Abstract
Mitotic Aurora-A is an oncogene, which undergoes a cell-cycle-dependent regulation of both its synthesis and degradation. Overexpression of Aurora-A leads to aneuploidy and cellular transformation in cultured cells. It has been shown that the cell-cycle-dependent turnover of Aurora-A is mediated by Cdh1 (CDC20 homologue 1) through the anaphase-promoting complex/cyclosome (APC/C)–ubiquitin–proteasome pathway. We have described previously the identification of an Aurora-A kinase interacting protein, AURKAIP1 (formerly described as AIP), which is also involved in the destabilization of Aurora-A through the proteasome-dependent degradation pathway. In an attempt to investigate the mechanism of AURKAIP1-mediated Aurora-A degradation, we report here that AURKAIP1 targets Aurora-A for degradation in a proteasome-dependent but Ub (ubiquitin)-independent manner. AURKAIP1 inhibits polyubiquitination of Aurora-A. A non-interactive AURKAIP1 mutant that cannot destabilize Aurora-A restores ubiquitination of Aurora-A. An A-box mutant of Aurora-A, which cannot be targeted for proteasome-dependent degradation by Cdh1, can still be degraded by AURKAIP1. Inhibition of cellular ubiquitination either by expression of dominant negative Ub mutants or by studies in ts-20 (temperature sensitive-20) CHO (Chinese-hamster ovary) cell line lacking the E1 Ub activating enzyme at the restrictive temperature, cannot abolish AURKAIP1-mediated degradation of Aurora-A. AURKAIP1 specifically decreases the stability of Aurora-A in ts-20 CHO cells at the restrictive temperature, while cyclinB1 and p21 are not affected. This demonstrates that there exists an Ub-independent alternative pathway for Aurora-A degradation and AURKAIP1 promotes Aurora-A degradation through this Ub-independent yet proteasome-dependent pathway.
Keywords: Aurora-A, Aurora-A kinase interacting protein (AURKAIP), cell cycle, proteasome-dependent degradation, ubiquitin-independent degradation
Abbreviations: APC/C, anaphase-promoting complex/cyclosome; AURKAIP, Aurora-A kinase interacting protein; CDC4, cell division cycle 4; Cdh1, CDC20 homologue 1; Chfr, checkpoint with forkhead and ring finger domains; CHO, Chinese-hamster ovary; CHX, cycloheximide; HRP, horseradish peroxidase; Mdm2, murine double minute 2; MG132, carbobenzoxy-L-leucyl-L-leucyl-L-leucinal; Ni-NTA, Ni2+-nitrilotriacetate; NQO1, NAD(P)H quinone oxidoreductase 1; PI, propidium iodide; Rb, retinoblastoma; TBS, Tris buffered saline; TG, 6-thioguanine; TR-AURKAIP, N-terminal truncated AURKAIP; ts-20, temperature sensitive-20; Ub, ubiquitin
INTRODUCTION
Protein degradation plays an essential role in the regulation of many cellular physiological processes, particularly in cell cycle control, where cell cycle proteins are periodically expressed. Aberrant protein degradation could lead to an uncontrolled cell cycle and subsequently cancer. The Ub (ubiquitin)–proteasome system has evolved as the key machinery in the selective degradation of most intracellular short-lived regulatory or abnormal proteins [1–4]. Target proteins are covalently tagged with multiple Ubs, forming the polyubiquitin chain, which not only serves as the recognition signal for the 26 S proteasome, but also assists in the unfolding of target proteins. Ubiquitination requires the Ub activating enzyme (E1), the Ub conjugating enzyme (E2) and the Ub ligase (E3), where E3 confers the substrate specificity. The Ub-dependent degradation pathway is presumed to be involved in the degradation of most protein. However, some proteins can also be degraded in the absence of detectable prior ubquitination either directly by 20 S proteasome or by 26 S proteasome in the presence of ATP [5].
Aurora-A represents one of the many mitotic proteins, whose protein levels are temporally regulated by Ub-dependent proteolysis at the end of mitosis, before cells progress into the G1 phase of the cell cycle. Aurora-A is ubiquitinated by the Cdh1 (CDC20 homologue 1)-activated APC/C (anaphase-promoting complex/cyclosome), an E3 Ub ligase, through the recognition of the C-terminal D-box (destruction box) and N-terminal A-Box. Dephosphorylation of the highly conserved Ser51 in the A-box during mitotic exit could control the timing of Aurora-A degradation [6–8]. Regulation of Aurora-A degradation is very important, as ectopic expression of Aurora-A in human and rodent cells induces centrosome amplification, aneuploidy, transformed phenotype and tumour formation in nude mice [9,10]. Aurora-A is overexpressed in many types of cancer and the gene has been mapped to chromosome 20q13 region, which is frequently amplified in many human cancers [11–13]. Overexpression of Aurora-A correlates significantly with induction of aneuploidy, centrosome anomaly, poor prognosis and invasiveness of primary human tumours, and of experimental tumours in animal model systems [14,15].
Previously, in our attempt to understand the negative regulation of Aurora-A, we isolated a novel, direct negative regulator of Aurora-A, AURKAIP1 (Aurora-A kinase interacting protein 1) [16]. AURKAIP1 targets Aurora-A for degradation in a proteasome-dependent manner. AURKAIP1–Aurora-A interaction is necessary for AURKAIP1-mediated Aurora-A degradation. The exact mechanism of AURKAIP1-mediated Aurora-A degradation is unclear. In the present paper, we explore the mechanism of Aurora-A degradation in the AURKAIP1-regulated pathway. The results presented here demonstrate that AURKAIP1 facilitates proteasome-dependent degradation of Aurora-A by an alternative mechanism that is independent of ubiquitination. This implies that Aurora-A can be delivered to the proteasome via two distinct Ub-dependent and Ub-independent pathways.
MATERIALS AND METHODS
Plasmids and cloning
HA-tagged p21 expression construct was a gift from Dr Michele Pagano (Department of Pathology, NYU Cancer Institute, New York University School of Medicine, New York, NY, USA). His6-tagged wild-type Ub and HA-tagged Ub K48R mutant expression construct was a gift from Dr Ivan Dikic (Institute of Biochemistry II, Goethe University Medical School, Frankfurt, Germany). Addition of a His6-tag to the Ub K48R mutant was carried out by PCR and cloned into pcDNA3 (Invitrogen); His6-tagged wild-type Ub plasmid was used as the template to generate the K48R/K63R double mutant using the GeneEditor™ in vitro site-directed mutagenesis system (Promega); FLAG- and HA-tagged human Aurora-A were PCR amplified and cloned into pcDNA3. The A-box mutant (S51D) of Aurora-A in pcDNA3 was also generated using the GeneEditor™ in vitro site-directed mutagenesis system. HA- and FLAG-tagged human AURKAIP1 and TR-AURKAIP1 (N-terminal truncated AURKAIP) were PCR amplified and cloned into pcDNA3. All cloned sequences were verified by sequencing.
