cyp4a12 protein level in C57BL/6 (to 600% of the untreated control) and to a lesser extent also in NMRI (to 200%). Renal microsomal AA hydroxylase activities were 23±4 pmol·min−1·mg−1 in untreated C57BL/6 and 128±14 pmol·min−1·mg−1 in untreated NMRI (Figure 3D). 5α-DHT treatment increased the activity to 90±7 in C57BL/6 and to 167±11 pmol·min−1·mg−1 in NMRI. These results show that 5α-DHT treatment significantly induced Cyp4a12a mRNA, protein and enzyme activity levels in C57BL/6, leading to Cyp4a12a expression and 20-HETE production similar to NMRI under basal conditions.
Figure 3. Effect of 5α-DHT on Cyp4a12a expression and renal microsomal 20-HETE production in male NMRI and C57BL/6.
(A, B) Cyp4a12a mRNA levels in total kidney and renal arterioles; (C) renal Cyp4a12 protein levels; (D) renal microsomal AA hydroxylase activities. Results are means±S.E.M. (n=6 each). Significant differences (P<0.05) between the groups are marked as follows: *, compared with untreated C57BL/6; #, compared with untreated NMRI; +, compared with testosterone-treated C57BL/6.
DISCUSSION
In the present study, we identified Cyp4a12a as the predominant AA ω-hydroxylase in the mouse kidney and demonstrated that Cyp4a12a expression levels determine the sex- and strain-specific differences in mouse renal 20-HETE production. We suggest that the differences in 20-HETE we identified among the strains could account for resistance to target organ damage exhibited by some strains, but not others. Furthermore, the sex differences we identified, coupled with the androgen effects we observed, may explain the relative resistance of female mice to target organ damage compared with males within strains.
Recombinant Cyp4a12a showed a high regioselectivity in favour of producing 20-HETE from AA similar to that observed previously with mouse renal microsomes [12]. Cyp4a12a did not form any detectable amounts of AA epoxides. However, with EPA, Cyp4a12a efficiently catalysed the epoxidation of the 17,18-double bond that distinguishes this ω-3 PUFA from AA. Thus Cyp4a12a generates very different sets of metabolites when either AA or EPA becomes accessible as substrate. Such a shift in CYP-dependent eicosanoids may have physiological significance. 20-HETE is a powerful endogenous vasoconstrictor that functions via inhibition of BK channels (calcium-activated potassium channels) in vascular smooth-muscle cells [4]. In contrast, 17,18-EETeTr shows vasodilator properties and is a very potent activator of BK channels [22,29]. BK channel activation is highly stereoselective [22] and we found that Cyp4a12a produces preferentially the active (R,S)-enantiomer. Alterations in cyclo-oxygenase- and lipoxygenase-dependent prostanoid and leukotriene biosynthesis have been generally assumed to contribute to the beneficial effects of fish oil ω-3 PUFAs in cardiovascular diseases [30]. Considering the enzymatic properties of Cyp4a12a described above, it appears highly probable that CYP-catalysed 20-HETE production can also be manipulated under in vivo conditions by ω-3 PUFA-rich diets. How this state-of-affairs affects 20-HETE-mediated signalling pathways regulating vascular and renal function will be an important research topic.
Cyp4a12a not only hydroxylated AA to 20-HETE but also catalysed the subsequent oxidation steps necessary to convert 20-HETE into 20-COOH-AA. The ability to catalyse an oxidation cascade from a given fatty acid to the corresponding dicarboxylic acid was shown earlier for several CYP enzymes, examining substrates other than AA [31]. Alcohol dehydrogenase 4 may be the responsible enzyme in vascular smooth-muscle and endothelial cells that converts 20-HETE into 20-COOH-AA [32]. This reaction may serve to overcome the vasoconstrictor action of 20-HETE [27,33]. Moreover, 20-COOH-AA is known to inhibit the Na,K,2Cl-co-transporter in mTALH (medullary thick ascending limb of the loop of Henle) [4,27] and to function as a dual activator of peroxisome-proliferator-activated receptors α and γ [34].
