Abstract
Protein chips have emerged as a promising approach for a wide variety of applications including the identification of protein–protein interactions, protein–phospholipid interactions, small molecule targets, and substrates of proteins kinases. They can also be used for clinical diagnostics and monitoring disease states. This article reviews current methods in the generation and applications of protein microarrays.
Keywords: Protein microarrays, Proteomics, High-throughput, Kinase substrates, DNA-binding proteins, Microarray
Understanding complex cellular systems will require the identification and analysis of each of its components and determining how they function together and are regulated. A critical step in this process is to determine the biochemical activities of the proteins and how these activities themselves are controlled and modified by other proteins. Traditionally, the biochemical activities of proteins have been elucidated by studying single molecules, one experiment at a time. This process is not optimal, as it is slow and labor intensive.
In contrast to this traditional approach, high-throughput scientific methods have been developed in the last decade to optimize the study of large numbers of molecules, including DNA, proteins and metabolites. DNA microarrays in particular have proved valuable in genomic research (Schena et al., 1995). They have been used to study gene expression patterns, to locate transcription factor binding sites, and to detect sequence mutations and deletions on a grand scale. However, DNA microarrays tell us only about the genes themselves and provide little information regarding the functions of the proteins they encode. More recently high throughput approaches have been developed for the study of proteins, including profiling of proteins using mass spectrometry (Gavin et al., 2006, Gavin et al., 2002, Ho et al., 2002, Krogan et al., 2006, Washburn et al., 2001), tagging and subcellular localization (Ghaemmaghami et al., 2003, Huh et al., 2003) and protein microarrays (MacBeath and Schreiber, 2000, Zhu et al., 2001, Zhu et al., 2000). This article reviews the developments in the production and application of protein microarrays.
1. Types of protein microarrays
Three types of protein microarrays are currently used to study the biochemical activities of proteins: analytical microarrays, functional microarrays, and reverse phase microarrays. Analytical microarrays are typically used to profile a complex mixture of proteins in order to measure binding affinities, specificities, and protein expression levels of the proteins in the mixture. In this technique, a library of antibodies, aptamers, or affibodies is arrayed on a glass microscope slide. The array is then probed with a protein solution. Antibody microarrays are the most common analytical microarray (Bertone and Snyder, 2005).
These types of microarrays can be used to monitor differential expression profiles and for clinical diagnostics. Examples include profiling responses to environmental stress and healthy versus disease tissues (Sreekumar et al., 2001).
Functional protein microarrays differ from analytical arrays in that functional protein arrays are composed of arrays containing full-length functional proteins or protein domains. These protein chips are used to study the biochemical activities of an entire proteome in a single experiment. They are used to study numerous protein interactions, such as protein–protein, protein–DNA, protein–RNA, protein–phospholipid, and protein–small molecule interactions (Hall et al., 2004, Zhu et al., 2001).
A third type of protein microarray, related to analytical microarrays, is known as a reverse phase protein microarray (RPA). In RPA, cells are isolated from various tissues of interest and are lysed. The lysate is arrayed onto a nitrocellulose slide using a contact pin microarrayer. The slides are then probed with antibodies against the target protein of interest, and the antibodies are typically detected with chemiluminescent, fluorescent, or colorimetric assays. Reference peptides are printed on the slides to allow for protein quantification of the sample lysates.
RPAs allow for the determination of the presence of altered proteins that may be the result of disease. Specifically, post-translational modifications, which are typically altered as a result of disease, can be detected using RPAs (Speer et al., 2005). Once it is determined which protein pathway may be dysfunctional in the cell, a specific therapy can be determined to target the dysfunctional protein pathway and treat the disease of interest.
2. Proteome libraries
Challenges to creating a proteome microarray include not only the creation of the necessary over-expression library, but also the development of a high-throughput expression and purification protocol necessary to efficiently produce thousands of pure, functional proteins that can be immobilized onto a solid surface. There are two major recombinational cloning strategies that are used to create libraries of open reading frames (ORFs) in expression vectors (Phizicky et al., 2003). The first makes use of recombination in yeast. Open reading frames of interest are amplified such that they all contain common 5′ and different common 3′ ends. The ORFs are mixed with a linearized vector that contains ends identical to the 5′ and 3′ ends of the amplified ORFs. The mixture is then transformed into yeast where gap-mediated recombination occurs (Phizicky et al., 2003). The first yeast proteome collection was produced in this fashion (Zhu et al., 2001).