Construction of Aurora-A deletion mutants
N-terminal and C-terminal truncations in Aurora-A were generated using a PCR-based approach. A 300- and a 600-bp deletion of the N- and C-terminus of Aurora-A respectively, were generated by PCR using primers flanking the desired regions of Aurora-A. All forward primers were designed to introduce a FLAG-tag with initiator codon into the N-terminus of truncated Aurora-A proteins. The PCR amplified fragments containing defined Aurora-A deletions were then cloned into pcDNA3. Expected deletions in all Aurora-A deletion constructs were subsequently verified by sequencing.
Antibodies
Mouse monoclonal anti-FLAG M2 antibody (diluted 1:2000; Stratagene); rabbit polyclonal anti-FLAG (diluted 1:2000 Sigma); mouse monoclonal anti-β tubulin antibody (diluted 1:1000; Sigma); mouse monoclonal anti-(HA tag) (diluted 1:2000; Sigma); mouse monoclonal anti-IAK1 (Aurora-A kinase) (diluted 1:1000; BD Transduction); rabbit polyclonal anti-(cyclin B1) antibody (diluted 1:3000; Santa Cruz Biotechnology); and mouse monoclonal anti-(His6 tag) antibody (diluted 1:1000; Sigma), were used. All HRP (horseradish peroxidase)-conjugated secondary antibodies (Pierce) were used at 1:6000 to 1:8000 dilutions.
Cell culture, transfection and drug treatment
ts20 (temperature sensitive-20) TG (6-thioguanine) mouse cells were obtained from Dr Harvey Ozer, Department of Microbiology and Molecular Genetics, International Center for Public Health, Newark, New Jersey, U.S.A. ts20-CHO (Chinese-hamster ovary) cells were obtained from Dr Ger J. Strous, Department of Cell Biology, University Medical Center Utrecht, Utrecht, The Netherlands. ts20TG mouse cells and COS7 cells were maintained in Dulbecco's modified Eagle's medium (Sigma) and HeLa cells were maintained in RPMI 1640 medium (Sigma) supplemented with 10% (v/v) foetal bovine serum (JRH). ts20 CHO cell line, which harbours the temperature-sensitive mutation in E1 Ub-activating enzyme was maintained in α-MEM (Sigma), supplemented with 4.5 g/l glucose and 10% (v/v) foetal bovine serum at 30 °C. Cells were incubated at 40 °C for 24 h to inactivate the E1 Ub-activating enzyme. Transfection of cultured cell lines was carried out using Lipofectamine™ 2000 (Invitrogen). Typically, cells were grown in their respective growth medium without antibiotic and transfected with expression plasmids using Lipofectamine™ 2000 and OPTIMEM (Invitrogen) according to the manufacturer's recommendations. To block the 26 S proteasome-mediated protein degradation, cells were treated with 20 μM MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal) or lactacystin for 16 h. To block the protein synthesis, the ts20-CHO and ts20TG mouse cells were treated with 50 μg/ml CHX (cycloheximide) (Sigma) for the indicated times.
Cell cycle synchronization and flow cytometry
To obtain cells arrested at G1/S and M phases of the cell cycle, asynchronously growing HeLa cells were treated with aphidicolin (1 μg/ml) for 24 h and nocodazole (0.1 μg/ml) for 16 h respectively. Cells treated similarly with the vehicle (DMSO) were used as the control. The extent of synchronization was assessed by PI (propidium iodide) staining and flow cytometry. Briefly, cells were harvested and fixed with 70% ethanol overnight at 4 °C. The fixed cells were washed twice in PBS containing 0.1% Triton X-100, resuspended in PI staining solution (50 μg/ml PI, 100 μg/ml RNase A and 0.1% Triton X-100) and incubated for 1 h at room temperature (25 °C) before analysis. DNA content was analysed using the FACSCalibur system (Becton Dickinson) and the data were analysed using the Modfit software (Verity Software House).
Cell lysis and immunoblotting
Typically, cells were lysed for 15 min on ice in lysis buffer [1× TBS (Tris-buffered saline; 50mM Tris/HCl, 150 mM NaCl, pH 7.6); 10% (v/v) glycerol and 1% (v/v) Nonidet P40] containing protease inhibitor cocktail (Roche). The lysates were then cleared by centrifugation at 16000 g for 10 min at 4 °C. Alternatively cells were also lysed in 1× Laemmli buffer (25 mM Tris base, 192 mM glycine and 0.1% SDS), followed by pulsed sonication (5×5 s pulses with a 10 s interval between pulses; Vibra Cell, Sonics) on ice and subsequently cleared by centrifugation at 16000 g at 4 °C. The protein concentration of the lysates were assayed using the Bio-Rad Protein Assay reagent (Bio-Rad Laboratories). The proteins (50–100 μg) were resolved by SDS/PAGE on 10 or 12% (v/v) gels. The proteins were subsequently transferred on to nitrocellulose membranes (Gelman Laboratory). After incubation in blocking buffer [5% (w/v) non-fat dried milk in TBS], the blots were incubated with various antibodies at their optimal dilutions overnight at 4 °C. The HRP-conjugated secondary antibodies [goat anti-rabbit–HRP and goat anti-mouse–HRP (Pierce)] were diluted at 1:8000 in blocking buffer and incubated with the blot for 1 h at room temperature. The conjugated secondary antibodies were detected by SuperSignal Pico or Dura Chemiluminescence detection system (Pierce).
In vivo ubiquitination assay
The in vivo ubiquitination assays were carried out essentially as described in [17]. HeLa cells were co-transfected with HA- tagged Aurora-A, His6-tagged wild-type, K48R or K48R/K63R Ub mutants and either pcDNA3 or AURKAIP1. The cells were treated, 36 h post-transfection, with 20 μM MG132 for a further 12 h, prior to harvesting. The cells were lysed in 1 ml Buffer G (6 M guanidinium chloride, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 10 mM imidazole) per 60-mm-diameter dish. The lysate was sonicated in pulses to reduce viscosity and incubated with 100 μl of 50% slurry of Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) with rotation for 75 min at room temperature. The beads were washed three times with 1 ml Buffer G, twice with 1 ml of Wash Buffer I (Buffer G diluted 1:4 in 25 mM Tris/HCl, pH 6.8 and 20 mM imidazole), and twice with 1 ml of Wash Buffer II (25 mM Tris/HCl, pH 6.8, and 20 mM imidazole). The bound proteins were eluted by boiling the beads in 2× SDS sample buffer [0.24 M Tris/HCl, pH 6.8, 2.5 % (w/v) SDS, 20% (v/v) glycerol, 8% (v/v) 2-mercaptoethanol and 0.02% Bromophenol Blue] supplemented with 100 mM EDTA and analysed by immunoblotting.
