Abstract
The mTOR (mammalian target of rapamycin) signalling pathway is a key regulator of cell growth and is controlled by growth factors and nutrients such as amino acids. Although signalling pathways from growth factor receptors to mTOR have been elucidated, the pathways mediating signalling by nutrients are poorly characterized. Through a screen for protein kinases active in the mTOR signalling pathway in Drosophila we have identified a Ste20 family member (MAP4K3) that is required for maximal S6K (S6 kinase)/4E-BP1 [eIF4E (eukaryotic initiation factor 4E)-binding protein 1] phosphorylation and regulates cell growth. Importantly, MAP4K3 activity is regulated by amino acids, but not the growth factor insulin and is not regulated by the mTORC1 inhibitor rapamycin. Our results therefore suggest a model whereby nutrients signal to mTORC1 via activation of MAP4K3.
Keywords: amino acid signalling, cell growth, mammalian target of rapamycin (mTOR), MAP4K3
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; DPBS, Dulbecco's PBS; dsRNA, double-stranded RNA; eIF4E, eukaryotic initiation factor 4E; 4E-BP1, eIF4E-binding protein; FCS, foetal calf serum; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; HA, haemagglutinin; HEK, human embryonic kidney; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MBP, myelin basic protein; MEM, minimal essential medium; Msn, misshapen; mTOR, mammalian target of rapamycin; p90RSK, p90 ribosomal S6 kinase; PI3K, phosphoinositide 3-kinase; PKB, protein kinase B; RNAi, RNA interfering; PTEN, phosphatase and tensin homologue deleted on chromosome 10; siRNA, small interference RNA; TSC, tuberous sclerosis complex; TSC1-2, TSC1–TSC2 complex
INTRODUCTION
Both growth factors such as insulin/IGF-1 (insulin growth factor 1) and ambient nutrients such as amino acids stimulate growth via signalling to the mTOR (mammalian target of rapamycin) pathway [1]. The pathway regulates growth primarily through regulation of ribosome biogenesis and protein translation, and has emerged as a promising target for therapies to combat diseases such as cancer and diabetes [2]. Recent work has indicated that the growth factor signalling pathway resulting in mTOR pathway activation involves the PI3K (phosphoinositide 3-kinase) or MAPK (mitogen-activated protein kinase) signalling cascades, which activate protein kinases such as PKB (protein kinase B), ERK (extracellular-signal-regulated kinase) and p90RSK (p90 ribosomal S6 kinase). These kinases phosphorylate TSC (tuberous sclerosis complex) 2 thereby functionally inactivating the TSC1-2 tumour suppressor complex through an undefined mechanism [3]. TSC1-2 functions as a GAP (GTPase-activating protein) inhibitor of the Rheb GTPase [4], which itself positively regulates the mTOR pathway, possibly through regulation of mTOR activity [1].
In contrast with the growth factor regulation of mTOR signalling, the role of nutrients in the regulation of mTOR signalling has been relatively understudied. Although amino acids have been identified as activators of mTOR signalling, and are required for maximal activation of mTOR signalling by growth factors such as insulin [5], the signalling pathway downstream of amino acids is poorly characterized. To identify new components in this pathway that may be amenable to future therapies, we performed an RNAi (RNA interference) screen of Drosophila protein kinases required for signalling to dS6K, a serine-threonine kinase downstream of mTOR. In the present study we report our identification of a Ste20-related kinase, MAP4K3, which we show is required for amino acids to activate S6K and also induces phosphorylation of the mTOR-regulated inhibitor of eIF4E (eukaryotic initiation factor 4E), 4E-BP1 (eIF4E-binding protein). Importantly, we show that the activity of MAP4K3 is itself regulated by amino acids but not by insulin, suggesting that MAP4K3 plays an important role primarily in the nutrient regulation of mTOR signalling.
MATERIALS AND METHODS
Chemicals and antibodies
Rapamycin and wortmannin (Calbiochem) were dissolved in DMSO, and used at final concentrations of 50 nM and 100 nM respectively. PD184352 (a gift from Dr Richard Marais, Signal Transduction Laboratory, Institute of Cancer Research, London, U.K.) was dissolved in DMSO, and used at a final concentration of 1 or 10 μM. The anti-MAP4K3 antibody was prepared at the Hybridoma Unit (Institute of Cancer Research, London, U.K.) by immunizing rabbits with a GST (glutathione S-transferase) fusion protein of amino acids 360–530 of human MAP4K3 after PCR cloning from a MAP4K3 cDNA (Origene) into pGEX6P-1. The antibody was subsequently affinity-purified using the immunogen chemically coupled to glutathione–agarose. A monoclonal antibody against Rheb (3H6) was isolated at the Cancer Research U.K. London Research Institute by immunizing mice with a GST-fusion protein against Rheb amino acids 1–167 and a hybridoma produced by standard protocols. The monoclonal antibody against S6K1 was from Transduction Laboratories. A polyclonal anti-dTsc1 antibody and anti-dS6K antibodies were provided by D. J. Pan (McKusick–Nathans Institute of Medical Genetics, Johns Hopkins University, Baltimore, Md, U.S.A.) and Professor D. Alessi (Department of Biochemistry, University of Dundee, Dundee, U.K.) respectively. All other antibodies were purchased from Cell Signaling Technologies.