The other major cloning strategy makes use of the Gateway recombinational cloning system (Phizicky et al., 2003). The Gateway system takes advantage of the integration and excision properties of phage λ in E. coli. λ phage integrates into its hosts DNA by recombination between the λ att P site and the host att B site. The recombination results in DNA with ends called att L and att R. Upon excision of the λ phage, recombination between att L and att R results in the original att P and att B sites. The Gateway system takes advantage of these unique properties of λ phage to create an expression vector with the ORF of interest via a two-step recombination reaction. First, components of λ integration are mixed with the amplified ORF that contains two different directional att B sites. This mixture is then combined, in vitro, with a vector that contains corresponding att P sites. Recombination occurs and the resulting vector then contains two different att L sites. This vector is then combined with an expression vector that contains appropriate att R sites. Recombination occurs in vitro, and the ORF of interest is then in a useful expression vector. The Gateway system is quite flexible as it allows for easy shuttling of ORFs between different expression vectors. The Gateway system has been used to create ORF collections for genes from yeast (Gelperin et al., 2005), C. elegans (Reboul et al., 2003), and humans (Rual et al., 2004b).
The generation of the first proteome library and its use in microarrays were performed for yeast. The first library contains greater than 5800 yeast proteins, which are tagged at their N-terminus with GST-HisX6 for ease of purification. After expressing and purifying the proteins in a high-throughput fashion in budding yeast, the proteins were spotted onto glass slides (Zhu et al., 2001). Protein signals on the chips were quantified by probing the GST-HisX6 tags with anti-GST antibodies, followed by incubation with fluorophore-conjugated secondary antibodies. Protein–protein and protein–lipid experiments conducted on the chip led to the discovery of new calmodulin and phospholipid binding proteins and were proof that the immobilized proteins on the array were indeed active.
More recently, Gelperin et al. (2005) have created a yeast proteome library of C-terminally TAP-tagged proteins which allows for the more efficient purification of transmembrane and secreted proteins. Gelperin and colleagues’ collection makes use of the Gateway recombinational technology, which allows for the easy transfer of ORFs into a variety of expression vectors.
In addition to existing proteome collections in yeast, efforts such as the ORFeome project are underway to create clone collections in higher eukaryotes (Brasch et al., 2004, Reboul et al., 2003, Rual et al., 2004a, Rual et al., 2004b). Vidal and colleagues define the ORFeome as the set of protein encoding open reading frames for an entire organism. They have used Gateway recombinational cloning technology (Hartley et al., 2000) to create the first C. elegans ORFeome (Reboul et al., 2003), and a first version of the human ORFeome (Rual et al., 2004b). Human ORFeome collections and microarrays have also been generated by Invitrogen. These ORFeomes are helpful in improving the annotation of genomes, and because they were created using Gateway technology, they exist in a flexible format that allows for the high-throughput expression of proteins in a number of different experimental systems.
Successful ORF cloning in higher eukaryotes is dependent on full length cDNA collections, and there are numerous public efforts underway to create human full length cDNA collections, such as the Unigene set (Wheeler et al., 2004), the Full Length Expression (FLEX) Gene repository (Brizuela et al., 2001), the Integrated Molecular Analysis of Genomes and their Expression (IMAGE) cDNA collection (Lennon et al., 1996), and the Mammalian Gene Collection (MGC) (Gerhard et al., 2004). Commercial collections are also available from companies such as Invitrogen, GeneCopoeia, OriGene, and Open Biosystems.
A protein microarray is only as useful as the quality of the proteins arrayed on the chip. The production of proteins in homologous systems (i.e. yeast protein expressed in yeast) is expected to greatly improve the quality of the proteins and produce them in an active state. Because the function of many if not most proteins are not known, it is impossible to completely determine whether every protein on the array is active and functional. It is likely, however, that for a large majority of the proteins on the yeast proteome chips at least some material is functional, because a number of successful biochemical activities have been confirmed and discovered using our proteome chips. These biochemical activities include protein–protein, protein–phospholipid, protein–DNA, and protein–small molecule interactions (Hall et al., 2004, Huang et al., 2004, Zhu et al., 2001), as well as enzymatic reactions (Ptacek et al., 2005, Zhu et al., 2000) (Fig. 1 ).