RESULTS
Degradation of Aurora-A by AURKAIP1 is cell cycle-independent
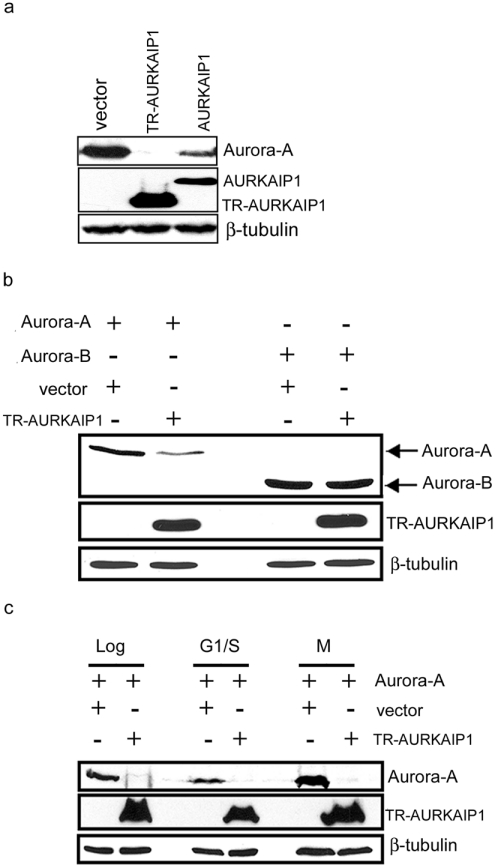
Multiple regulators of Aurora-A kinase stability such as Cdh1 [CDC (cell division cycle)20 homologue 1], CDC4 and Chfr (checkpoint with forkhead and ring finger domains), which have been described recently [18,19], target Aurora-A through the Ub- and proteasome-dependent degradation pathway. Previously, we have described the isolation of AURKAIP1, a novel interacting partner and negative regulator of Aurora-A, which also targets Aurora-A for degradation through proteasome-dependent pathway [16]. As a further step towards understanding the role of AURKAIP1 in Aurora-A degradation, we investigated the mechanism by which AURKAIP1 destabilizes Aurora-A. Although the full-length AURKAIP1 was capable of interacting and destabilizing Aurora-A, TR-AURKAIP1, which was originally isolated in a yeast dosage suppressor screen [16], was more stable and potent than the full length AURKAIP1 in the destabilization of Aurora-A (Figure 1a). This suggests that the N-terminal 87 amino acids of AURKAIP1 might harbour putative negative elements, which render the full-length protein less stable and thus less effective. To verify the specificity of the truncated AURKAIP1 in destabilizing Aurora-A, in vivo degradation assays were carried out with Aurora-B. The results presented in Figure 1(b) show that the truncated from of AURKAIP1 did not have any destabilizing effect on Aurora-B, whereas it could destabilize Aurora-A more effectively, suggesting that the truncated AURKAIP1 contains all the necessary elements to specifically destabilize Aurora-A. Hence all the subsequent experiments designed to understand the mechanism of Aurora-A degradation were performed with the truncated form of AURKAIP1. To address the role of AURKAIP1 in cell-cycle-dependent turnover of Aurora-A, we performed in vivo degradation assays in cells arrested at different phases of the cell cycle. Cells were co-transfected with Aurora-A and TR-AURKAIP1 or empty vector as a control, and synchronized with cell-cycle phase-specific inhibitors before analysis. Monitoring cell cycle distribution by flow cytometry indicated that both aphidicolin and nocodazole arrested more than 80% of the cells in the G1/S and G2/M phases of the cell cycle respectively (see Supplementary Figure S1a at http://www.BiochemJ.org/bj/403/bj4030119add.htm). TR-AURKAIP1 was able to target Aurora-A for degradation in both aphidicolin and nocodazole treated cells, suggesting that the AURKAIP1-mediated degradation of Aurora-A is cell cycle-independent (Figure 1c). To exclude the possibility that the missing N-terminus of AURKAIP1 in the truncated version used above might be involved in the specification of cell-cycle-dependent degradation of Aurora-A, identical experiments were performed with the full-length AURKAIP1 also (see Supplementary Figure S1b and S1c). The results suggested that both full-length and TR-AURKAIP1 could target Aurora-A for degradation independent of the cell cycle stage. The observation that AURKAIP1 targets Aurora-A for degradation independently of cell cycle stage is in contrast with the reported differential effect of hCdh1 (human Cdh1) on the steady-state levels of Aurora-A in S- and M-phase arrested cells. hCdh1 showed a cell cycle-specific differential effect on the steady-state levels of Aurora-A, with no significant effect in M-phase cells, while the control untreated and thymidine treated (S-phase) cells showed significant decrease of Aurora-A protein [20].
Figure 1. Degradation of Aurora-A by AURKAIP1 is cell-cycle-independent.
(a) N-terminal truncated AURKAIP1 is more effective in destabilizing Aurora-A than the full length AURKAIP1. COS7 cells were co-transfected with Aurora-A and FLAG-tagged full-length AURKAIP1 or TR-AURKAIP1 at a ratio of 1:9 for 36 h before being harvested for Western Blot analysis of Aurora-A and AURKAIP1 using anti-IAK1 and anti-FLAG M2 mouse monoclonal antibodies respectively. A vector control was been included in which the AURKAIP1 plasmid has been replaced with pcDNA3. β-tubulin was used as the loading control. (b) TR-AURKAIP1 specifically targets Aurora-A for degradation. ts20 CHO cells were co-transfected with plasmids expressing FLAG-tagged human Aurora-A or Aurora-B and HA-tagged TR-AURKAIP1 at a 1:5 ratio respectively. The vector control was as described for (a). The effect of TR-AURKAIP1 overexpression on Aurora-A or Aurora-B kinase stability was assessed at 36 h post-transfection by immunoblot analysis. Cell extracts were analysed for Aurora-A, Aurora-B and TR-AURKAIP1 proteins using the anti-FLAG M2 and anti-HA antibodies. The blot was probed with mouse anti-β-tubulin as the loading control. (c) TR-AURKAIP1-mediated Aurora-A degradation is not cell cycle-dependent. COS7 cells were co-transfected with HA-tagged human Aurora-A and FLAG-tagged TR-AURKAIP1 at a ratio of 1:9. The vector control was been included as described for (a). At 24 h post-transfection, the transfected cells were collected at different phases of the cell cycle by treatment with either DMSO (Log), aphidicolin (G1/S) or nocodazole (M) for a further 16 h. Cell extracts were analysed for Aurora-A and TR-AURKAIP1 proteins using the anti-HA and anti-FLAG M2 antibodies respectively. The blot was probed with anti-β tubulin as the loading control.
AURKAIP1 can target Cdh1-resistant Aurora-A mutant protein for degradation
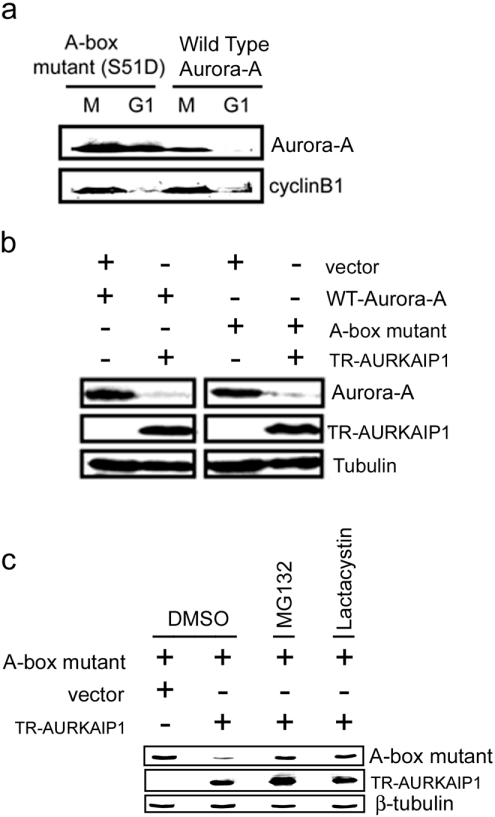
To compare further the nature of Cdh1-dependent and AURKAIP1-dependent Aurora-A degradation, the effect of AURKAIP1 on the degradation of the A-box mutant, an Aurora-A mutant that cannot be targeted for degradation by Cdh1 was studied. Mutation of Ser51, located within the A-box of human Aurora-A, to aspartic acid leads to stabilization of Aurora-A, through inhibition of degradation [6–8]. As shown in Figure 2(a), wild-type Aurora-A was degraded rapidly upon exit from mitosis into G1, whereas the A-box mutant was stabilized. In vivo Aurora-A degradation assays with the A-box mutant showed that it was targeted for degradation with the same efficiency as the wild-type Aurora-A in the presence of TR-AURKAIP1 (Figure 2b). These results further reinforced the mechanistic differences in the Cdh1-dependent and AURKAIP1-dependent degradation of Aurora-A. However, reversal of this AURKAIP1-mediated degradation of the A-box mutant by proteasome inhibitors MG132 and lactacystin (Figure 2c) confirmed the proteasome-dependent nature of this process.