Plasmid constructs
pRK5 S6K1–GST and pcDNA 3xHA (haemagluttinin) 4E-BP1 have been described previously [6]. pRK5mycMAP4K3 was constructed using a full-length cDNA clone (Origene, accession number NM_003618). A mutation of the DFG motif (to AFG) was made using the QuikChange® site-directed mutagenesis kit (Stratagene) and sequenced to ensure only the correct mutation had been introduced. pRK5Msn (misshapen) and pCINeo HA-TNIK were provided by Dr Edward Skolnik (New York University Medical Centre, New York, NY, U.S.A.) and Dr K. Kariya (Division of Cell Biology, University of Ryukyus, Okinawa, Japan) respectively. pGEX GST-S6 was provided by Dr Andrew Tee (MSI/WTB Complex, University of Dundee, U.K.) and GST–S6 was produced by standard purification on glutathione-agarose.
Production of dsRNA (double-stranded RNA) for RNAi of kinases in Drosophila S2 cells
Using database searches (www.kinase.com/kinbase), 239 protein kinases in the Drosophila genome were identified, and the position of each kinase dsRNA template in a whole-genome Drosophila RNAi library (MRC Geneservices, U.K.) was determined. Each kinase template (1 μl) was diluted in 50 μl of TE buffer [10 mM Tris/HCl (pH 8.0) and 1 mM EDTA], 5 μl of which was amplified by the Expand Hi-Fidelity PCR kit (Roche Diagnostics) using T7 primers. PCR products were then analysed by agarose gel electrophoresis, purified by ethanol precipitation, and 1 μg of DNA was used as a template for dsRNA synthesis using an in vitro transcription kit (Ambion). dsRNAs were purified by lithium chloride precipitation according to the manufacturer's instructions, before quantity and quality were determined by agarose gel electrophoresis; subsequently 202 kinase dsRNAs were used for screening. For addition of dsRNA, Drosophila S2 cells were diluted to a final concentration of 1×106 cells/ml in standard Schneider's medium (Invitrogen). For the primary screen dsRNA additions were scaled down to a 24-well format and kinase dsRNAs added in equal combination with dsRNA targeting dTsc1 [7] (total of 30 μg/ml dsRNA). The cDNAs for positives identified in a primary screen were subsequently cloned into pGEM-T (Promega) and sequenced to verify correct annotation of the gene. Positives were confirmed in a secondary screen in 12-well dishes; 1 ml of cells were plated per well of a 12-well cell culture dish, and allowed to settled for at least 2 h. dsRNAs were added directly to the medium to a final concentration of 15 μg/ml for each dsRNA, followed immediately by vigorous agitation, with addition of water serving as control. Following incubation at room temperature (20 °C) for 1 h, 1 ml of Drosophila SFM (serum-free medium) containing 10% FCS (foetal calf serum; PAA laboratories) was added and cells were incubated for 5 days to allow for turnover of the target protein, before cell extracts were prepared in lysis buffer [50 mM Tris/HCl (pH 7.5), 1% (v/v) Triton X-100, 1 mM EDTA, 1 mM EGTA, 50 mM NaF, 50 mM β-glycerophosphate, 1 mM Na3VO4, 0.27 M sucrose, 0.1% (v/v) 2-mercaptoethanol, and complete protease inhibitors (Roche)].
Mammalian cell treatments, transfections, siRNAs (small interfering RNAs)
HeLa and HEK (human embryonic kidney)-293T cells were grown in DMEM (Dulbecco's modified Eagle's medium) containing 10% FCS (TCS Laboratories) and antibiotics. For plasmid transfections of HEK-293T cells, cells were plated at a density of 8×105 cells per well in 6-well plates, and incubated overnight before transfection. Plasmid transfections were performed using Lipofectamine™ 2000 (Invitrogen) according to the manufacturer's instructions, and cell extracts prepared 40 h post-transfection in lysis buffer. For siRNA transfections HeLa cells were plated at 1×105 cells/dish in six-well plates and transfected with Lipofectamine™ 2000 with the following siRNAs purchased from Dharmacon: control siRNA (scrambled S6K1 control [8]), Rheb (siRNA-2 [4]), TSC2 human siRNA Smartpool, M4K3-1 (nucleotides 847–865 of NM_003618 human MAP4K3). M4K3-2 corresponds to duplex 2 (MAP4K3 targeting Duopack, Invitrogen). All siRNAs were used in single transfections at a final concentration of 66 nM, whereas for double transfections the final concentrations were 33 nM. For amino acid deprivation experiments in HeLa or HEK-293T cells, cells in 6-well plates were serum-starved for 1 h (HeLa) or 16 h (HEK-293T), washed once in DPBS (Dulbecco's PBS) containing 1×MEM (minimal essential medium) vitamins and 10 mM glucose, and maintained in this medium for 5–60 min. To restimulate with amino acids DPBS was replaced with serum-free DMEM (also containing 10 mM glucose).
