Abstract
Candida albicans cells of opposite mating types are thought to conjugate during infection in mammalian hosts, but paradoxically, the mating-competent opaque state is not stable at mammalian body temperatures. We found that anaerobic conditions stabilize the opaque state at 37°C, block production of farnesol, and permit in vitro mating at 37°C at efficiencies of up to 84%. Aerobically, farnesol prevents mating because it kills the opaque cells necessary for mating, and as a corollary, farnesol production is turned off in opaque cells. These in vitro observations suggest that naturally anaerobic sites, such as the efficiently colonized gastrointestinal (GI) tract, could serve as niches for C. albicans mating. In a direct test of mating in the mouse GI tract, prototrophic cells were obtained from auxotrophic parent cells, confirming that mating will occur in this organ. These cells were true mating products because they were tetraploid, mononuclear, and prototrophic, and they contained the heterologous hisG marker from one of the parental strains.
Candida albicans is a common, typically benign mammalian commensal that can exploit defects in host defenses to become an effective opportunistic pathogen. Untreated, over 95% of AIDS patients will be infected by C. albicans, and this fungal pathogen has become the fourth leading nosocomial isolate, causing mortality rates between 30 and 70% in systemically infected patients (8). C. albicans is a diploid organism that exploits genetic and phenotypic variability created through yeast-hypha transitions (4), phase transitions (30), and mating (2) to maintain commensalism and facilitate pathogenesis.
Like most dimorphic fungal pathogens, C. albicans exhibits an inoculum size effect in that the cells develop as budding yeasts when inoculated at ≥106 cells per ml and as mycelia when inoculated at <106 cells per ml (23). The active principle mediating this effect is E,E-farnesol (10). In keeping with the precedent established by homoserine lactone-based signaling in gram-negative bacteria (7), the inoculum size effect in fungi is also called quorum sensing (10, 23) and the extracellular cell density-dependent signals are called quorum-sensing molecules (QSMs). Farnesol produced by C. albicans was the first QSM identified in a eukaryotic organism, and this compound was found to prevent the yeast-to-mycelium conversion (10), resulting in actively budding yeasts without otherwise altering cellular growth rates (10, 25). Farnesol is produced when the cells are grown in the presence of oxygen, but during anaerobic growth the cells neither produce nor respond to exogenous farnesol, even at concentrations of up to 1.2 mM (6).
Although C. albicans was not thought to have a sexual cycle until recently (2, 11), molecular and genomic investigations have established that a signaling pathway, very similar to that characterized in the well-studied yeast Saccharomyces cerevisiae, controls mating. A unique component of C. albicans mating is the role of white-opaque switching; for a or α cells of C. albicans to be competent to mate, they must first undergo a phenotypic switch from the white form to the opaque form (19). These opaque-phase a and α cells mate ca. 106 times more efficiently than the white-phase cells do (19). Paradoxically, this mating-competent opaque form is unstable at temperatures compatible with growth in a mammalian host (29). Because at 37°C the opaque cells switch back to the white phase en masse in only 3 h (26, 31), how can mating occur at human body temperature? Aerobic mating in C. albicans has been extensively investigated (5, 11, 17), but nothing is known about anaerobic mating. In this paper we show that mating in C. albicans can occur in vitro anaerobically even at 37°C and efficient mating is permitted in these conditions because anaerobic opaque cells are stable at 37°C for at least 1 week. Additionally, we have recovered true mating products from a direct mating test in the mouse gastrointestinal (GI) tract. Because in disseminated candidiasis, C. albicans cells often originate from the anaerobic gastrointestinal tract where they reside as commensals, this observation suggests a potential niche for Candida mating (15).
MATERIALS AND METHODS
Strains and media.