Figure 2. AURKAIP1 can target Cdh1-resistant Aurora-A mutant protein for degradation.
(a) An A-box stabilizing mutant of Aurora-A is not degraded at the G1 phase. HeLa cells were transfected with either HA-tagged wild-type or A-box mutant of Aurora-A. At 24 h post-transfection, the cells were treated with 0.1 μg/ml nocodazole for 16 h to arrest them at M phase. The floating mitotic cells were collected by shake-off and replated in the presence of 50 μg/ml CHX. The cells were harvested for analysis at 4 h post-mitotic release. Stability of the wild-type and A-box mutant of Aurora-A at the M/G1 transition was detected by immunoblotting using the anti-HA antibody. Endogenous cyclin B1 levels were used as the positive control to verify M/G1 transition. (b) TR-AURKAIP1 can degrade mutant Aurora-A. For in vivo degradation assay, COS7 cells were co-transfected with HA-tagged Aurora-A [wild-type (WT) or A-box mutant] and FLAG-tagged TR-AURKAIP1 at a ratio of 1:9. A vector control was been included in which the TR-AURKAIP1 plasmid was replaced by empty pcDNA3. The cells were harvested and analysed 36 h post-transfection by immunoblot analysis. Aurora-A and TR-AURKAIP1 proteins were detected with anti-HA and FLAG M2 antibodies respectively. β tubulin was used as the loading control. (c) AURKAIP1-mediated degradation of A Box Mutant is proteasome-dependent. HeLa cells were co-transfected with HA-tagged A-box mutant and FLAG-tagged TR-AURKAIP1 at a ratio of 1:9. The vector control was as described for (b). At 24 h post-transfection, one set of cells was treated with DMSO (control), while the other set was treated with 20 μM MG132 or lactacystin for 16 h before harvest for Western blotting. The A-Box mutant and TR-AURKAIP1 were detected using the anti-HA and FLAG M2 antibodies respectively. β-tubulin was used as the loading control.
AURKAIP1 inhibits polyubiquitination of Aurora-A
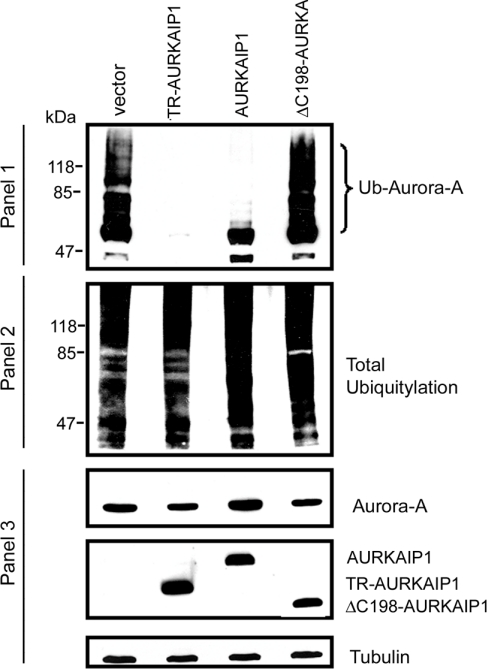
Ubiquitination represents one of the essential modifications to target the protein(s) for recognition by 26 S proteasome and subsequent degradation. It is known that Aurora-A is poly-ubiquitinated before cell-cycle-dependent degradation by APC/C [17]. In order to understand the mechanism of AURKAIP1-mediated degradation, we asked whether AURKAIP1 plays any role in the ubiquitination of Aurora-A. To determine the possibility that AURKAIP1 might potentiate the polyubiquitination of Aurora-A and therefore enhance its degradation similar to Chfr, in vivo Aurora-A ubiquitination assays were performed as described in the Materials and methods section. HeLa cells were co-transfected with wild-type Aurora-A and Ub in the presence of either AURKAIP1 expression constructs or empty vector. As shown in Figure 3, wild-type Aurora-A can be ubiquitinated readily. However, co-expression of TR-AURKAIP1 totally abolished the polyubiquitination of Aurora-A (Figure 3, panel 1). The decreased polyubiquitination of Aurora-A in the presence of TR-AURKAIP1 was not due to the alteration of total cellular polyubiquitination (Figure 3, panel 2). Under the experimental conditions, which were carried out in the presence of the proteasome inhibitor MG132, Aurora-A levels were maintained even in the presence of TR-AURKAIP1 (Figure 3, panel 3), suggesting that the decreased polyubiquitination observed in the presence of AURKAIP1 is mainly due to inhibition of the polyubiquitination of Aurora-A per se, rather than due to decreased Aurora-A protein levels. ΔC198-AURKAIP1 mutant, an Aurora-A non-interactive AURKAIP1 mutant, was also used as a control to verify whether the interaction between AURKAIP1 and Aurora-A is essential for ubiquitination. Interestingly, ΔC198-AURKAIP1 mutant, which does not interact with Aurora-A and is less efficient in mediating Aurora-A degradation, lacked this inhibitory effect and restored Aurora-A polyubiquitination to a level similar to the empty vector control. These observations suggest that AURKAIP1 inhibits polyubiquitination of Aurora-A, and its interaction with Aurora-A is essential for the inhibitory effect on polyubiquitination.
Figure 3. AURKAIP1 inhibits polyubiquitination of Aurora-A.
HeLa cells were transiently transfected with HA-tagged Aurora-A in combination with either empty vector pcDNA3 or AURKAIP1 constructs (TR-AURKAIP1, AURKAIP1 and ΔC198-AURKAIP1 mutant) at a ratio of 1:9, in the presence of an expression construct encoding His6-tagged wild-type Ub. In vivo ubiquitination assays were performed as described in the Materials and methods section. Total ubiquitinated proteins were pulled down with Ni-NTA–agarose and the ubiquitinated HA-tagged Aurora-A was detected with anti-HA antibodies. The polyubiquitinated, HA-tagged Aurora-A appears as a ladder (Panel 1). The total cellular polyubiquitination was determined by Western blot analysis of the total lysates with anti-His antibodies (Panel 2). The protein levels of Aurora-A in the total lysates under different conditions were analyzed using anti-HA antibodies (Panel 3). The blot used for the experiment described in Panel 3 was reprobed with anti-tubulin antibody as a loading control.