Fig. 1.
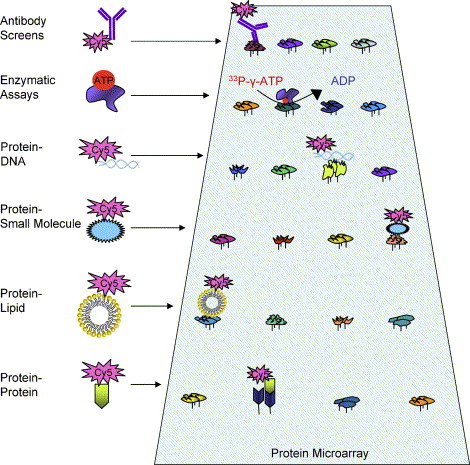
Applications of functional protein microarrays. A representative sample of the different assays that have been performed on functional protein microarrays. Proteins are immobilized at high spatial density onto a microscope slide and the slide can then be probed for various interactions. While Cy5 is the fluorophore shown, many other fluorophores can be used for detection.
It is possible that the position of the affinity tags used for purification may interfere with the functionality of the proteins. The use of two yeast proteome collections, one with C-terminal tags, and one with a different N-terminal tag is expected to complement each other (Gelperin et al., 2005, Zhu et al., 2001). If one version of the tagged protein exhibits a loss of functionality due to the location of the tag, then the other tagged version of the protein can be used on the chip.
Recently a method has been described for the direct production of proteins on chips. DNA is spotted on a microscope slide and subjected to an in vitro transcription and translation system. The proteins are produced as GST fusions and adhere to glutathione on the surface of the slide. Using this system LaBaer and coworkers have produced a number of human proteins involved in DNA metabolism and demonstrated protein–protein interactions (Ramachandran et al., 2004). This method is advantageous because it produces protein directly on the slide without requiring purification and that proteins do not need to be stored.
3. Protein chips
Typically, protein chips are prepared by immobilizing proteins onto a treated microscope slide using a contact spotter (MacBeath and Schreiber, 2000, Zhu et al., 2001) or a non-contact microarrayer (Delehanty, 2004, Delehanty and Ligler, 2003, Jones et al., 2006). It is critical that the proteins remain in a wet environment. Thus, sample buffers contain a high percent of glycerol, and the printing process is carried out in a humidity-controlled environment (MacBeath and Schreiber, 2000, Zhu et al., 2001). Because equipment and procedures developed for DNA microarrays are easily adaptable to the development of protein microarrays, the choice of using treated microscope slides follows from the ready availability of robotic arrayers and laser scanners that have become commonplace in the world of DNA microarrays (Bertone and Snyder, 2005).
A number of different slide surfaces can be used for protein chips. In choosing a slide surface, the goals should be immobilizing the protein on the chip, maintaining the conformation and the functionality of the protein, and achieving maximum binding capacity (Zhu et al., 2003). It is also important to consider whether a random or a uniform orientation of proteins on the slide surface is desired (Fig. 2 ). For random attachment of proteins through amines, aldehyde- and epoxy-derivatized glass surfaces can be used (Kusnezow et al., 2003, MacBeath and Schreiber, 2000). Coating the glass surface with nitrocellulose, gel pads, or poly-l-lysine also achieves a random orientation of the proteins as the proteins are passively adsorbed onto the surface (Angenendt et al., 2002, Charles et al., 2004, Kramer et al., 2004, Stillman and Tonkinson, 2000, Zhu et al., 2003).
Fig. 2.
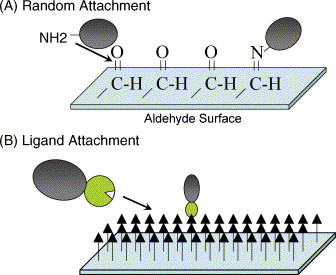
Protein attachment methods. (A) Proteins can be attached randomly via different chemistries including aldehyde- and epoxy-treated slides that covalently attach protein by their primary amines or by adsorpition onto slides coated with nitrocellulose or acrylamide gel pads. (B) Proteins can be uniformly orientated onto slides coated with a ligand. For example, His6X-tagged proteins can be bound to nickel-derivatized slides and biotinylated proteins can be attached to streptavidin-coated slides. This leads to attachment through the tag and presumably orientates the protein away from the slide surface.