All the strains used in this study are described in Table 1. Glucose-phosphate-proline (GPP) is the growth medium for fungi described by Kulkarni and Nickerson (14). For the anaerobic growth of Candida albicans, regular GPP medium (10 ml) was supplemented with 200 μl of 1 mM oleic acid in methanol, 200 μl of 4 mM nicotinic acid, and 1 ml of 50 mM NH4Cl (6). Synthetic complete (SC) medium was used for all the experiments with cells in the opaque phase. It is composed of 2% (wt/vol) dextrose, 0.67% (wt/vol) yeast nitrogen base without amino acids, and is supplemented when needed with arginine at 70 μg/ml; after sterilization, it is supplemented with uridine at 100 μg/ml, tryptophan at 60 μg/ml, lysine at 60 μg/ml, histidine at 60 μg/ml, and 0.1 μM zinc sulfate. The progenies were recovered by plating the cells onto glucose-phosphate-ammonium sulfate plates with a 5-amino-acid dropout (uridine, arginine, histidine, tryptophan, and lysine) (GPA−5aa medium). GPA medium contains the following (per 900 ml of distilled water): 4.0 g of KH2PO4, 3.2 g of NaH2PO4, 4.5 g ammonium sulfate, and 0.7 g of MgSO4 · 7H2O. After the medium was autoclaved, 100 ml of 20% (wt/vol) glucose, vitamin mix, and mineral mix (14) were added.
TABLE 1.
Strains used in this study
| C. albicans strain | Relevant genotype or description | Reference(s) |
|---|---|---|
| 3294 (CNC43) | MTLa/ahis1/his1 ura3/ura3 arg5,6/arg5,6 | 17, 22 (from P. T. Magee) |
| 3315 (A505) | MTLα/α trp1/trp1 lys2/lys2 | 12, 17 (from P. T. Magee) |
| 3740 (CNC43) | MTLα/α his1/his1 ura3/ura3 arg5,6/arg5,6 | 17, 22 (from P. T. Magee) |
| 3745 (A505) | MTLa/atrp1/trp1 lys2/lys2 | 12, 16, 17 |
| DDCA45 | Aerobic mating zygote between strains 3740 and 3745 | This work |
Farnesol production.
Candida albicans CNC43 (3740) white and opaque cultures were grown aerobically in 50 ml of GPP medium (10) supplemented with histidine, arginine, and uridine and anaerobically in 50 ml of the defined anaerobic GPP medium (6) supplemented with the required amino acids. Microscopically, the opaque cultures appeared to have 100% opaque cells at the time of inoculation and at the time of harvesting. The white and opaque anaerobic cultures were grown at 37°C for 72 h, whereas the aerobic white and opaque cultures were grown at 25°C for 24 h. The cell-free culture supernatants were extracted with ethyl acetate and analyzed by gas chromatography/mass spectrometry as described previously (10, 21).
In vitro aerobic mating.
Opaque cells of both C. albicans strains CNC43 (3740) MTLα/α and A505 (3745) MTLa/a (Table 1) were obtained from white-phase cells that had been grown for 24 h at 30°C in liquid SC medium (28). A 1-μl aliquot was diluted 100-fold with sterile water and plated at a very low density on SC plates containing 0.0005% phloxine B (1). The plates were incubated for 1 week at 25°C to isolate mature opaque colonies (dark pink colonies). The cells from an opaque colony were analyzed by microscopy to verify the cells had the opaque phase-specific elongated shape. Opaque starter cultures of both strains 3740 (MTLα/α) and 3745 (MTLa/a) were inoculated from the SC plates into fresh SC liquid medium (28) and grown aerobically at 25°C overnight with shaking at 200 rpm. The cells were harvested by centrifugation and washed three times in phosphate-buffered saline (PBS) (pH 6.5). Opposite mating types were adjusted to optical density at 600 nm (OD600) values so that inoculation of 100 μl per parent into 5 ml of liquid medium gave final OD600 values of 0.1, corresponding to 2.5 × 106 cells/ml per parent, or 0.5, corresponding to 1.2 × 107 cells/ml per parent. The liquid glucose-phosphate-proline medium was supplemented with oleic acid, nicotinic acid, ammonium chloride (6), arginine at 70 μg/ml; after sterilization, the medium was supplemented with uridine at 100 μg/ml, tryptophan at 60 μg/ml, lysine at 60 μg/ml, histidine at 60 μg/ml, and 0.1 μM zinc sulfate. The cells were incubated at 25°C in two 25-ml flasks, one with shaking at 250 rpm and the other without shaking. After 24 h of incubation, cells were harvested, washed three times in PBS, and spread on two types of agar plates: GPA−5aa on which only mating products are able to form colonies and yeast extract-peptone-dextrose (YPD) on which both parents and the mating products can grow. To determine the mating efficiency in the presence of farnesol, the same conditions were used with 20 μM farnesol added to the GPP medium.
Evans blue and propidium iodide staining of aerobic cells.