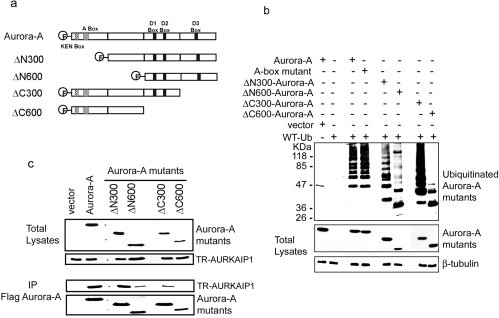
To further verify the interaction-dependent inhibition of ubiquitination of Aurora-A, mapping of the regions of Aurora-A protein essential for the ubiquitination and binding of AURKAIP1 was performed. Both N-terminal and C-terminal truncated overlapping fragments of Aurora-A were generated (Figure 4a) and subjected to in vivo ubiquitination and AURKAIP1 interaction assays as described in the Materials and methods section. The results presented in Figure 4(b) show that both N-terminal deletions (ΔN300 and ΔN600) of Aurora-A lead to decreased polyubiquitination, but were still capable of being polyubiquitinated. Surprisingly, in contrast with the general belief that the A-box mutant is ubiquitination-defective, we were able to observe ubiquitylation of the A-box mutant to an extent similar to that of wild-type Aurora-A under the given experimental conditions. The difference in the effect of polyubiquitination by mutations in the A-box and N-terminal deletion mutants of Aurora-A could be due to the nature of the mutations, as one is a point mutation compared with deletion of larger sections of the protein in the others. It is noteworthy that, despite the prediction that the S51D mutation in the A-box negatively influences ubiquitination and subsequent degradation of Aurora-A [6,7], a formal demonstration of the effect of Aurora-A S51D mutation on the ubiquitination of Aurora-A is still lacking. On the other hand, the C-terminal deletion, ΔC300, showed increased polyubiquitination of Aurora-A. This observation is in agreement with the results published previously [17], which also showed an increase in the polyubiquitination of an Aurora-A protein lacking the extreme C-terminal region. However, further deletion of the C-terminus (ΔC600) suppressed polyubiquitination completely, suggesting that this region is essential for the efficient polyubiquitination of Aurora-A. Analysis of the Aurora-A regions necessary for interaction with AURKAIP1 (Figure 4c) showed that the ΔN300 mutant could interact with AURKAIP1 with an efficiency similar to the full-length Aurora-A. The other deletions, ΔN600 and ΔC300, were capable of interacting with AURKAIP1, albeit with lower efficiency than the wild-type protein. Interestingly, the polyubiquitination defective ΔC600 was incapable of binding AURKAIP1, suggesting that there could be an overlap between the regions of Aurora-A protein that are necessary for polyubiquitination and its interaction with AURKAIP1.
Figure 4. Mapping of regions of Aurora-A essential for ubiquitination.
(a) Aurora-A kinase and its various deletion mutants. The diagram illustrates the size and location of the deletions of all the Aurora-A deletion mutant proteins compared with full length Aurora-A protein. All of the Aurora-A mutants contain a FLAG-tag at the N-terminus. The locations of the KEN-, A-, and D- (D1, D2, D3) boxes are indicated. (b) Domain mapping for efficient ubiquitination of Aurora-A. HeLa cells were transfected with His-tagged wild-type Ub and FLAG-tagged wild-type Aurora-A, the A-box mutant or various deletion mutants of Aurora-A. At 24 h post-transfection, the transfected cells were treated with 20μM MG132 for an additional 16 h before harvesting for immunoprecipitation with anti-His antibody. The ubiquitinated wild-type Aurora-A and deletion mutants were detected by the anti-FLAG M2 antibody. (c) Mapping of the AURKAIP1-interacting domain in Aurora-A. HeLa cells were transfected with HA-tagged TR-AURKAIP1 and FLAG-tagged Aurora-A or its various deletion mutants at 1:1 ratio. At 24 h post-transfection, the transfected cells were harvested for immunoprecipitation with anti-FLAG M2 antibody. The interacting TR-AURKAIP1 was detected using the anti-HA antibody.
AURKAIP1 targets Aurora-A for degradation in the presence of dominant negative ubiquitin mutants
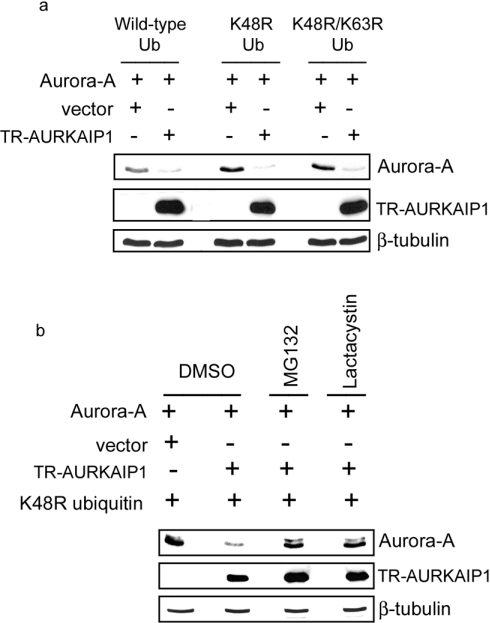
Destabilization of Aurora-A, despite the inhibition of its polyubiquitination by AURKAIP1, prompted us to investigate its ubiquitin-independent degradation. To this end, a dominant negative ubiquitin mutant, K48R, was used to suppress cellular polyubiquitination. Ub-dependent degradation involves the attachment of multiple Ubs to the lysine residue of the target protein(s), facilitating the substrate recognition by the 26 S proteasome. Incorporation of the K48R dominant negative mutant Ub has been shown to have a chain terminating effect, blocking further Ub chain extension [21]. The lysine residue at position 63 of Ub is also implicated in the polyubiquitination of target proteins. However, unlike Lys48, its role in protein degradation is minimal. Regardless, to exclude a role for Lys63-mediated polyubiquitination in the degradation of Aurora-A, a K48R/K63R double mutant was also generated. To verify whether the AURKAIP1-mediated Aurora-A degradation is affected under the conditions where the cellular polyubiquitination is suppressed, AURKAIP1-mediated in vivo degradation of Aurora-A was carried out in the presence of K48R and K48R/K63R dominant negative Ub mutants. It was observed that the AURKAIP1-mediated Aurora-A degradation was unaffected even in the presence of K48R or K48R/K63R mutant Ubs, and was as efficient as observed with wild-type Ub (Figure 5a). Taken together, efficient degradation of Aurora-A even in the presence of the K48R/K63R double mutant, and the inhibition of Aurora-A polyubiquitination by AURKAIP1 without compromising its effect on degradation indicated that Aurora-A could be targeted for degradation even in the absence of polyubiquitination. To verify that the AURKAIP1-mediated degradation of Aurora-A in the presence of mutant Ub involves proteasomal function, a similar experiment to that described above was carried out in the presence and absence of proteasomal inhibitors MG132 and lactacystin. The results shown in Figure 5(b) show that the observed AURKAIP1-mediated degradation of Aurora-A in the presence of dominant negative mutant Ub is also proteasome-dependent.