In vitro assay of S6K and MAP4K3 activity
S6K–GST was purified from cell extracts using 10 μl glutathione-agarose (Sigma–Aldrich) for 2 h at 4 °C. Beads were washed three times with lysis buffer plus 0.5 M NaCl, and once with S6K kinase buffer [50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 0.1% (v/v) 2-mercaptoethanol, 10 mM magnesium acetate and 2.5 μM PKI (a protein kinase A inhibitor; Sigma–Aldrich)]. Kinase assays were performed in 50 μl of S6K kinase buffer supplemented with 100 μM ATP, 3 μCi of [γ-32P]ATP and 3 μg of GST–S6. Reactions were carried out at 30 °C and terminated after 30 min by the addition of 2X SDS/PAGE sample buffer. Proteins were separated by SDS/PAGE, transferred to PVDF membranes whereupon 32P incorporation into GST–S6 was determined by autoradiography. Myc-tagged MAP4K3 was immuno-precipitated from cell extracts using 5 μg of anti-Myc 9E10 monoclonal antibody plus 20 μl of Protein G-Sepharose for 2 h at 4 °C. Immunoprecipitates were washed three times with lysis buffer plus 0.5 M LiCl, and once with Ste20 kinase buffer [20 mM Tris/HCl (pH 7.2), 25 mM β-glycerophosphate, 5 mM EGTA, 1 mM Na3VO4, 0.1% (v/v) 2-mercaptoethanol and 15 mM MgCl2)]. Kinase assays were carried out in 50 μl of Ste20 kinase buffer supplemented with 100 μM ATP, 3 μCi of [γ-32P]ATP and 5 μg of MBP (myelin basic protein; Sigma–Aldrich). Reactions were carried out at 30 °C and terminated after 10 min by the addition of 2X SDS/PAGE sample buffer. Proteins were separated by SDS/PAGE, transferred to PVDF membranes whereupon 32P incorporation into MBP was determined by autoradiography and subsequently analysed with a phosphorimager. Phosphorimager quantitation of S6K1–GST and Myc-MAP4K3 kinase assays were performed on three parallel dishes from a single experiment using ImageQuant software (Amersham Biosciences).
FACS analysis
HeLa cells transfected with siRNAs and maintained in DMEM containing 10% FCS (one 6-well plate per condition) were harvested after 72 h by trypsinization and washed twice in PBS. Cells were fixed with 70% ethanol for 30 mins at 4 °C. The fixed cells were centrifuged at 400 g for 3 min, washed twice with PBS and treated with 100 μg/ml ribonuclease (Roche) at room temperature for 15 min. Fixed cells were stained with propidium iodide at a final concentration of 50 μg/ml, and subjected to FACS analysis on a Becton Dickonson LSRII to determine the forward scatter of G1-, S- and G2/M-phase populations. Analysis of FACS and overlays of forward scatter were performed using CellQuest Pro software.
RESULTS
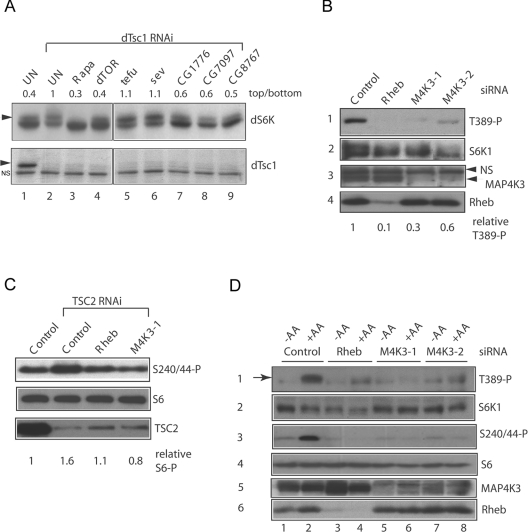
To activate dS6K in Drosophila S2 cells we depleted dTsc1, which activates dS6K [7] via Rheb, a small GTPase inhibited by TSC1-2 [4], and monitored dS6K phosphorylation by SDS/PAGE. As in other cell types, activation of dS6K led to a slower-migrating hyperphosphorylated species that was diminished by treating cells with the mTORC1 inhibitor rapamycin or depleting dTOR by RNAi (Figure 1A, lanes 3 and 4). Using dsRNA to co-suppress approx. 200 Drosophila protein kinases, as well as dTsc1 we identified three kinases not implicated previously in mTOR signalling that are required for dS6K phosphorylation in this setting (Figure 1A, lanes 7–9). Suppression of CG1776, CG7097 or CG8767, decreased dS6K phosphorylation in S2 cells, whereas co-depletion of 196/202 of the other kinases tested (such as sev and tefu, Figure 1A, lanes 5 and 6) did not.
Figure 1. Identification of MAP4K3 as an activator of mTOR signalling to S6K.