Affinity tag surfaces can be used for the uniform orientation of proteins on the chip surface. One popular slide choice is the nickel coated slide for use with HisX6 tagged proteins (Zhu et al., 2003). Another is streptavidin coated slides (Lesaicherre et al., 2002). The display of the proteins away from the chip surface should give reagents easier access to the active sites of the proteins (Zhu et al., 2003).
Finally, microwells can also be used for protein assays. Zhu et al. (2000) developed a slide surface which contains microwells made of a silicone elastomer and attached protein to the epoxy-treated microwells. The benefit of microwells is the ability to perform experiments in an aqueous environment while preventing cross contamination.
4. Detection methods
To locate reactive proteins on a proteome chip, small molecule probes are labeled with either fluorescent, affinity, photochemical, or radioisotope tags. Fluorescent labels are generally preferred, as they are safe and effective and are compatible with readily available microarray laser scanners. However, probes can also be labeled with affinity tags or photochemical tags (Colca and Harrigan, 2004, Mitsopoulos et al., 2004). Huang et al. (2004) used biotin labeled small molecules to probe proteome chips for small molecule inhibitors of rapamycin. The reactive proteins were identified with Cy3 labeled streptavidin.
Regardless of the type of label used, there are problems associated with labeling the molecules used to probe a proteome chip. Chief among these problems is the possibility that the label itself may interfere with the probe's ability to interact with the target protein. To overcome this problem, a number of label-free detection methods have recently been developed. Label-free detection methods not only overcome the problem of steric hindrance of a label, but also allow for the collection of kinetic binding data (reviewed in Ramachandran et al., 2005).
The current leading technology for label-free detection of protein interactions is surface plasmon resonance (SPR), which probes the local index of refraction. Other choices include carbon nanotubes, carbon nanowires, and microelectromechanical systems cantilevers. While these technologies are still in their infancy and are not suitable for high-throughput protein interaction detections, they do offer much promise (Ramachandran et al., 2005).
5. Applications of protein chips
The biochemistries of thousands of proteins can be characterized and quantified in a parallel format through the use of protein microarrays. Not only have protein chips been used to characterize the functions of previously uncharacterized proteins, they have also been used to discover new functionalities for previously characterized proteins. Proteome chips have been used to study protein–protein interactions (Zhu et al., 2001), protein–DNA interactions (Hall et al., 2004), protein–lipid interactions (Zhu et al., 2001), protein–drug interactions (Huang et al., 2004), protein–receptor interactions (Jones et al., 2006), and antigen–antibody interactions (Michaud et al., 2003). In addition, proteome chips have been used to study kinase activities (Ptacek et al., 2005, Zhu et al., 2000) and have been used for serum profiling (Zhu et al., 2006).
With respect to protein–protein interactions, yeast proteome chips have been used to study calmodulin binding proteins (Zhu et al., 2001). Calmodulin is a calcium binding protein involved in many calcium-regulated cellular pathways (Hook and Means, 2001). Zhu et al. (2001) biotinylated the calmodulin probes, and detected the protein–protein interactions using Cy3-labeled streptavidin. Their study found 6 known calmodulin binding proteins, and 33 additional potential binders. Their study also revealed a novel consensus binding motif that was related to a previously known calmodulin binding motif.
Hall et al. (2004) used yeast proteome chips to identify previously unrecognized DNA binding activities (Fig. 3 ). Both single and double stranded Cy3 labeled yeast genomic DNA was used to probe a yeast proteome array. Over 200 DNA binding proteins were identified, but only about half of those were expected to bind DNA based on their known function. One of the unexpected targets we found was Arg5,6, a mitochondrial enzyme involved in arginine biosynthesis. Chromatin immunoprecipitation experiments revealed that Arg5,6 associates with specific nuclear and mitochondrial loci in vivo. Gel shift assays revealed that Arg5,6 binds to specific DNA fragments in vitro, and a common binding motif was found. Real time PCR experiments with Arg5,6 indicated that it may have a role in regulating gene expression. Thus, a yeast proteome chip was used to discover a novel DNA binding protein, and a metabolic enzyme was found that appears to have a role in directly regulating eukaryotic gene expression.
Fig. 3.