Our study indicates that both Cyp4a12 genes [14] are functional but differ significantly in the tissue specificity of expression. Considering the very high degree of homology between the two Cyp4a12 variants, the differences that we observed when comparing their enzymatic properties are remarkable. In contrast with Cyp4a12a, Cyp4a12b showed (i) no preference for ω- over ω-1 hydroxylation when converting LA, (ii) no stereoselectivity when epoxidizing the 17,18-double bond of EPA, and (iii) a higher EPA epoxygenase/EPA hydroxylase ratio. These findings indicate that the positioning of the substrate alkyl chain is probably less strict in the active site of Cyp4a12b compared with that in Cyp4a12a. Interestingly, there are also similar pairs of highly homologous CYP4A enzymes in human (CYP4A11 and CYP4A22; [8,35]) and rat (CYP4A2 and CYP4A3; [36,37]). However, the sequence differences in these CYP pairs concern other amino acid positions and have different functional consequences. Thus the significance of the CYP4A gene duplications and of the subsequently evolved gene variants is not readily obvious and may differ in human, rat and mouse.
Cyp4a10 is the murine Cyp isoform showing the highest homology to CYP4A1 (92% amino acid sequence identity), the most active rat AA hydroxylase [36]. However, our results show that Cyp4a10 displays only a very weak AA hydroxylase activity. Assuming that 10 μM is a reasonable physiological AA concentration, the activity of Cyp4a10 would be approx. 70-fold lower than that of Cyp4a12a. Cyp4a10 gene disruption did not alter mouse renal AA hydroxylase activities, confirming that the Cyp4a10 enzyme does not contribute to 20-HETE production [16]. Cyp4a12a and Cyp4a12b have 78% sequence identity with CYP4A1 and are most closely related to CYP4A8 (87%), which showed, however, the lowest AA hydroxylase activity among the rat CYP4A isoforms [36]. CYP4A8 shares with Cyp4a12a the inducibility by androgens [38]. Cyp4a14 is most similar to CYP4A2 (88%) and CYP4A3 (89%). The latter are highly expressed in the male rat kidney and display a significant 20-HETE synthase activity [36]. In contrast, Cyp4a14 shows a female-specific expression and we could not detect any 20-HETE synthase activity with this isoform.
Our data on the sex specificity of renal Cyp4a gene expression in five different mouse strains are in agreement with previous reports [10,11] that Cyp4a10 is highly expressed in both sexes, whereas Cyp4a12a is clearly a male- and Cyp4a14 a female-specific isoform. Moreover, we found that the expression of Cyp4a12a is correlated to the capacity of mouse renal microsomes to hydroxylate AA thus providing further evidence for the role of this isoform in 20-HETE production. Comparing the males of the different strains, NMRI mice had clearly the highest Cyp4a12a expression levels, whereas C57BL/6 appears as a naturally occurring mouse model with a specific knock-down in Cyp4a12a/20-HETE expression. Our results and those of other authors [11] demonstrate that Cyp4a12 is inducible by androgens. The androgen effect was much more pronounced with C57BL/6 compared with NMRI. Providing a possible explanation, mouse strains have been long known to differ in blood testosterone levels and C57BL/6 is considered as being chronically testosterone-deficient [39]. The androgen-dependent expression also explains the low levels of Cyp4a12a in the females of all strains. We noticed, however, that the gender differences in Cyp4a12a mRNA and protein expression levels were apparently more pronounced than in the AA hydroxylase activities. Therefore it remains to be analysed whether the low but still significant renal microsomal AA hydroxylase activities in the females are in part due to different Cyp isoforms possibly including Cyp4F subfamily members [13,40].