(A) Co-depletion of dTsc1 with Drosophila kinases by dsRNA addition to S2 cells. Upper panels, immunoblots of dS6K; lower panels, immunoblots of dTsc1 following dsRNA addition. Depletion of dTsc1 leads to increased abundance of a hyperphosphorylated species of dS6K (arrowhead) compared with control untreated cells (UN) that is reversed by treatment with 50 nM rapamycin for 1 h (Rapa) or co-depletion of dTOR, CG1776, CG7097 or CG8767. The ratio of hyperphosphorylated dS6K (upper panel) to hypophosphorylated dS6K (lower panel) is indicated above the immunoblots following quantitation of the autoradiographs with ImageQuant software. NS indicates a non-specific band detected with the dTsc1 antibody. BLAST homology of the predicted Drosophila proteins indicates that the closest human orthologues of CG1776, CG7097 and CG8767 respectively are: myosin light chain kinase (E value 3e -77), MAP4K3 (E value 4e -131) and c-Mos (E value 1e -18). (B) Suppression of S6K1 Thr389 by RNAi of MAP4K3 in HeLa cells. HeLa cells were transfected in DMEM containing 10 mM glucose and 10% FCS with control siRNA, Rheb siRNA and two distinct siRNA duplexes (M4K3-1 and M4K3-2) targeting MAP4K3. After 72 h cells were serum-starved for 1 h and lysates analysed by immunoblotting. Panel 1, phosphorylation of S6K1 at Thr389 detected with a phospho-specific antibody; panel 2, reprobing of panel 1 with a monoclonal antibody to total S6K1; panel 3, MAP4K3 levels detected with a polyclonal anti-MAP4K3 antibody; panel 4, Rheb levels detected with a monoclonal anti-Rheb antibody. Note: the blots in these panels were spliced to remove a lane corresponding to treatment with a MAP4K3 siRNA that did not effectively knockdown MAP4K3 expression. The ratio of Thr389 phosphorylated (T389-P) to total S6K following siRNA treatments is shown and was quantified from scanned autoradiographs using ImageQuant software, relative to control siRNA treatment which was assigned a value of 1. (C) Depletion of MAP4K3 by RNAi inhibits phosphorylation of S6 induced by depletion of TSC2. HeLa cells were transfected with control siRNA alone or TSC2 siRNA together with control siRNA or siRNAs targeting Rheb or MAP4K3 (M4K3-1) and lysates prepared as in (B). Upper panel, phosphorylation of S6 at Ser240/244 detected with a phospho-specific antibody; middle panel, duplicate gel probed with an antibody to S6; lower panel, TSC2 detected with a polyclonal anti-TSC2 antibody. The ratio of Ser240/244 phosphorylated (S240/44-P) total S6 following siRNA treatments is shown and was quantified from scanned autoradiographs using ImageQuant software, relative to control siRNA treatment which was assigned a value of 1. (D) Depletion of MAP4K3 suppresses S6K1 Thr389 phosphorylation induced by amino acid restimulation. HeLa cells were transfected and serum-starved for 1 h as in (B). For amino acid depletion/restimulation, duplicate wells transfected with control siRNA, Rheb siRNA or MAP4K3 siRNAs were transfered into DPBS containing 10 mM glucose and 1×MEM with vitamins (−AA) for 30 min or the same treatment followed by transfer into serum-free DMEM (containing 10 mM glucose) for 30 min (+AA). Panel 1, phosphorylation of S6K1 at Thr389 (arrow) detected with a phospho-specific antibody; panel 2, duplicate gel probed with an antibody to total S6K1; panel 3, phosphorylation of S6 at Ser240/244 detected with a phospho-specific antibody; panel 4, duplicate gel probed with an antibody to total S6; panel 5, MAP4K3 levels detected with a polyclonal anti-MAP4K3 antibody; panel 6, Rheb levels detected with a monoclonal anti-Rheb antibody.
BLAST homology searches revealed that the closest human orthologue of Drosophila CG7097 is MAP4K3 (also known as GLK (germinal centre-like kinase [9]), a Ste20-related MAP4K. We therefore tested whether MAP4K3 suppression by RNAi in mammalian cells also inhibits S6K1 phosphorylation as occurs in Drosophila cells. S6K1 activity is regulated by multi-site phosphorylation, including Thr389 in the hydrophobic motif, a site that may be directly phosphorylated by mTORC1 [10] and is essential for enzyme activity. Using RNAi we suppressed Rheb or MAP4K3 in HeLa cells and measured phosphorylation of S6K1 at Thr389 in DMEM containing amino acids or following amino acid removal and restimulation. In DMEM medium, phosphorylation of S6K1 at Thr389 is strongly inhibited by RNAi of Rheb, and partially by two RNAi sequences that suppress MAP4K3 expression (Figure 1B). We consistently observe that a more complete knockdown of MAP4K3 expression is required to elicit effective suppression of S6K1 phosphorylation at Thr389 (results not shown), suggesting that MAP4K3 may be a very active or abundant kinase that must be very effectively suppressed to fully inhibit signalling to S6K1. To verify that the effect of MAP4K3 on maintaining S6K1 activity is not via inhibition of TSC1-2, we suppressed TSC2 by RNAi in HeLa cells and co-depleted Rheb or MAP4K3. Depleting TSC2 alone stimulated S6 phosphorylation at the site phosphorylated only by S6K (Ser240/244), and this activation is suppressed by co-depletion of Rheb or MAP4K3, indicating that the effect of MAP4K3 on promoting S6K1 activity is independent of effects on TSC1-2 (Figure 1C).
Since amino acid activation of S6K1 also appears to be independent of TSC1-2 [11,12], we asked whether MAP4K3 was involved in the activation of S6K1 by amino acids. As previously reported [12], stimulation of HeLa cells with amino acids induces phosphorylation of S6K1 Thr389 and Ser240/244 phosphorylation of S6 that is strongly suppressed by an RNAi sequence targeting Rheb (Figure 1D, lanes 2 and 4), but also by two siRNAs targeting MAP4K3 (Figure 1D, lanes 6 and 8). Since MAP4K3 has been proposed to be involved in activation of JNK (c-Jun N-terminal kinase) [9], we also tested whether it is required for JNK activation in response to two common stresses. However, both osmotic stress and treatment with the protein synthesis inhibitor anisomycin induced equivalent phosphorylation of JNK in control HeLa cells or in cells in which MAP4K3 was suppressed (results not shown). Thus MAP4K3 is the likely mammalian orthologue of Drosophila CG7097, and plays an unexpected role in the phosphorylation of S6K1.