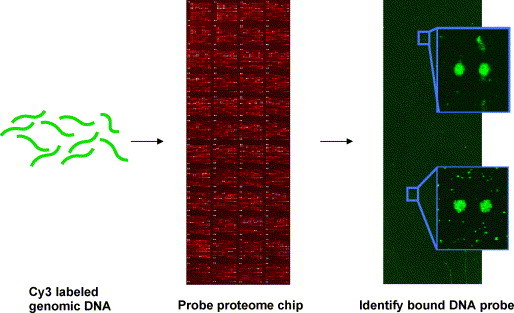
Identification of DNA-binding proteins using a functional protein microarray. Genomic DNA is purified, fragmented, and labeled with Cy3-dCTP. A yeast proteome array with the majority of the yeast proteins was incubated with the labeled DNA to identify novel DNA-binding proteins, including Arg5,6, a mitochondrial enzyme.
Zhu et al. (2001) also used yeast proteome chips to study phosphoinositide (PI) binding proteins. Phosphoinositides are constituents of cellular membranes and also regulate a number of diverse cellular processes (Fruman et al., 1998, Odorizzi et al., 2000). Biotinylated PI liposomes were used to probe the chip. They were detected using Cy3 labeled streptavidin. Of the 150 novel lipid binding proteins that were identified, 52 corresponded to uncharacterized proteins, and 45 were membrane associated proteins.
Proteome microarrays are also an excellent way to discover drug targets. Entire proteomes printed on a chip can be probed with small molecules in one experiment to discover interactions. Huang et al. (2004) used yeast proteome microarrays to study protein–drug interactions. They probed the array with a biotinylated small molecule inhibitor of rapamycin (SMIR) to find protein targets that may be involved in the target-of-rapamycin (TOR) dependent nutrient response network. They discovered a protein of previously unknown function to be a target of the SMIR. They followed up the array probing experiment with gene deletion experiments to prove that the target protein was indeed a legitimate target of the SMIR.
Jones et al. (2006) have used protein microarrays to study protein recruitment to receptors in a high-throughput fashion. Using the data obtained from the microarrays, they also calculated the dissociation constants of the protein–receptor binding. Specifically, they created protein microarrays containing most of the human Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains. These binding domains are known to interact with different epidermal growth factor receptors (EGFR), which themselves are involved in a wide variety of cellular responses. The activated receptors become phosphorylated on their tyrosine residues, which then become the sites for the SH2 and PTB domain binding. The human binding domain microarray of 159 proteins was probed with 66 fluorescently labeled peptides representing the binding sites of the epidermal growth factor receptors. The microarrays were probed with eight concentrations of each peptide, and the resulting fluorescence data was used to calculate dissociation constants to determine the binding affinities of the peptide-binding domain interactions. The accuracy of a sample of the dissociation constants calculations was confirmed with surface plasmon resonance experiments. From their experimental data, they constructed a quantitative EGFR interaction map. Their data not only confirmed previously known interactions, but also showed novel biophysical interactions of SHT2 and PTB domains with the epidermal growth factor receptor network. With their quantitative data, they found that different receptor tyrosine kinases differ in their level of selectivity when overexpressed, which they suggest may be a clue as to why some receptors have more oncogenic potential than others.
There have been multiple yeast protein kinase studies using protein chips. Zhu et al. (2000) used silicone elastomer nanowell sheets placed onto glass slides to study the activity of 119 yeast kinases using 17 different substrates. The 119 kinases were overexpressed and covalently attached to the nanowell chip. The kinases were incubated with 17 different substrates along with radiolabeled ATP to test for in vitro kinase activity. They obtained many novel results, including that 27 yeast kinases can act in vitro as tyrosine kinases. This is roughly triple the number of tyrosine kinases originally thought to exist in yeast.
Ptacek et al. (2005) have also studied protein phosphorylation in yeast using proteome chips. Their goal was to develop a global kinase–substrate map for yeast. To this end, they incubated 87 different yeast protein kinases or kinase complexes separately with radiolabeled ATP on a yeast proteome chip containing 4400 unique proteins (Fig. 4 ). They found about 4200 phosphorylation events affecting 1325 different proteins, and from this data assembled an in vitro phosphorylation network.
Fig. 4.