20-HETE plays an important role in the regulation of various cardiovascular processes. It acts as a second messenger in signalling pathways modulating vascular tone, salt excretion, cell proliferation and angiogenesis [4]. Since the intensity of these signalling pathways may depend on the extent of 20-HETE production, the question arises whether sex- and strain-specific differences in the response to pro-hypertensive and pro-hypertrophic stimuli are partially due to differences in Cyp AA ω-hydroxylase expression levels. Substantiating this possibility, androgen-induced overproduction of 20-HETE in mice [11] and rats [38] is associated with the development of hypertension. Moreover, rats transduced with an adenovirus for vascular overexpression of CYP4A2 developed endothelial dysfunction and hypertension [41]. Female mice, which have very low renal 20-HETE production, are more resistant to Ang II-induced hypertension [18] and to ischaemia/reperfusion-induced kidney injury [42], compared with males. C57BL/6 males, which have lower renal 20-HETE production than the males of other strains, show reduced susceptibilities to the development of DOCA-salt hypertension [17], glomerulosclerosis [43] and injury-induced neointimal hyperplasia [44]. Recently, CYP-dependent AA metabolism has also been implicated in cardiac function and ischaemia/reperfusion-induced myocardial injury [45]. Clearly, there are many other factors potentially involved in mediating gender-related differences in the manifestation of cardiovascular disease [46], and the relative importance of differences in CYP-dependent AA metabolism remains to be determined.
Our study provides comprehensive information on the expression and enzymatic properties of the Cyp4a subfamily members in the mouse kidney, identifies Cyp4a12a as the predominant 20-HETE synthase, and suggests that differences in Cyp4a12a/20-HETE expression may be involved in determining sex- and strain-specific differences in susceptibility to hypertension and other cardiovascular diseases. Transgenic and gene-deletion approaches to alter Cyp4a12a expression in selected tissues may help to prove directly the various physiological and pathophysiological functions attributed to 20-HETE in the regulation of renal, vascular, and cardiac function. In any such studies, strain and sex selection will be a highly important aspect of study design.
Online data
Acknowledgments
We thank Christel Andrée, Ramona Zummach and Ilona Kamer for excellent technical assistance. This study was supported by grants-in-aid to D.N. M., F.C. L. and W.-H. S. from the Deutsche Forschungsgemeinschaft.
References
- 1.Okita R. T., Okita J. R. Cytochrome P450 4A fatty acid omega hydroxylases. Curr. Drug Metab. 2001;2:265–281. doi: 10.2174/1389200013338423. [DOI] [PubMed] [Google Scholar]
- 2.Johnson E. F., Hsu M. H., Savas U., Griffin K. J. Regulation of P450 4A expression by peroxisome proliferator activated receptors. Toxicology. 2002;181–182:203–206. doi: 10.1016/s0300-483x(02)00282-2. [DOI] [PubMed] [Google Scholar]
- 3.Capdevila J. H., Falck J. R., Harris R. C. Cytochrome P450 and arachidonic acid bioactivation. Molecular and functional properties of the arachidonate monooxygenase. J. Lipid Res. 2000;41:163–181. [PubMed] [Google Scholar]
- 4.McGiff J. C., Quilley J. 20-HETE and the kidney: resolution of old problems and new beginnings. Am. J. Physiol. 1999;277:R607–R623. doi: 10.1152/ajpregu.1999.277.3.R607. [DOI] [PubMed] [Google Scholar]