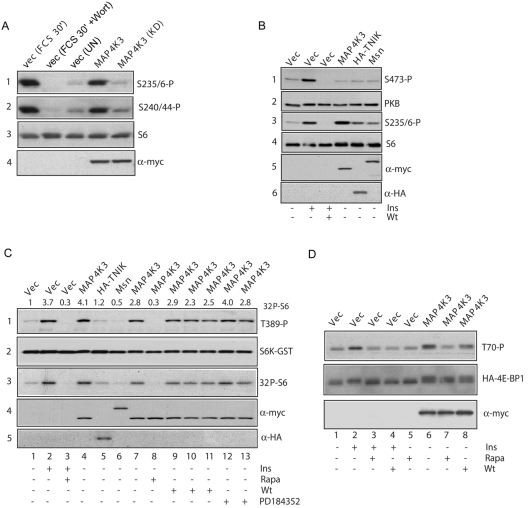
To lend support to the loss-of-function analyses we explored whether overexpression of MAP4K3 in HEK-293T cells increased mTOR signalling resulting in phosphorylation of S6. Overexpression of Myc epitope-tagged wild-type MAP4K3 but not a kinase-inactive mutant (of the conserved DFG motif) stimulated phosphorylation of S6 at both Ser235/236 and Ser240/244 to a level similar to stimulation with serum (Figure 2A). As a control for the specificity of phosphorylation, we similarly overexpressed MAP4K3 and determined the phosphorylation of the related AGC family kinase PKB at its hydrophobic motif Ser473 site, since increased phosphorylation of this site generally correlates with increased PKB activity. Unlike S6K, PKB activity is not regulated by amino acids [5] and is regulated by an mTOR complex (mTORC2) that is insensitive to rapamycin [2], unlike the mTORC1 complex upstream of S6K1. However, MAP4K3 overexpression does not induce PKB Ser473 phosphorylation under conditions where it increases phosphorylation of S6 to a level similar to stimulation with insulin, whereas expression of related control Ste20 family kinases TNIK and Msn poorly stimulate S6 phosphorylation (Figure 2B), indicating that MAP4K3 is unlikely to be a general regulator of mTOR activity.
Figure 2. MAP4K3 activates mTOR signalling to S6K and 4E-BP1.
(A) Overexpression of MAP4K3 activates phosphorylation of S6 and is dependent upon kinase activity. HEK-293T cells were transfected with 2.5 μg of pRK5myc vector or pRK5myc MAP4K3 wild-type or AFG kinase-dead MAP4K3 (KD). After 24 h, cells were serum-starved for 16 h and controls stimulated with 10% FCS for 30 min (FCS 30′) or pretreated with 100 nM wortmannin for 60 min prior to stimulation (FCS 30′+Wort) and lysates prepared. Panel 1, endogenous S6K phosphorylation detected with a phospho-specific antibody against Ser235/236; panel 2, S6 phosphorylation detected with a phospho-specific antibody against Ser240/244; panel 3, total S6; panel 4, an anti-Myc (9E10) antibody to detect expression of Myc-epitope tagged kinase. (B) Overexpression of MAP4K3 does not activate Ser473 phosphorylation of PKB. HEK-293T cells were transfected as in (A) with pRK5myc vector, pRK5mycMAP4K3, pRK5mycMsn or pCI HA TNIK and lysates were probed with antibodies as in (A), or with antibodies to PKB Ser473 (panel 1), total PKB (panel 2) or a HA epitope to detect HA-TNIK (panel 6). As a control for PKB activation, vector-transfected cells were stimulated with 1 μM insulin (Ins) for 30 min or pretreated with 100 nM wortmannin (Wt) for 60 min prior to stimulation. (C) Activation of S6K1 by MAP4K3 is rapamycin-sensitive, but independent of MEK and PI3K signalling. HEK-293T cells were transfected with the indicated plasmids as in (A) and (B) together with 0.1 μg of pRK5 S6K–GST. Prior to lysis, cells were pretreated with 50 nM rapamycin for 60 min where indicated (Rapa) or treated with rapamycin and stimulated with 1 μM insulin (Ins) for 30 min. For wortmannin and PD184352 treatments, cells were treated with 50 nM (lane 9), 100 nM (lane 10) or 200 nM (lane 11) wortmannin, or 1 μM (lane 12) or 10 μM (lane 13) PD184352 for 60 min prior to lysis. Panel 1, Thr389 phosphorylation of the S6K–GST reporter; panel 2, S6K–GST reporter detected with a monoclonal antibody to S6K1; panel 3, autoradiograph of an in vitro kinase assay measuring S6K1–GST activity with S6 substrate; panels 4 and 5, detection of kinase expression with the 9E10 antibody to the Myc epitope (panel 4) and HA epitope (panel 5). Values of S6K activity were determined by quantitation of the phosphorimage using ImageQuant software. (D) Overexpression of MAP4K3 activates phosphorylation of 4E-BP1. HEK-293T cells were transfected as in (A) with 2.5 μg of pRK5myc vector or pRK5mycMAP4K3 together with 1 μg of pcDNA-3xHA-4E-BP1 reporter. Treatments with 100 nM wortmannin (Wt) and 50 nM rapamycin (Rapa) and insulin stimulation were as in (C). Upper panel, HA–4E-BP1 phosphorylation at Thr70 detected with a phospho-specific antibody; middle panel, level of HA–4E-BP1 detected with an antibody to 4E-BP1; lower panel, level of MAP4K3 detected with an anti-Myc 9E10 antibody.