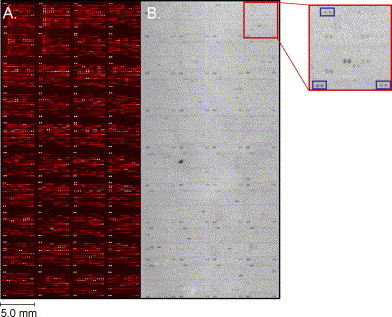
(A) A yeast proteome microarray containing ∼4400 GST-tagged yeast proteins printed in duplicate. The slide was probed with anti-GST antibodies followed by Cy5-labeled anti-rabbit antibodies. Forty of 48 blocks are shown. (B) A kinase assay with 33P-γ-ATP and active kinase on a yeast proteome microarray. Dark spots represent radiolabeled phosphorylated substrates on the array. Kinases that autophosphorylate are printed in each of the 40 blocks (shown above in blue boxes) and serve as reference points on the slide.
Proteome chips have also been used successfully to screen patient's sera for the presence of autoantibodies (Hueber et al., 2005, Kattah et al., 2006) or viral specific antibodies (Zhu et al., 2006). Zhu et al. (2006) created a coronavirus protein microarray and used the array to rapidly screen patient's sera for the presence of antibodies against the SARS-CoV coronavirus. They fabricated a protein microarray using coronavirus proteins overexpressed in yeast. The proteins were spotted in duplicate onto microscope slides. Human serum samples from SARS infected or healthy subjects were screened using the protein chips, and bound antibodies were detected fluorescently using Cy3-labelled anti-human IgG or IgM antibodies. They found that their coronavirus protein arrays were able to accurately diagnose the presence of antibodies against coronaviruses in greater than 90% of the patient serum samples they tested. Accordingly, the proteome array they created was an excellent way to quickly diagnose those patients displaying symptoms of SARS infection.
Another set of antigen–antibody experiments using proteome microarrays was performed by Michaud et al. (2003). To analyze antibody specificity, they screened 11 monoclonal and polyclonal antibodies against a yeast proteome microarray made up of approximately 5000 different yeast proteins. Fluorescent detection was performed using secondary antibodies labeled with Cy5. They found varying degrees of cross-reactivity among the antibodies that could not necessarily be predicted according to the amino acid sequences of the proteins in question. Their data should prove useful in evaluating assays using antibodies included in their study.
6. Conclusion
Proteome chip technology is an excellent high-throughput method used to probe an entire collection of proteins for a specific function or biochemistry. It is an exceptional new way to discover previously unknown multifunctional proteins, and to discover new functionalities for well-studied proteins.
Acknowledgement
We thank Heng Zhu for comments on the manuscript. This work was funded by grants from the NIH.
References
- Angenendt P., Glokler J., Murphy D., Lehrach H., Cahill D.J. Toward optimized antibody microarrays: a comparison of current microarray support materials. Anal. Biochem. 2002;309:253–260. doi: 10.1016/s0003-2697(02)00257-9. [DOI] [PubMed] [Google Scholar]
- Bertone P., Snyder M. Advances in functional protein microarray technology. FEBS J. 2005;272:5400–5411. doi: 10.1111/j.1742-4658.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- Brasch M.A., Hartley J.L., Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- Brizuela L., Braun P., LaBaer J. FLEXGene repository: from sequenced genomes to gene repositories for high-throughput functional biology and proteomics. Mol. Biochem. Parasitol. 2001;118:155–165. doi: 10.1016/s0166-6851(01)00366-8. [DOI] [PubMed] [Google Scholar]
- Charles P.T., Goldman E.R., Rangasammy J.G., Schauer C.L., Chen M.S., Taitt C.R. Fabrication and characterization of 3D hydrogel microarrays to measure antigenicity and antibody functionality for biosensor applications. Biosens. Bioelectron. 2004;20:753–764. doi: 10.1016/j.bios.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Colca J.R., Harrigan G.G. Photo-affinity labeling strategies in identifying the protein ligands of bioactive small molecules: examples of targeted synthesis of drug analog photoprobes. Comb. Chem. High Throughput Screen. 2004;7:699–704. doi: 10.2174/1386207043328337. [DOI] [PubMed] [Google Scholar]
- Delehanty J.B. Printing functional protein microarrays using piezoelectric capillaries. Methods Mol. Biol. 2004;264:135–143. doi: 10.1385/1-59259-759-9:135. [DOI] [PubMed] [Google Scholar]