Both white and opaque starter cultures were grown aerobically for 24 h at 25°C in 5 ml of SC medium. After growth, the cells were washed three times in PBS, resuspended at a cell concentration of 1.2 × 107 cells/ml in 5 ml of fresh SC medium supplemented with three different concentrations (2, 20, and 40 μM) of farnesol from a stock solution of 10 mM farnesol in methanol, and shaken (250 rpm) at 25°C for 4 h. Cells were assessed for viability by Evans blue (13) and propidium iodide (27) staining after every hour. In each case, 1 ml of cell culture was harvested, washed in PBS three times, and stained with either 1 mg/ml Evans blue or 50 μg/ml propidium iodide for 5 min. The control cells were grown for 4 h under identical conditions in two flasks, one containing SC medium without farnesol and another containing SC medium with methanol. Cell staining was assessed with a confocal fluorescence microscope, using an excitation wavelength of 620 nm and an emission wavelength of 680 nm for Evans blue and an excitation wavelength of 488 nm and an emission wavelength of 585 nm for propidium iodide.
In vitro anaerobic mating.
Opaque starter cultures of both MTLa/a and MTLα/α were inoculated separately in 5 ml of fresh SC medium and grown aerobically at 25°C overnight. The cells were harvested and washed three times in PBS. Opposite mating types adjusted to give three different final OD600 values of 0.1, 0.3, and 0.5, corresponding to 2. 5 × 106 cells/ml per parent, 4.2 × 106 cells/ml per parent, and 1.2 × 107 cells/ml per parent, respectively, were inoculated into triplicate tubes each containing 10 ml of anaerobic GPP medium (6) supplemented with the required amino acids and incubated at three different temperatures, 25°C, 30°C, and 37°C, for 3 days. After 24, 48, and 72 h of incubation, the cells in individual tubes were harvested and washed, and serial dilutions were plated onto two types of agar plates: GPA−5aa on which only mating products are able to form colonies and YPD on which both parents and mating products can grow. To compare these results with the mating efficiency in the presence of farnesol, the same conditions were used except that 10 or 20 μM farnesol was added to 10 ml of anaerobic medium. Because the farnesol stock solution was dissolved in methanol, for a control, the opaque cells were also grown in 10 ml of GPP medium supplemented with 20 μl of methanol.
Stability of opaque cells at 37°C under anaerobic conditions.
Our goal was to determine whether the opaque cells used in the in vitro anaerobic mating experiments maintained an opaque cell-specific gene expression pattern. The MTLa/a and MTLα/α opaque cells were grown separately in 10 ml of anaerobic GPP medium (6) supplemented with the respective required amino acids for 7 days at 37°C. White MTLa/a and MTLα/α cells were used as controls. The white cells were obtained by growing opaque MTLa/a and MTLα/α cells in the same medium aerobically overnight at 37°C. Under these conditions, a high percentage of cells (≥80%) switched to the more stable white phase. All the cells were pelleted by centrifugation at 4,000 × g in a microcentrifuge for 5 min. The supernatant was discarded, and the pellet was processed according to yeast protocol I of the QIAGEN RNeasy mini kit (QIAGEN). RNA integrity and quantification were assessed using standard protocols (27). The reverse transcription reaction was performed by the two-step protocol of the RETROscript reverse transcription-PCR (RT-PCR) kit (Ambion), using a 20-μl volume with a final concentration of 100 ng of total RNA. The same PCR primers used by Miller and Johnson (19) were used to detect WH11 (5′-CAATTAAACATGTCCGACTTAGG-3′ and 5′-ATTGAGGTTACTCACTCATTG-3′) and OP4 (5′-CACAAGCCACCATCTTAGCC-3′ and 5′-CACTAGCAGCAGCTGGAGTG-3′). We used the mRNA for β-tubulin (TUB mRNA) as the loading control. The primers used for TUB were 5′-CAACTGGTCAATGTGGTAATCA-3′ and 5′-GTAATGACCTTTAGCCCAAACATTG-3′. The PCR thermal cycling conditions were as follows: an initial step at 94°C for 3 min; followed by 30 cycles, with 1 cycle consisting of 30 seconds at 94°C, 30 seconds at each specific annealing temperature (46°C for WH11, 52°C for OP4, and 49°C for TUB), and 1 min and 30 seconds at 72°C; and a final step at 72°C for 7 min.