To examine whether MAP4K3 regulates phosphorylation of the hydrophobic motif Thr389 site in S6K1 and/or promotes S6K1 activity, we compared Thr389 phosphorylation and activity of a co-expressed S6K1 reporter induced by MAP4K3 with that of two related Ste20 kinases. Overexpression of MAP4K3, but not TNIK or Msn, led to increased reporter S6K1–GST Thr389 phosphorylation and activity similar to insulin stimulation (Figure 2C, lanes 2 and 4). The activation of reporter S6K1 by MAP4K3 is rapamycin-sensitive (Figure 2C, lane 8), but is relatively insensitive to concentrations of an inhibitor of PI3K (wortmannin) up to 200 nM (Figure 2C, lanes 9–11) or an inhibitor of MEK (MAPK/ERK kinase) 1-2 (PD184352) up to concentrations of 10 μM (Figure 2C, lanes 12 and 13), under conditions where these concentrations of inhibitors efficiently block activation of S6K1 by insulin and ERK1/2 by PMA (results not shown).
Lastly, in an overexpression experiment in HEK-293T cells we assessed the effect of MAP4K3 on 4E-BP1, an inhibitor of the translation initiation factor eIF4E which is similarly regulated by nutrients and mTORC1 [13,14]. Stimulation of cells with insulin (Figure 2D, lane 2) or overexpression of MAP4K3 also led to an accumulation of slower-migrating forms of 4E-BP1 that were hyperphosphorylated at Thr70, an inhibitory site thought to be regulated by mTORC1 [15] (Figure 2D, lane 6). Similar to the results for S6K1, increased phosphorylation of 4E-BP1 induced by MAP4K3 overexpression was inhibited by treatment with rapamycin (Figure 2D, lane 7), and was relatively insensitive to 100 nM wortmannin (Figure 2D, lane 8).
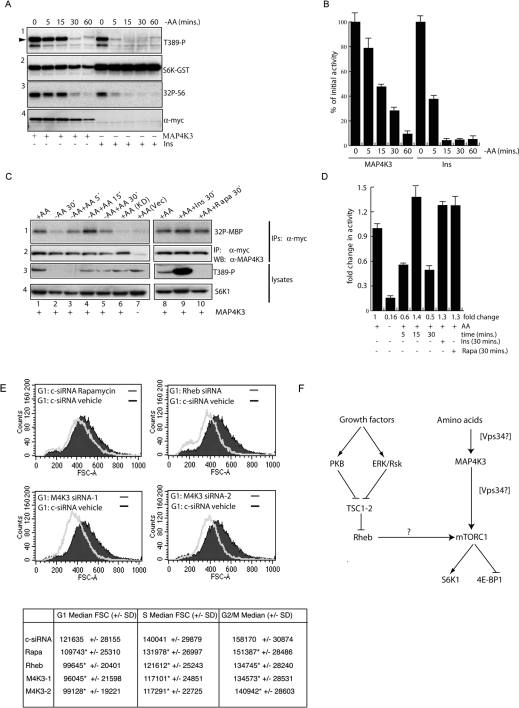
Because MAP4K3 is required for the activation of S6K1 by amino acids, we next sought to determine whether ectopic MAP4K3 overexpression rescued S6K1 from inactivation during amino acid withdrawal. Amino acid withdrawal leads to a rapid inactivation of S6K1 activity [5] associated with dephosphorylation at Thr389. Following stimulation of serum-starved HEK-293T cells with insulin or overexpression of MAP4K3 to activate S6K1, we removed amino acids and at varying times measured S6K1 activity and phosphorylation at the Thr389 site. We found that, when overexpressed, MAP4K3 significantly delayed dephosphorylation of Thr389 and inhibition of S6K1 activity (S6K1 remained approx. 50% active in cells overexpressing MAP4K3 compared with >10% for control insulin-stimulated after 15 min of amino acid withdrawal, Figures 3A and 3B), indicating that ectopic MAP4K3 overexpression partly uncoupled S6K1 from the inhibitory effect of amino acid withdrawal.
Figure 3. Involvement of MAP4K3 in amino acid signalling and growth.