Figure 5. AURKAIP1 targets Aurora-A for degradation in the presence of dominant negative ubiquitin mutants.
(a) Overexpression of K48R and K48R/K63R dominant negative Ub mutants does not affect AURKAIP1-mediated Aurora-A degradation. HeLa cells were co-transfected with FLAG-tagged Aurora-A and FLAG-tagged TR-AURKAIP1 at a ratio of 1:9 in the presence of His-tagged wild-type or K48R or K48R/K63R mutant Ub expression constructs. A vector control was been included in which the TR-AURKAIP1 plasmid was replaced with pcDNA3. At 36 h post-transfection, the cells were harvested and analysed for Aurora-A and TR-AURKAIP1 using the anti-FLAG M2 antibody. β-tubulin was detected as the loading control. (b) AURKAIP1-mediated Ub-independent degradation of Aurora-A is proteasome-dependent. HeLa cells were co-transfected with HA-tagged Aurora-A and FLAG-tagged TR-AURKAIP1 at a ratio of 1:9 in the presence of K48R ubiquitin mutant overexpression. The vector control was as described for (a). At 24 h post-transfection, one set of cells was treated with DMSO (control), while the other set was treated with 20 μM MG132 or lactacystin for 16 h before harvesting for Western blot analysis. Aurora-A and TR-AURKAIP1 were detected using the anti-HA and anti-FLAG M2 antibodies respectively. β-tubulin was used as the loading control.
Lack of polyubiquitination does not completely stabilize Aurora-A
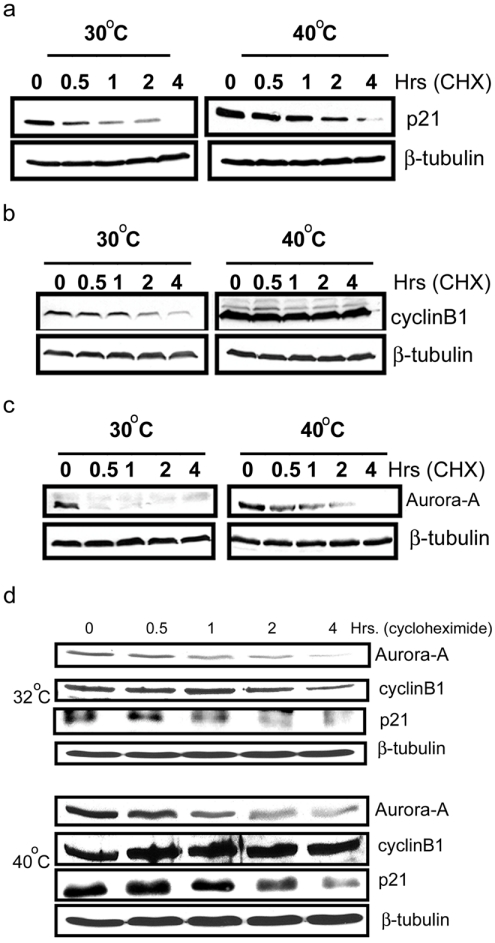
As an alternative approach to verify the existence of an Ub-independent pathway for Aurora-A degradation, the turnover of Aurora-A, p21 and cyclin B1 was assessed in the presence of CHX in ts20-CHO cells harbouring a temperature-sensitive mutation in E1-ubiquitin activating enzyme [22]. The turnover of these proteins was assessed at either permissive conditions (30 °C) where both Ub-dependent and independent pathways are functional, or at non-permissive conditions (40 °C) where only the Ub-independent pathway is functional. p21 has previously been demonstrated to be a target for Ub-independent degradation [23], whereas cyclin B1 has been a prototype target for Ub-dependent degradation. Results presented in Figure 6(a) show that the temperature shift to 40 °C did not completely stabilize p21, supporting previous findings that p21 can be degraded in the absence of ubiquitination and hence act as the target for Ub-independent degradation. In contrast, cyclin B1 level was completely stabilized upon temperature shift to 40 °C, indicating that cyclin B1 can only be targeted by Ub-dependent degradation (Figure 6b). It was evident that even within 30 min of CHX treatment, there was sharp decline in Aurora-A steady state level, indicating that Aurora-A is normally an unstable protein. However, its steady state level was increased but not stabilized when the Ub-dependent pathway was blocked at 40 °C (Figure 6c). The lack of complete stabilization implies that Aurora-A can also be targeted for degradation in the absence of Ub, similar to p21.
Figure 6. Lack of polyubiquitination does not completely stabilize Aurora-A.
To determine the turnover of Aurora-A, cyclin B1 and p21 in ts20 CHO cells in the presence and absence of polyubiquitination, cells were transfected with FLAG-tagged Aurora-A, HA-tagged p21 and cyclin B1 expression plasmids at 30 °C. At 24 h post-transfection, the cells were divided into two sets; one set was maintained at the permissive temperature (30 °C), while the other set was shifted to the non-permissive temperature (40 °C) for 16 h. After 16 h, the cells were treated with 50 μg/ml CHX and both sets of cells were harvested at the indicated time points. The levels of p21 (a), cyclin B1 (b) and Aurora-A (c) were analysed by immunoblot analysis by using anti-FLAG, anti-(cyclin B1) and anti-HA-tag antibodies respectively. β tubulin was detected as the loading control. (d) Ub-independent degradation of endogenous Aurora-A kinase: Mouse ts20 cells were incubated at 32 °C or 40 °C for 18 h followed by CHX treatment for the indicated times. The protein levels of endogenous Aurora-A, p21 and cyclin B1 were analysed by immunoblot analysis using the anti-IAK1, anti-p21 and anti-(cyclin B1) antibodies respectively. β-tubulin was detected as the loading control
To address the question whether endogenous Aurora-A also follows the same fate as the exogenous protein, with respect its the stabilization in the absence of ubiquitination, we carried out the experiment in the temperature sensitive mouse cell line ts20-TG, which has a mutation in the E1 Ub activating enzyme, to facilitate detection of endogenous proteins with available antibodies. Identical experiments as described above for ts20-CHO cells to investigate the turnover of exogenous Aurora-A, p21 and cyclin B1 were carried out in this mouse cell line. The results presented in Figure 6(d) demonstrate that the endogenous Aurora-A, p21 and cyclin B1 behave in a similar manner to their exogenous counterparts with respect to their turnover in the absence of polyubiquitination.