Growth curves for aerobic and anaerobic cells in GPP medium.
Opaque MTLa/a cells and MTLα/α cells were inoculated separately at a cell density of 2.5 × 106 cells/ml in two Hungate tubes, each containing 10 ml of anaerobic GPP (6) liquid medium supplemented with the auxotrophic requirements and incubated at 25°C and 37°C for 7 days. The OD600 was read every 24 h. The aerobic growth curves were monitored for the same two strains grown in 10 ml of aerobic GPP (obtained without flushing nitrogen and without the addition of resazurin) incubated at 25°C. The OD600 was recorded every hour.
In vivo mating experiment.
For the experimental oral administration of C. albicans, the opaque MTLa/a and MTLα/α cells were grown for 24 h in 50 ml of SC medium supplemented with amino acids at 25°C with aeration. Cells were harvested by centrifugation at 4,750 × g for 10 min and washed three times with 50 ml of sterile phosphate-buffered saline before adjusting to the proper concentrations using a Petroff-Hausser counter. Outbred, 4- to 6-week-old (20 to 25 g), CF-1 female mice obtained from a commercial supplier (Charles River Laboratories, Wilmington, MA) were randomly allocated to groups of two to four animals, placed in polycarbonate cages with stainless steel wire tops, using aspen shavings as bedding material (Harlan Teklad Laboratory Grade Sano-Chips, Madison, WI), and maintained on a 12-hour light/12-hour dark cycle in heated, thermostatically controlled rooms for the duration of the studies. The mice were fed a commercial rodent diet (4% Mouse/Rat Diet 7001; Harlan Teklad, Madison, WI) ad libitum. Tap water was provided in glass bottles fitted with stainless steel nipples mounted in rubber corks. After a 14-day observation period, mice were not allowed to have food or water for 3 hours prior to the experimental oral C. albicans administration. A group of four mice was given MTLa/a cells in their drinking bottles at a concentration of 105 cells/ml for 3 hours, followed by MTLα/α cells for 3 hours. Subsequently, the water bottles with MTLa/a cells only and MTLα/α cells only were switched every 3 hours for 24 h. After this oral exposure to C. albicans-containing water, mice were allowed to rest at normal conditions for 3 days before euthanasia and the removal of their colons and ceca. In control experiments, one group of three mice was exposed to MTLa/a cells only, and another group of three mice was exposed to MTLα/α cells only. A final group of two mice, the negative-control group, were not provided with either MTLa/a or MTLα/α. The entire colon and cecum were harvested from each animal, dissected longitudinally, and immersed in 5 ml of phosphate-buffered saline followed by vigorous vortexing. Four replicates (0.2 ml) from each tube were then plated at 30°C for 48 h on defined synthetic dextrose (SD) medium (28) without added amino acids but with ampicillin (1.4 mg/ml) and streptomycin (125 μg/ml) to prevent bacterial growth. The experimental protocol, housing, and care of the mice were in accordance with the approved guidelines of the University of Nebraska—Lincoln Institutional Animal Care and Use Committee.
Genetic analysis of the in vivo mating progenies.
Ten randomly picked prototrophic colonies recovered from the large intestines of the mice were restreaked onto YPD plates, and PCR was done on individual colonies to identify whether hisG sequence from Salmonella enterica serovar Typhimurium is present. This specific sequence is found in one of the parents, CNC43 (ura3Δ his1Δ arg5,6Δ). The construction of this strain has been described by Negredo et al. (22). The primers used for this sequence were 5′-TCCTCAAACGCTACCTCGACC-3′ and 5′-ATCTTCTCGATCGGCAGTACCAG-3′. The PCR thermal cycling conditions were an initial denaturation step at 93°C for 5 min; followed by 35 cycles, with 1 cycle consisting of 30 seconds at 94°C, 45 seconds at 50°C, and 1 min at 72°C; and the last step at 72°C for 10 min. The control used was the tubulin (TUB) gene, and the same primers and conditions were used as for the experiment describing the stability of the opaque cells anaerobically.
FACS analysis.