(A) Ectopic MAP4K3 overexpression delays inactivation of S6K1 induced by amino acid withdrawal. HEK-293T cells were transfected as in Figure 2(A) with 2.5 μg of pRK5myc vector or pRK5mycMAP4K3 and 0.1 μg of pRK5 S6K-GST reporter and serum-starved for 16 h. Vector-transfected cells were subsequently stimulated for 30 min with 1 μM insulin (Ins, time 0) and transferred to DPBS as in Figure 1(D) for 5–60 min prior to lysis, or if expressing MAP4K3 left unstimulated and transferred to DPBS for 5–60 min (MAP4K3). Panel 1, Thr389 phosphorylation (arrowhead) of S6K–GST reporter; panel 2, S6K–GST reporter detected with a monoclonal antibody to S6K1; panel 3, autoradiograph of an in vitro kinase assay measuring S6K1–GST activity with S6 substrate; panel 4, detection of MAP4K3 expression with the 9E10 antibody against the Myc epitope. (B) Quantitation of [32P]GST-S6 shown in (A) from triplicate assays of a single experiment repeated three times with similar results. Arbitrary values for [32P]GST-S6 were measured using a phosphorimager and results expressed separately for the MAP4K3 and insulin-stimulated samples as a percentage (means±S.D.) of the initial activity (100% at time 0, prior to amino acid withdrawal) of each group. (C) MAP4K3 is regulated by amino acids, but not insulin or mTORC1. HEK-293T cells were transfected with 0.1 μg of pRK5myc vector (+AA Vec), 0.1 μg of pRK5mycMAP4K3 AFG kinase-dead mutant (+AA KD) or 0.1 μg of pRK5mycMAP4K3 wild-type (MAP4K3) and serum-starved for 16 h. Right panels: serum-starved cells (+AA) were either stimulated for 30 mins with 1 μM insulin (+AA+ Ins 30′) or treated with 50 nM rapamycin for 30 min (+AA+Rapa 30′); left panels: serum-starved cells were transferred to DPBS (−AA) for 30 min and lysates prepared or were subsequently restimulated with amino acids by transferring to DMEM lacking serum for 5–30 minutes (−AA+AA). Panel 1, autoradiograph of an in vitro kinase assay of anti-Myc immunoprecipitates with MBP substrate; panel 2, MAP4K3 levels in one-fifth of the 9E10 immunoprecipitate prior to the in vitro kinase assay detected with a polyclonal anti-MAP4K3 antibody; panel 3, phosphorylation of S6K1 at Thr389 detected with a phospho-specific antibody; panel 4, reprobing of panel 2 with a monoclonal antibody against total S6K1. (D) Quantitation of [32P]MBP shown in (C) from triplicate assays of a single experiment repeated three times with similar results. Arbitrary values for [32P]MBP were measured using a phosphorimager and processed with ImageQuant software. Results are expressed as fold difference of the activity in the presence of amino acids (+AA). Quantitation reveals an 6.3-fold inhibition of MAP4K3 activity following 30 min of amino acid withdrawal and an 8.8-fold activation after 15 min of amino acid re-addition to amino acid-deprived cells. In the presence of amino acids both insulin stimulation and treatment with rapamycin led to a 1.3-fold activation of MAP4K3 activity. (E) MAP4K3 regulates cell size in HeLa cells. HeLa cells maintained in DMEM with 10% FCS were transfected with control siRNA and left untreated or treated with 50 nM rapamycin for 48 h, or transfected with siRNAs against Rheb or MAP4K3 (M4K3-1 or M4K3-2) and analysed by FACS analysis after propidium iodide staining. G1 cells analysed by forward scatter from untreated control siRNA-transfected cells (black) are shown overlaid with the rapamycin-treated cells, and Rheb siRNA- or MAP4K3 siRNA-transfected cells (grey). In the Table the differences in values of median FSC (forward scatter) of G1-, S- and G2/M-phase cells (marked by *) are highly statistically significant by z-test (P<0.001) compared with control siRNA vehicle-treated cells. (F) Model for the involvement of MAP4K3 in mTOR signalling. Growth factors activate the mTORC1 pathway primarily via activation of protein kinases such as PKB [3] and/or ERK/Rsk [17,18] leading to inactivation of the TSC1-2 tumour suppressor and increased Rheb.GTP levels. In turn Rheb.GTP has been proposed to activate mTORC1 kinase activity, although uncertainty exists on this mechanism as binding of Rheb to mTOR is independent of bound nucleotide [20], unlike other small GTPase/effector interactions (indicated by the question mark). Since Rheb.GTP levels are insensitive to amino acid withdrawal [12], but both MAP4K3 and S6K activities are sensitive (the present study and [5]), we suggest that MAP4K3 may provide a specific link between amino acids and S6K1 activity. The involvement of Vps34 in amino acid signalling to S6K is also indicated, and since it is currently unclear whether this lipid kinase acts upstream or downstream of MAP4K3, both possibilities are indicated. Since phosphorylation of 4E-BP1 (at Thr70, a site regulated by mTORC1 [15]) is also regulated by MAP4K3, and MAP4K3 activity is insensitive to rapamycin, we suggest that MAP4K3 may regulate both S6K and 4E-BP1 activities by acting upstream of mTORC1.
These data suggested to us that amino acid sufficiency might itself regulate MAP4K3 activity. To test this possibility, we transiently transfected Myc-tagged MAP4K3 into HEK-293T cells at a level of expression that did not significantly activate S6K1, and removed amino acids, or re-added them after deprivation for 30 min. Like S6K1, we found that MAP4K3 activity measured against the generic substrate MBP was strongly inhibited (to approx. 16% of initial activity) after 30 min of amino acid withdrawal (Figure 3C, lane 2). MAP4K3 was rapidly restimulated by re-addition of amino acids and was activated 3.8-fold within 5 min, with peak activity observed 15 min after amino acid re-addition (an 8.8-fold activation compared with the amino acid-deprived condition, Figure 3D). Interestingly, phosphorylation of S6K1 at Thr389 was somewhat delayed relative to MAP4K3 activation and was detectable only 15–30 min after amino acid re-addition (Figure 3C, compare lanes 3 and 4 of panels 1 and 3). As controls, we assessed MAP4K3 activity following either insulin stimulation or mTORC1 inhibition with rapamycin. However, unlike S6K1, Myc-MAP4K3 was not activated by insulin or inhibited by rapamycin (Figure 3C, panel 1, lanes 9 and 10) under conditions where these treatments respectively stimulate or inhibit S6K1 Thr389 phosphorylation (Figure 3C, panel 3, lanes 9 and 10). Thus, like S6K1, MAP4K3 activity is regulated by amino acid sufficiency, but unlike S6K1 is independent of mTORC1 and not activated by insulin stimulation.