AURKAIP1 specifically targets Aurora-A for degradation through the Ub-independent pathway
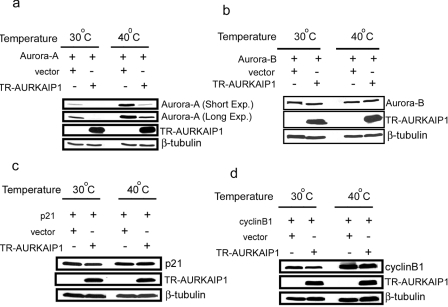
To further confirm the Ub-independent nature of Aurora-A degradation in the presence of AURKAIP1, in vivo degradation assays were performed in ts20-CHO cells. In support of the results from the first approach using dominant negative Ub mutants, suppression of polyubiquitination by temperature-sensitive mutation of the E1 enzyme increased the levels of Aurora-A, but still did not abolish the AURKAIP1-mediated Aurora-A degradation (Figure 7a). These results confirm that AURKAIP1 mediates Aurora-A degradation even in the absence of polyubiquitination. Therefore Aurora-A can be targeted by both Ub-dependent as well as Ub-independent degradation pathways. To verify the specificity of AURKAIP1-mediated Ub-independent Aurora-A degradation, the effect of overexpression of TR-AURKAIP1 on human Aurora-B, p21 and cyclin B1 stability was investigated in ts-20- CHO cells. In contrast to the effect on Aurora-A, overexpression of TR-AURKAIP1 did not influence the stability of either Aurora-B (Figure 7b), p21 (Figure 7c) or cyclin B1 (Figure 7d). Interestingly, in the absence of AURKAIP1, inhibition of polyubiquitination leads to a higher basal level of Aurora-A. This implies that under normal conditions, the Ub-dependent degradation is a major pathway operational for Aurora-A turnover, and AURKAIP1 promotes Aurora-A degradation through a Ub-independent but proteasome-dependent pathway.
Figure 7. AURKAIP1 specifically targets Aurora-A for degradation in an Ub-independent manner.
ts20-CHO cells were co-transfected with FLAG-tagged TR-AURKAIP1 and the constructs expressing the targets genes (Aurora-A, Aurora-B, p21 and cyclin B1) at a ratio of 9:1. A vector control was included in which the TR-AURKAIP1 plasmid was replaced with pcDNA3. The transfected cells were divided into two sets; both sets were initially incubated at the permissive temperature (30 °C) for 24 h. One set was maintained at the permissive temperature and the other set was incubated at the non-permissive temperature (40 °C) for 16 h. The cells were harvested and the steady state levels of Aurora-A (a), Aurora-B (b), p21 (c) and cyclin B1 (d) proteins were analysed using their respective antibodies. The TR-AURKAIP1 expression was also monitored. The blots were probed with mouse anti-(β tubulin) as the loading control.
DISCUSSION
The abundance of Aurora-A protein is tightly controlled by synthesis and degradation, as its overexpression leads to disruption of checkpoints [24,25], induction of aneuploidy and transformation [9,10]. Cell-cycle-dependent degradation of Aurora-A is mediated by Cdh1 through the Ub-dependent proteasomal degradation pathway [6,7]. Previously, two other candidate regulators, Hcdc4 and Chfr, involved in the destabilization of Aurora-A have been described [18,19]. All these candidate regulators, however, target Aurora-A for proteasome-dependent degradation with prior ubiquitination. Herein, we provide the first demonstration that an alternative Ub-independent pathway for Aurora-A degradation exists, and that AURKAIP1 facilitates the degradation of Aurora-A through this alternative pathway.
Although ubiquitination is a pre-requisite for the majority of the extralysosomal proteolysis by the 26 S proteasome, both 20 S and 26 S proteasomes can degrade many proteins in an Ub-independent manner [5]. Ornithine decarboxylase was the first example of a protein degraded by the 26 S proteasome using this alternative pathway [26]. The other proteins, which are degraded by 26 S proteasome in an Ub-independent manner, include c-jun [27], p21 [28], p53 [29] and calmodulin [30]. The 26 S/20 S proteasome can degrade proteins in an Ub-independent manner, provided the substrate is targeted to the proteasome machinery by another protein, or by a degradation signal present in the substrate itself. For example, Tax, a protein encoded by human T cell leukaemic virus promotes the binding of IκBα to the HsN3 subunit of 20 S proteasome and facilitates the constitutive degradation of IκBα in a Ub- and phosphorylation-independent manner [31]. Similarly, hyperphosphorylated forms of members of the Rb (retinoblastoma) protein family were targeted by the viral protein pp71 for Ub-independent, proteasome-dependent degradation [32]. On the other hand, NQO1 (NAD(P)H quinone oxidoreductase 1), which is capable of binding both p53 and 20 S proteasome, functions as a gatekeeper of the 20 S proteasome and negatively regulates the degradation of p53 [29,33]. Dicoumarol, an inhibitor of NQO1, has been shown to induce p53 degradation by the 20 S proteasome-dependent, Ub-independent pathway [34]. p21, a transcriptional target of p53, constitutes an example of a protein that targets itself for Ub-independent degradation by directly binding to the 20 S proteasome [34]. It has been shown that p21 interacts directly with the C8 subunit of 20 S proteasome in vitro and the turnover of mutant p21 in vivo correlates directly with its affinity for the C8 subunit in vitro [35]. Thus it is apparent that the interaction of the target proteins with the proteasomal machinery is a prerequisite for Ub-independent degradation.
Using dominant-negative ubiquitin mutants and temperature-sensitive mutant cell lines defective in E1 Ub activating enzyme at the restrictive temperature, we have shown that Aurora-A can be degraded in the absence of polyubiquitination. We further show that AURKAIP1, which constitutively targets Aurora-A for degradation in a proteasome-dependent manner [16], can obviate the need for polyubiquitination. The results of the present study conform to the trend of proteins being degraded by both Ub-dependent and -independent pathways [27–29]. It has been shown recently that Mdm2 (murine double minute 2) can target Rb protein for degradation through a similar Ub-independent pathway [36]. It is intriguing to note that, although Mdm2 promotes the degradation of p53 through the Ub-dependent pathway by its Ub ligase activity, the degradation of Rb protein by Mdm2 is Ub-independent and accompanies suppression of polyubiquitination. We also observed the suppression of polyubiquitination of Aurora-A in the presence of AURKAIP1 suggesting that the AURKAIP1-mediated suppression of polyubiquitination might be one of the determinants to switch between these alternative pathways. Studies carried out to understand the mechanism by which AURKAIP1 could suppress polyubiquitination of Aurora-A suggest that the binding of AURKAIP1 to Aurora-A might inhibit the interaction of the ubiquitination machinery with Aurora-A. This speculation was supported by the observation that the non-interactive AURKAIP1 mutant ΔC198 restored ubiquitination of Aurora-A. Further studies with Aurora-A deletion mutants showed that there is an overlap of the AURKAIP1 binding region and the region essential for proper polyubiquitination of Aurora-A. Thus the binding of AURKAIP1 to Aurora-A could mask the region essential for ubiquitination, thereby inhibiting polyubiquitination.
The next interesting question is how AURKAIP1 targets Aurora-A to the proteasome in the absence of ubiquitination? Generally, marking of the substrates with Ub is thought to serve two functions, unfolding of the protein and targeting it for degradation [31,32,37]. It is not yet clear whether binding of AURKAIP1 can unfold Aurora-A so that it can be a better substrate for the 20 S proteasome. Similarly, it has not yet been shown that AURKAIP1 is capable of targeting Aurora-A directly to the proteasome. In this context, it will be interesting to investigate whether AURKAIP1 can interact directly with proteasome and if Aurora-A can be degraded by the 26 S/20 S proteasome in vitro in the presence of AURKAIP1. On the other hand, in the AURKAIP1-mediated Aurora-A degradation pathway, other possibilities such as modification of Aurora-A by other small molecules [38] or the involvement of other secondary proteins that can co-operate with AURKAIP1 in the targeting of Aurora-A to the proteasome also cannot be ruled out.