- Delehanty J.B., Ligler F.S. Method for printing functional protein microarrays. Biotechniques. 2003;34:380–385. doi: 10.2144/03342mt02. [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Meyers R.E., Cantley L.C. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L.J., Bastuck S., Dumpelfeld B., Edelmann A., Heurtier M.A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A.M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J.M., Kuster B., Bork P., Russell R.B., Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., Remor M., Hofert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M.A., Copley R.R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gelperin D.M., White M.A., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Gerstein M., Dumont M.E., Phizicky E.M., Snyder M., Grayhack E.J. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard D.S., Wagner L., Feingold E.A., Shenmen C.M., Grouse L.H., Schuler G., Klein S.L., Old S., Rasooly R., Good P., Guyer M., Peck A.M., Derge J.G., Lipman D., Collins F.S., Jang W., Sherry S., Feolo M., Misquitta L., Lee E., Rotmistrovsky K., Greenhut S.F., Schaefer C.F., Buetow K., Bonner T.I., Haussler D., Kent J., Kiekhaus M., Furey T., Brent M., Prange C., Schreiber K., Shapiro N., Bhat N.K., Hopkins R.F., Hsie F., Driscoll T., Soares M.B., Casavant T.L., Scheetz T.E., Brown-stein M.J., Usdin T.B., Toshiyuki S., Carninci P., Piao Y., Dudekula D.B., Ko M.S., Kawakami K., Suzuki Y., Sugano S., Gruber C.E., Smith M.R., Simmons B., Moore T., Waterman R., Johnson S.L., Ruan Y., Wei C.L., Mathavan S., Gunaratne P.H., Wu J., Garcia A.M., Hulyk S.W., Fuh E., Yuan Y., Sneed A., Kowis C., Hodgson A., Muzny D.M., McPherson J., Gibbs R.A., Fahey J., Helton E., Ketteman M., Madan A., Rodrigues S., Sanchez A., Whiting M., Madari A., Young A.C., Wetherby K.D., Granite S.J., Kwong P.N., Brinkley C.P., Pearson R.L., Bouffard G.G., Blakesly R.W., Green E.D., Dickson M.C., Rodriguez A.C., Grimwood J., Schmutz J., Myers R.M., Butterfield Y.S., Griffith M., Griffith O.L., Krzywinski M.I., Liao N., Morin R., Palmquist D. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hall D.A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Hartley J.L., Temple G.F., Brasch M.A. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A.R., Sassi H., Nielsen P.A., Rasmussen K.J., Andersen J.R., Johansen L.E., Hansen L.H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sorensen B.D., Matthiesen J., Hendrickson R.C., Gleeson F., Pawson T., Moran M.F., Durocher D., Mann M., Hogue C.W., Figeys D., Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hook S.S., Means A.R. Ca(2+)/CaM-dependent kinases: from activation to function. Annu. Rev. Pharmacol. Toxicol. 2001;41:471–505. doi: 10.1146/annurev.pharmtox.41.1.471. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhu H., Haggarty S.J., Spring D.R., Hwang H., Jin F., Snyder M., Schreiber S.L. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber W., Kidd B.A., Tomooka B.H., Lee B.J., Bruce B., Fries J.F., Sonderstrup G., Monach P., Drijfhout J.W., van Venrooij W.J., Utz P.J., Genovese M.C., Robinson W.H. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jones R.B., Gordus A., Krall J.A., MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- Kattah M.G., Alemi G.R., Thibault D.L., Balboni I., Utz P.J. A new two-color Fab labeling method for autoantigen protein microarrays. Nat. Methods. 2006;3:745–751. doi: 10.1038/nmeth910. [DOI] [PubMed] [Google Scholar]
- Kramer A., Feilner T., Possling A., Radchuk V., Weschke W., Burkle L., Kersten B. Identification of barley CK2alpha targets by using the protein microarray technology. Phytochemistry. 2004;65:1777–1784. doi: 10.1016/j.phytochem.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Punna T., Peregrin-Alvarez J.M., Shales M., Zhang X., Davey M., Robinson M.D., Paccanaro A., Bray J.E., Sheung A., Beattie B., Richards D.P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M.M., Vlasblom J., Wu S., Orsi C., Collins S.R., Chandran S., Haw R., Rilstone J.J., Gandi K., Thompson N.J., Musso G., St Onge P., Ghanny S., Lam M.H., Butland G., Altaf-Ul A.M., Kanaya S., Shilatifard A., O'Shea E., Weissman J.S., Ingles C.J., Hughes T.R., Parkinson J., Gerstein M., Wodak S.J., Emili A., Greenblatt J.F. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kusnezow W., Jacob A., Walijew A., Diehl F., Hoheisel J.D. Antibody microarrays: an evaluation of production parameters. Proteomics. 2003;3:254–264. doi: 10.1002/pmic.200390038. [DOI] [PubMed] [Google Scholar]