The mTOR pathway regulates cell growth, at least in part, by regulation of S6K and eIF4E activities [16]. To determine whether MAP4K3 regulates cell growth we again used RNAi to down-regulate MAP4K3 or Rheb in HeLa cells growing in serum, or treated cells with rapamycin for a sustained (48 h) period to block mTORC1 activity, and analysed the size of populations in G1, S and G2/M cell cycle phases by FACS analysis. The results (Figure 3E) showed that suppression of either MAP4K3 (using either the M4K3-1 or M4K3-2 siRNAs, lower panels) or Rheb (upper-right panel) markedly decreased cell size in G1-phase and at all cell cycle phases (Figure 3E, table) in a similar manner to sustained mTORC1 inhibition caused by treating cells with rapamycin. Thus MAP4K3 function promotes both mTOR signalling to S6K1 and 4E-BP1 and cell growth in HeLa cells. On the basis of these observations we propose a model in which MAP4K3 is down-stream of a signal provided by amino acids, but upstream of mTORC1 (Figure 3F).
DISCUSSION
In metazoans both nutrient amino acids and mitogens are required to promote mTOR signalling and cell growth [1]. Activation of mTOR signalling by mitogens involves activation of protein kinases such as PKB [3], p90RSK [17] and ERK [18] that have been reported to inhibit the function of the TSC1-2 complex which is then thought to lead to increased GTP loading of the Ras-like GTPase Rheb [4]. In contrast, amino acids do not appear to regulate Rheb GTP loading [11,12], and, although a class III PI3K has been implicated upstream of mTORC1 [12,19], no analogous protein kinases have been identified that respond to amino acids and activate mTOR signalling. We found, both in Drosophila and in mammalian cells, that suppression of MAP4K3 inhibited mTOR signalling to S6K in the context of activation of the pathway induced by deficiency in TSC1-2. Since a variety of oncogenic signalling pathways inhibit TSC1-2 [3,17,18], and TSC1-2 is predicted also to be inactivated by loss of function of the tumour suppressor PTEN (phosphatase and tensin homologue deleted on chromosome 10) via activation of PKB [3], MAP4K3 may represent a promising new candidate for inhibition of the pathway in diseases of TSC1-2 or PTEN loss-of-function. Although we have not defined the mechanism of MAP4K3 action on mTOR signalling, our data on the positive regulation of both S6K1 activity and phosphorylation of 4E-BP1 by MAP4K3 overexpression points to the mTORC1 complex as the likely target of MAP4K3 action. Interestingly, addition of excess amino acids (relative to the normal concentrations found in DMEM) has been reported to fully activate S6K in the absence of growth factors [5], which is similar to our finding with overexpressed MAP4K3, suggesting that MAP4K3 may be activated further by amino acid excess. Future studies will be required to clarify these points. However, having defined the action of a new protein kinase in the mTOR pathway, delineating the signals downstream of amino acids that activate MAP4K3, and determining its mechanism of action will likely shed further light on nutrient regulation of cell growth.
In conclusion, our data indicate that amino acids stimulate the activity of a Ste20-family kinase, MAP4K3, with maximal kinase activation occurring concordant with phosphorylation of the Thr389 site of S6K1. In HeLa cells MAP4K3 activity is required for amino acid-induced activation of S6K1 and mediates rapamycin-senstive signalling to two effectors of mTORC1, but is not itself stimulated by insulin or inhibited by rapamycin. Lastly, MAP4K3 promotes cell growth in human HeLa cells in culture in a similar manner to Rheb and mTORC1.
Acknowledgments
We thank Dr E. Skolnick, Dr K. Kariya and Dr A. Tee for plasmids, Dr Buzz Baum (Ludwig Institute for Cancer Research, University College London) for advice on RNAi screening in Drosophila cells, and Pascal Meier and Chris Marshall for critical reading of the manuscript. Work in this study was funded by grants from Cancer Research UK and the Tuberous Sclerosis Association (UK), and an Institute of Cancer Research Studentship to G.M.F.
References
- 1.Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 2.Sarbassov D. D., Ali S. M., Sabatini D. M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 4.Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 5.Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 6.Yan L., Findlay G. M., Jones R., Procter J., Cao Y., Lamb R. F. Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J. Biol. Chem. 2006;281:19793–19797. doi: 10.1074/jbc.C600028200. [DOI] [PubMed] [Google Scholar]
- 7.Gao X., Zhang Y., Arrazola P., Hino O., Kobayashi T., Yeung R. S., Ru B., Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 8.Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener K., Wang X. S., Chen C., Meyer C. F., Keesler G., Zukowski M., Tan T. H., Yao Z. Activation of the c-Jun N-terminal kinase pathway by a novel protein kinase related to human germinal center kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9687–9692. doi: 10.1073/pnas.94.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnett P. E., Barrow R. K., Cohen N. A., Snyder S. H., Sabatini D. M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith E. M., Finn S. G., Tee A. R., Browne G. J., Proud C. G. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J. Biol. Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 12.Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., Gulati P., Byfield M. P., Backer J. M., Natt F., Bos J. L., Zwartkruis F. J., Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara K., Yonezawa K., Kozlowski M. T., Sugimoto T., Andrabi K., Weng Q. P., Kasuga M., Nishimoto I., Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 14.Gingras A. C., Raught B., Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 15.Gingras A. C., Raught B., Gygi S. P., Niedzwiecka A., Miron M., Burley S. K., Polakiewicz R. D., Wyslouch-Cieszynska A., Aebersold R., Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux P. P., Ballif B. A., Anjum R., Gygi S. P., Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P. P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 19.Byfield M. P., Murray J. T., Backer J. M. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 20.Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]