- 5.Roman R. J. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol. Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 6.Sacerdoti D., Balazy M., Angeli P., Gatta A., McGiff J. C. Eicosanoid excretion in hepatic cirrhosis. Predominance of 20-HETE. J. Clin. Invest. 1997;100:1264–1270. doi: 10.1172/JCI119640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ward N. C., Rivera J., Hodgson J., Puddey I. B., Beilin L. J., Falck J. R., Croft K. D. Urinary 20-hydroxyeicosatetraenoic acid is associated with endothelial dysfunction in humans. Circulation. 2004;110:438–443. doi: 10.1161/01.CIR.0000136808.72912.D9. [DOI] [PubMed] [Google Scholar]
- 8.Gainer J. V., Bellamine A., Dawson E. P., Womble K. E., Grant S. W., Wang Y., Cupples L. A., Guo C. Y., Demissie S., O'Donnell C. J., et al. Functional variant of CYP4A11 20-hydroxyeicosatetraenoic acid synthase is associated with essential hypertension. Circulation. 2005;111:63–69. doi: 10.1161/01.CIR.0000151309.82473.59. [DOI] [PubMed] [Google Scholar]
- 9.Henderson C. J., Bammler T., Wolf C. R. Deduced amino acid sequence of a murine cytochrome P-450 Cyp4a protein: developmental and hormonal regulation in liver and kidney. Biochim. Biophys. Acta. 1994;1200:182–190. doi: 10.1016/0304-4165(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 10.Heng Y. M., Kuo C. S., Jones P. S., Savory R., Schulz R. M., Tomlinson S. R., Gray T. J., Bell D. R. A novel murine P-450 gene, Cyp4a14, is part of a cluster of Cyp4a and Cyp4b, but not of CYP4F, genes in mouse and humans. Biochem. J. 1997;325:741–749. doi: 10.1042/bj3250741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holla V. R., Adas F., Imig J. D., Zhao X., Price E., Jr, Olsen N., Kovacs W. J., Magnuson M. A., Keeney D. S., Breyer M. D., et al. Alterations in the regulation of androgen-sensitive Cyp 4a monooxygenases cause hypertension. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5211–5216. doi: 10.1073/pnas.081627898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honeck H., Gross V., Erdmann B., Kargel E., Neunaber R., Milia A. F., Schneider W., Luft F. C., Schunck W. H. Cytochrome P450-dependent renal arachidonic acid metabolism in desoxycorticosterone acetate-salt hypertensive mice. Hypertension. 2000;36:610–616. doi: 10.1161/01.hyp.36.4.610. [DOI] [PubMed] [Google Scholar]
- 13.Stec D. E., Flasch A., Roman R. J., White J. A. Distribution of cytochrome P-450 4A and 4F isoforms along the nephron in mice. Am. J. Physiol. Renal Physiol. 2003;284:F95–F102. doi: 10.1152/ajprenal.00132.2002. [DOI] [PubMed] [Google Scholar]
- 14.Nelson D. R., Zeldin D. C., Hoffman S. M., Maltais L. J., Wain H. M., Nebert D. W. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Vera T., Taylor M., Bohman Q., Flasch A., Roman R. J., Stec D. E. Fenofibrate prevents the development of angiotensin II-dependent hypertension in mice. Hypertension. 2005;45:730–735. doi: 10.1161/01.HYP.0000153317.06072.2e. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa K., Holla V. R., Wei Y., Wang W. H., Gatica A., Wei S., Mei S., Miller C. M., Cha D. R., Price E., Jr, et al. Salt-sensitive hypertension is associated with dysfunctional Cyp4a10 gene and kidney epithelial sodium channel. J. Clin. Invest. 2006;116:1696–1702. doi: 10.1172/JCI27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartner A., Cordasic N., Klanke B., Veelken R., Hilgers K. F. Strain differences in the development of hypertension and glomerular lesions induced by deoxycorticosterone acetate salt in mice. Nephrol. Dial. Transplant. 2003;18:1999–2004. doi: 10.1093/ndt/gfg299. [DOI] [PubMed] [Google Scholar]
- 18.Xue B., Pamidimukkala J., Hay M. Sex differences in the development of angiotensin II-induced hypertension in conscious mice. Am. J. Physiol. Heart Circ. Physiol. 2005;288:H2177–H2184. doi: 10.1152/ajpheart.00969.2004. [DOI] [PubMed] [Google Scholar]
- 18a.Lowry O. H., Rosenbrough N. J., Farr A. L., Randall R. J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.Schwarz D., Kisselev P., Cascorbi I., Schunck W. H., Roots I. Differential metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-dihydrodiol by human CYP1A1 variants. Carcinogenesis. 2001;22:453–459. doi: 10.1093/carcin/22.3.453. [DOI] [PubMed] [Google Scholar]