Log-phase cells (approximately 107 cells/ml) grown at 25°C were fixed for 4 h in 37% formaldehyde that contained 11% methanol, washed with phosphate-buffered saline six times, treated with RNase (1 mg/ml, 37°C, 75 min), and washed again with PBS. Before cell sorting, cells were stained with propidium iodide (50 μg/ml). Fluorescence-activated cell sorting (FACS) analysis was performed on a FACSCalibur using an excitation wavelength of 488 nm. Emission from propidium iodide was collected through a 585/42 band pass filter. Data were collected in the logarithmic mode at rates of a few hundred cells per second.
RESULTS
Farnesol blocks aerobic mating of C. albicans.
Farnesol is a regulator of C. albicans morphogenesis (10, 23), and a morphogenetic switch from white to opaque cells is required for mating (2). However, the role of farnesol in mating has not been investigated previously. Farnesol, the extracellular QSM for C. albicans, reduces the aerobic mating efficiency (Table 2). MTLa/a opaque cells were mixed with MTLα/α opaque cells in 5 ml of glucose-phosphate-proline medium (6) supplemented with amino acids to fulfill the parental auxotrophic markers under four conditions (with and without farnesol and with and without shaking). Cells were incubated for 24 h at 25°C, and the mating efficiency was established by spreading the mating mixes on minimal agar plates to detect mated cells and on YPD to identify the input cells. The highest mating efficiency obtained was 8.7% for a nonshaking culture with a starting cell density per strain of 1.2 × 107 cells/ml (Table 2). This value compares favorably with previously published mating efficiencies in other growth media (3). However, when 20 μM farnesol was included, the mating efficiencies dropped to negligible levels (Table 2). Under these conditions, no prototrophic colonies were obtained. We conclude that farnesol blocks aerobic mating of C. albicans.
TABLE 2.
Aerobic mating efficiency at 25°Ca
| Condition | Farnesol (20 μM)b | Cell density (OD600) | Mating efficiency after 24 h (%) |
|---|---|---|---|
| With shaking | − | 0.05 | <0.2d |
| − | 0.25c | <0.2 | |
| + | 0.05 | <0.2 | |
| + | 0.25 | <0.2 | |
| Without shaking | − | 0.05 | 2.7 |
| − | 0.25 | 8.73 | |
| + | 0.05 | <0.2 | |
| + | 0.25 | <0.2 |
A quantitative mating assay was used to determine the aerobic mating efficiency for two Candida strains, strains 3740 (MTLα/α) and 3745 (MTLa/a). The cells were grown at 25°C with and without shaking and with and without the addition of 20 μM farnesol. The results presented are averages of three independent experiments.
−, farnesol not added; +, farnesol added.
Roughly 1.2 × 107 cells/ml. The aerobic mating efficiency in liquid medium is improved at higher cell densities (3).
Based on zero prototrophic colonies and ca. 500 auxotrophic colonies.
Farnesol kills opaque cells under aerobic conditions.
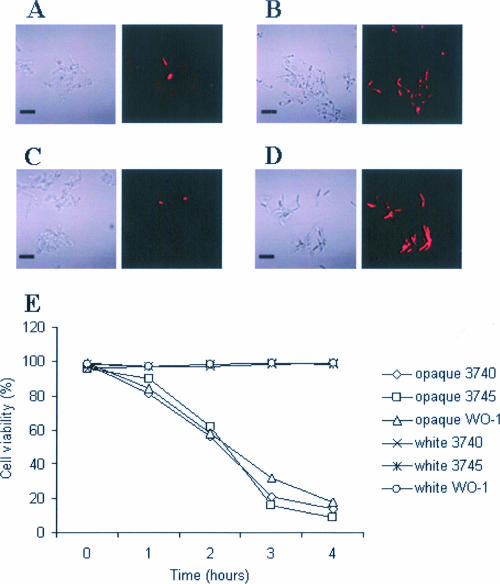
Three opaque cell types (strains 3740, 3745, and WO-1) were grown separately in liquid medium supplemented with farnesol (2, 20, or 40 μM) and assessed for viability by Evans blue (13) and propidium iodide staining (27) at hourly intervals (Fig. 1). The opaque cell cultures grown with 40 μM farnesol had >80% stained cells after 4 h (Fig. 1B and D), whereas opaque cells grown without farnesol (Fig. 1A and C) or with methanol as a control (not shown) had ≤2% stained cells. Lower concentrations of farnesol killed the opaque cells more slowly (data not shown). Significantly, white cells of strains 3740, 3745, and WO-1 were not killed by these levels of farnesol (Fig. 1E). This latter finding is not surprising because white cells of most strains of C. albicans are able to tolerate up to 250 to 300 μM farnesol without any change in their growth rates (10, 25). These observations suggest that farnesol lowers the mating efficiency under aerobic conditions because it kills the opaque cells necessary for mating.