- Lennon G., Auffray C., Polymeropoulos M., Soares M.B. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Lesaicherre M.L., Lue R.Y., Chen G.Y., Zhu Q., Yao S.Q. Intein-mediated biotinylation of proteins and its application in a protein microarray. J. Am. Chem. Soc. 2002;124:8768–8769. doi: 10.1021/ja0265963. [DOI] [PubMed] [Google Scholar]
- MacBeath G., Schreiber S.L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Michaud G.A., Salcius M., Zhou F., Bangham R., Bonin J., Guo H., Snyder M., Predki P.F., Schweitzer B.I. Analyzing antibody specificity with whole proteome microarrays. Nat. Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- Mitsopoulos G., Walsh D.P., Chang Y.T. Tagged library approach to chemical genomics and proteomics. Curr. Opin. Chem. Biol. 2004;8:26–32. doi: 10.1016/j.cbpa.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S.D. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- Phizicky E., Bastiaens P.I., Zhu H., Snyder M., Fields S. Protein analysis on a proteomic scale. Nature. 2003;422:208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R.R., Schmidt M.C., Rachidi N., Lee S.J., Mah A.S., Meng L., Stark M.J., Stern D.F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P.F., Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A.Y., Walter J.C., LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Ramachandran N., Larson D.N., Stark P.R., Hainsworth E., LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005;272:5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- Reboul J., Vaglio P., Rual J.F., Lamesch P., Martinez M., Armstrong C.M., Li S., Jacotot L., Bertin N., Janky R., Moore T., Hudson J.R., Jr., Hartley J.L., Brasch M.A., Vandenhaute J., Boulton S., Endress G.A., Jenna S., Chevet E., Papasotiropoulos V., Tolias P.P., Ptacek J., Snyder M., Huang R., Chance M.R., Lee H., Doucette-Stamm L., Hill D.E., Vidal M. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Hill D.E., Vidal M. ORFeome projects: gateway between genomics and omics. Curr. Opin. Chem. Biol. 2004;8:20–25. doi: 10.1016/j.cbpa.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Rual J.F., Hirozane-Kishikawa T., Hao T., Bertin N., Li S., Dricot A., Li N., Rosenberg J., Lamesch P., Vidalain P.O., Clingingsmith T.R., Hartley J.L., Esposito D., Cheo D., Moore T., Simmons B., Sequerra R., Bosak S., Doucette-Stamm L., Le Peuch C., Vandenhaute J., Cusick M.E., Albala J.S., Hill D.E., Vidal M. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Speer R., Wulfkuhle J.D., Liotta L.A., Petricoin E.F., 3rd Reverse-phase protein microarrays for tissue-based analysis. Curr. Opin. Mol. Ther. 2005;7:240–245. [PubMed] [Google Scholar]
- Sreekumar A., Nyati M.K., Varambally S., Barrette T.R., Ghosh D., Lawrence T.S., Chinnaiyan A.M. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res. 2001;61:7585–7593. [PubMed] [Google Scholar]
- Stillman B.A., Tonkinson J.L. FAST slides: a novel surface for microarrays. Biotechniques. 2000;29:630–635. doi: 10.2144/00293pf01. [DOI] [PubMed] [Google Scholar]
- Washburn M.P., Wolters D., Yates J.R., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Wheeler D.L., Church D.M., Edgar R., Federhen S., Helmberg W., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., Suzek T.O., Tatusova T.A., Wagner L. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Mitchell T., Miller P., Dean R.A., Gerstein M., Snyder M. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Snyder M. Proteomics. Annu. Rev. Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- Zhu H., Hu S., Jona G., Zhu X., Kreiswirth N., Willey B.M., Mazzulli T., Liu G., Song Q., Chen P., Cameron M., Tyler A., Wang J., Wen J., Chen W., Compton S., Snyder M. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Klemic J.F., Chang S., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]