- 20.Omura T., Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 21.Falck J. R., Yadagiri P., Capdevila J. Synthesis of epoxyeicosatrienoic acids and heteroatom analogs. Methods Enzymol. 1990;187:357–364. doi: 10.1016/0076-6879(90)87042-2. [DOI] [PubMed] [Google Scholar]
- 22.Lauterbach B., Barbosa-Sicard E., Wang M. H., Honeck H., Kargel E., Theuer J., Schwartzman M. L., Haller H., Luft F. C., Gollasch M., Schunck W. H. Cytochrome P450-dependent eicosapentaenoic acid metabolites are novel BK channel activators. Hypertension. 2002;39:609–613. doi: 10.1161/hy0202.103293. [DOI] [PubMed] [Google Scholar]
- 23.Falck J. R., Lumin S., Blair I., Dishman E., Martin M. V., Waxman D. J., Guengerich F. P., Capdevila J. H. Cytochrome P-450-dependent oxidation of arachidonic acid to 16-, 17-, and 18-hydroxyeicosatetraenoic acids. J. Biol. Chem. 1990;265:10244–10249. [PubMed] [Google Scholar]
- 24.Barbosa-Sicard E., Markovic M., Honeck H., Christ B., Muller D. N., Schunck W. H. Eicosapentaenoic acid metabolism by cytochrome P450 enzymes of the CYP2C subfamily. Biochem. Biophys. Res. Commun. 2005;329:1275–1281. doi: 10.1016/j.bbrc.2005.02.103. [DOI] [PubMed] [Google Scholar]
- 25.Ortiz de Montellano P. R., Reich N. O. Specific inactivation of hepatic fatty acid hydroxylases by acetylenic fatty acids. J. Biol. Chem. 1984;259:4136–4141. [PubMed] [Google Scholar]
- 26.Schwarz D., Kisselev P., Ericksen S. S., Szklarz G. D., Chernogolov A., Honeck H., Schunck W. H., Roots I. Arachidonic and eicosapentaenoic acid metabolism by human CYP1A1: highly stereoselective formation of 17(R),18(S)- epoxyeicosatetraenoic acid. Biochem. Pharmacol. 2004;67:1445–1457. doi: 10.1016/j.bcp.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 27.Carroll M. A., Sala A., Dunn C. E., McGiff J. C., Murphy R. C. Structural identification of cytochrome P450-dependent arachidonate metabolites formed by rabbit medullary thick ascending limb cells. J. Biol. Chem. 1991;266:12306–12312. [PubMed] [Google Scholar]
- 28.Capdevila J. H., Wei S., Helvig C., Falck J. R., Belosludtsev Y., Truan G., Graham-Lorence S. E., Peterson J. A. The highly stereoselective oxidation of polyunsaturated fatty acids by cytochrome P450BM-3. J. Biol. Chem. 1996;271:22663–22671. doi: 10.1074/jbc.271.37.22663. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Oltman C. L., Lu T., Lee H. C., Dellsperger K. C., VanRollins M. EET homologs potently dilate coronary microvessels and activate BK(Ca) channels. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H2430–H2440. doi: 10.1152/ajpheart.2001.280.6.H2430. [DOI] [PubMed] [Google Scholar]
- 30.Kris-Etherton P. M., Harris W. S., Appel L. J. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Circulation. 2002;106:2747–2757. doi: 10.1161/01.cir.0000038493.65177.94. [DOI] [PubMed] [Google Scholar]
- 31.Scheller U., Zimmer T., Becher D., Schauer F., Schunck W. H. Oxygenation cascade in conversion of n-alkanes to alpha,omega-dioic acids catalyzed by cytochrome P450 52A3. J. Biol. Chem. 1998;273:32528–32534. doi: 10.1074/jbc.273.49.32528. [DOI] [PubMed] [Google Scholar]
- 32.Collins X. H., Harmon S. D., Kaduce T. L., Berst K. B., Fang X., Moore S. A., Raju T. V., Falck J. R., Weintraub N. L., Duester G., et al. Omega-oxidation of 20-hydroxyeicosatetraenoic acid (20-HETE) in cerebral microvascular smooth muscle and endothelium by alcohol dehydrogenase 4. J. Biol. Chem. 2005;280:33157–33164. doi: 10.1074/jbc.M504055200. [DOI] [PubMed] [Google Scholar]
- 33.Kaduce T. L., Fang X., Harmon S. D., Oltman C. L., Dellsperger K. C., Teesch L. M., Gopal V. R., Falck J. R., Campbell W. B., Weintraub N. L., Spector A. A. 20-hydroxyeicosatetraenoic acid (20-HETE) metabolism in coronary endothelial cells. J. Biol. Chem. 2004;279:2648–2656. doi: 10.1074/jbc.M306849200. [DOI] [PubMed] [Google Scholar]