In summary, we have shown that Aurora-A can be targeted for degradation even in the absence of ubiquitination, and AURKAIP1 facilitates the degradation of Aurora-A through this Ub-independent pathway. At this juncture, it is unclear why there should be two pathways for the degradation of the same protein or what is the cellular context for AURKAIP1-mediated Ub-independent degradation of Aurora-A. The physiological relevance of the Aurora-A–AURKAIP1 interaction can be appreciated better when we have adequate information on the biology of AURKAIP1. Preliminary studies carried out on the expression of AURKAIP1 using an antibody raised against the C-terminal AURKAIP1 peptide, showed that while transfected and in vitro translated AURKAIP1 can be easily detected, the endogenous AURKAIP1 was undetectable even in cells that expressed very high levels of AURKAIP1 transcripts (results not shown). This suggests that AURKAIP1 could be regulated post-transcriptionally and/or the AURKAIP1 protein might be expressed only under a specific cellular context. Future studies on the identification of this as yet unidentified cellular context, as well as the mechanism by which AURKAIP1 promotes Ub-independent degradation of Aurora-A, will definitely throw more light on the physiological significance of this alternative pathway.
Online data
Acknowledgments
We thank Dr Ger. J. Strous and Dr Harvey Ozer for providing us with the ts20-CHO and ts20TG mouse cell lines respectively. We also thank Dr Michele Pagano, Dr Ivan Dikic and Dr Prochownik (Section of Hematology/Oncology, Children's Hospital of Pittsburgh, Pittsburgh, PA, U.S.A.) for the p21, K48R Ub and cyclin B1 expression plasmids respectively. This work was supported by the National Medical Research Council of Singapore in the form of a research grant (NMRC/0815/2003) to G.G. and as an Institutional Block Grant to the National Cancer Centre, Singapore.
References
- 1.King R. W., Deshaies R. J., Peter J. M., Kirschner M. W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 2.Peters J. M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 3.Koepp D. M., Harper J. W., Elledge S. J. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski M., Wilk S. Ubiquitin-independent proteolytic functions of the proteasome. Arch. Biochem. Biophys. 2003;415:1–5. doi: 10.1016/s0003-9861(03)00197-8. [DOI] [PubMed] [Google Scholar]
- 6.Castro A., Arlot-Bonnemains Y., Vigneron S., Labbe J. C., Prigent C., Lorca T. APC/Fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep. 2002;3:457–462. doi: 10.1093/embo-reports/kvf095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Littlepage L. E., Ruderman J. V. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro A., Vigneron S., Bernis C., Labbe J. C., Prigent C., Lorca T. The D-Box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-Box sequence of Aurora-A. EMBO Rep. 2002;3:1209–1214. doi: 10.1093/embo-reports/kvf241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H., Kuang J., Zhong L., Kuo W. L., Gray J. W., Sahin A., Brinkley B. R., Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff J. R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C., et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bischoff J. R., Plowman G. D. The Aurora/Ipl1p kinase family: regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 12.Katayama H., Brinkley W. R., Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer and Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka T., Kimura M., Matsunaga K., Fukada D., Mori H., Okano Y. Centrosomal kinase AIK1 is overexpressed in invasive ductal carcinoma of the breast. Cancer Res. 1999;59:2041–2044. [PubMed] [Google Scholar]
- 14.Miyoshi Y., Iwao K., Egawa C., Noguchi S. Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int. J. Cancer. 2001;92:370–373. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 15.Goepfert T. M., Adigun Y. E., Zhong L., Gay J., Medina D., Brinkley W. R. Centrosome amplification and overexpression of Aurora A are early events in rat mammary carcinogenesis. Cancer Res. 2002;62:4115–4122. [PubMed] [Google Scholar]
- 16.Kiat L. S., Hui K. M., Gopalan G. Aurora-A kinase interacting protein (AIP), a novel negative regulator of human Aurora-A kinase. J. Biol. Chem. 2002;277:45558–45565. doi: 10.1074/jbc.M206820200. [DOI] [PubMed] [Google Scholar]
- 17.Honda K., Mihara H., Kato Y., Yamaguchi A., Tanaka H., Yasuda H., Furukawa K., Urano T. Degradation of human Aurora2 protein kinase by the anaphase-promoting complex-ubiquitin-proteasome pathway. Oncogene. 2000;19:2812–2819. doi: 10.1038/sj.onc.1203609. [DOI] [PubMed] [Google Scholar]
- 18.Mao J. H., Perez-Losada J., Wu D., Delrosario R., Tsunematsu R., Nakayama K. I., Brown K., Bryson S., Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 19.Yu X., Minter-Dykhouse K., Malureanu L., Zhao W. M., Zhang D., Merkle C.J., Ward I. M., Saya H., Fang G., van Deursen J., Chen J. Chfr is required for tumor suppression and Aurora A regulation. Nat. Genet. 2005;37:401–406. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 20.Taguchi S., Honda K., Sugiura K., Yamaguchi A., Furukawa K, Urano T. Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS Lett. 2002;519:59–65. doi: 10.1016/s0014-5793(02)02711-4. [DOI] [PubMed] [Google Scholar]
- 21.Ward C. L., Omura S., Kopito R. R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 22.Strous G. J., van Kerkhof P., Govers R., Ciechanover A., Schwartz A. L. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- 23.Sheaff R. J., Singer J. D., Swanger J., Smitherman M., Roberts J. M., Clurman B. E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 24.Marumoto T., Hirota T., Morisaki T., Kunitoku N., Zhang D., Ichikawa Y., Sasayama T., Kuninaka S., Mimori T., Tamaki N., et al. Roles of Aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;2:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 25.Anand S., Penrhyn-Lowe S., Venkitaraman A. R. Aurora-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 26.Bercovich Z., Rosenberg-Hasson Y., Ciechanover A., Kahana C. Degradation of ornithine decarboxylase in reticulocyte lysate is ATP-dependent but ubiquitin-independent. J. Biol. Chem. 1989;264:15949–15952. [PubMed] [Google Scholar]
- 27.Jariel-Encontre I., Pariat M., Martin F., Carillo S., Salvat C., Piechaczyk M. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J. Biol. Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 28.Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 2003;22:6365–6377. doi: 10.1093/emboj/cdg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G., Tsevetkov P., Kahana C., Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarcsa E., Szymanska G., Lecker S., O'Connor C. M., Goldberg A. L. Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26 S proteasomes without ubiquitination. J. Biol. Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- 31.Krappmann D., Wulczyn F. G., Scheidereit C. Different mechanisms control signal-induced degradation and basal turnover of the NF-κB inhibitor IκBα in vivo. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 32.Kalejta R. F., Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3263–3268. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asher G., Lotem J., Tsvetkov P., Reiss V., Sachs L., Shaul Y. p53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15065–15070. doi: 10.1073/pnas.2436329100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asher G., Lotem J., Sachs L., Kahana C., Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 α-subunit of the 20 S proteasome. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sdek P., Ying H., Chang D. L. F., Qiu W., Zheng H., Touitou R., Allday M. J., Xiao Z. J. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell. 2005;20:699–708. doi: 10.1016/j.molcel.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Ghislain M., Dohmen R. J., Levy F., Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 38.Jentsch S., Pyrowolakis G. Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.