- 34.Fang X., Dilon J. S., Hu S., Harmon S. D., Yao J., Anjaiah S., Falck J. R., Spector A. A. 20-Caboxy-arachidonic acid is a dual activator of peroxisome proliferator-activated receptors α and γ. Prostaglandins Other Lipid Mediat. 2006;82:175–184. doi: 10.1016/j.prostaglandins.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Bellamine A., Wang Y., Waterman M. R., Gainer J. V., III, Dawson E. P., Brown N. J., Capdevila J. H. Characterization of the CYP4A11 gene, a second CYP4A gene in humans. Arch. Biochem. Biophys. 2003;409:221–227. doi: 10.1016/s0003-9861(02)00545-3. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen X., Wang M. H., Reddy K. M., Falck J. R., Schwartzman M. L. Kinetic profile of the rat CYP4A isoforms: arachidonic acid metabolism and isoform-specific inhibitors. Am. J. Physiol. 1999;276:R1691–R1700. doi: 10.1152/ajpregu.1999.276.6.R1691. [DOI] [PubMed] [Google Scholar]
- 37.Hoch U., Falck J. R., de Montellano P. R. Molecular basis for the omega-regiospecificity of the CYP4A2 and CYP4A3 fatty acid hydroxylases. J. Biol. Chem. 2000;275:26952–26958. doi: 10.1074/jbc.M004841200. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa K., Marji J. S., Schwartzman M. L., Waterman M. R., Capdevila J. H. Androgen-mediated induction of the kidney arachidonate hydroxylases is associated with the development of hypertension. Am. J. Physiol. Regulatory Integrative Comp. Physiol. 2003;284:R1055–R1062. doi: 10.1152/ajpregu.00459.2002. [DOI] [PubMed] [Google Scholar]
- 39.Brouillette J., Rivard K., Lizotte E., Fiset C. Sex and strain differences in adult mouse cardiac repolarization: importance of androgens. Cardiovasc. Res. 2005;65:148–157. doi: 10.1016/j.cardiores.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Lasker J. M., Chen W. B., Wolf I., Bloswick B. P., Wilson P. D., Powell P. K. Formation of 20-hydroxyeicosatetraenoic acid, a vasoactive and natriuretic eicosanoid, in human kidney. Role of Cyp4F2 and Cyp4A11. J. Biol. Chem. 2000;275:4118–4126. doi: 10.1074/jbc.275.6.4118. [DOI] [PubMed] [Google Scholar]
- 41.Wang J. S., Singh H., Zhang F., Ishizuka T., Deng H., Kemp R., Wolin M. S., Hintze T. H., Abraham N. G., Nasjletti A., Laniado-Schwartzman M. Endothelial dysfunction and hypertension in rats transduced with CYP4A2 adenovirus. Circ. Res. 2006;98:962–969. doi: 10.1161/01.RES.0000217283.98806.a6. [DOI] [PubMed] [Google Scholar]
- 42.Park K. M., Kim J. I., Ahn Y., Bonventre A. J., Bonventre J. V. Testosterone is responsible for enhanced susceptibility of males to ischemic renal injury. J. Biol. Chem. 2004;279:52282–52292. doi: 10.1074/jbc.M407629200. [DOI] [PubMed] [Google Scholar]
- 43.Ma L. J., Fogo A. B. Model of robust induction of glomerulosclerosis in mice: importance of genetic background. Kidney Int. 2003;64:350–355. doi: 10.1046/j.1523-1755.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 44.Kuhel D. G., Zhu B., Witte D. P., Hui D. Y. Distinction in genetic determinants for injury-induced neointimal hyperplasia and diet-induced atherosclerosis in inbred mice. Arterioscler. Thromb. Vasc. Biol. 2002;22:955–960. doi: 10.1161/01.atv.0000017994.77066.75. [DOI] [PubMed] [Google Scholar]
- 45.Gross G. J., Falck J. R., Gross E. R., Isbell M., Moore J., Nithipatikom K. Cytochrome P450 and arachidonic acid metabolites: role in myocardial ischemia/reperfusion injury revisited. Cardiovasc. Res. 2005;68:18–25. doi: 10.1016/j.cardiores.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Regitz-Zagrosek V. Therapeutic implications of the gender-specific aspects of cardiovascular disease. Nat. Rev. Drug Discov. 2006;5:425–438. doi: 10.1038/nrd2032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.