FIG. 1.
Evans blue staining of aerobically grown opaque MTLa/a and MTLα/α C. albicans. MTLα/α (strain 3740) (A and B), MTLa/a (strain 3745) (C and D), and WO-1 opaque cells were grown separately at 25°C in SD medium supplemented with the necessary amino acids, either with 40 μM farnesol (B and D) or without farnesol (A and C). Cells were assessed for viability after 4 h with 1 mg/ml Evans blue. The cells were analyzed by fluorescence microscopy; cells that stain are dead (12). Both phase-contrast (left panels) and fluorescence (right panels) microscopy pictures are shown. Bar, 10 μm. (E) Graph with the percentage of cell viability for opaque and white cells after treatment with 40 μM farnesol. Data presented are averages for two independent experiments. Similar values were obtained when the cells were treated with 50 μg/ml propidium iodide.
Farnesol production is turned off in opaque cells.
C. albicans 3740 was grown in triplicate liquid cultures as both white and opaque cells under both aerobic and anaerobic conditions. Only the aerobic white cells produced farnesol (37.4 ± 4.8 μg/g [dry weight]). The aerobic opaque cells and both anaerobic cultures did not produce detectable farnesol levels (<0.5 μg/g [dry weight]). The absence of farnesol production by both anaerobic cultures agrees with our previous report (6) that four wild-type strains of C. albicans (A72, 10261, MEN, and SC5314) did not produce detectable farnesol levels when grown anaerobically.
In vitro mating under anaerobic conditions.
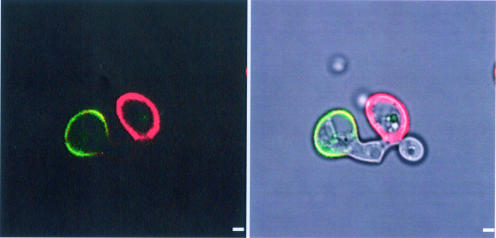
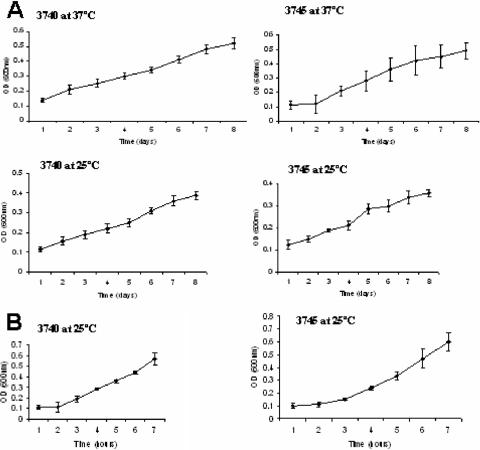
C. albicans cells do not produce farnesol under anaerobic conditions (6), suggesting that mating could be more efficient in the absence of oxygen. To investigate whether mating occurs anaerobically in vitro, we mixed opaque cells from two pairs of C. albicans strains carrying complementary auxotrophic markers and monitored the mating efficiency every 24 h for 3 days (Table 3). Significantly, anaerobic mating occurred at both 25 and 37°C: this mating reached 74 to 85% efficiency by the third day, it was cell density dependent, and it was not influenced by the presence of 20 μM farnesol. Figure 2 shows an anaerobic mating pair at 25°C as examined by fluorescence microscopy. The 3745 MTLa/a cells were prestained with rhodamine-concanavalin A (ConA), and the 3740 MTLα/α cells were prestained with fluorescein isothiocyanate (FITC)-ConA. The unstained conjugation tube shows that mating occurred after mixing. Also, anaerobic mating, like anaerobic growth (6), appears to be a comparatively slow process. At both 25 and 37°C, the percentage of mated cells increased steadily over the first 3 days (Table 3). This lengthy time frame probably reflects the altered metabolism operative under stringently anaerobic conditions, as the doubling time for both mating types growing anaerobically was ca. 3 days at both 25 and 37°C (Fig. 3A), instead of the 3 h observed for the same strains growing aerobically (Fig. 3B).
TABLE 3.
Mating efficiency of C. albicans under anaerobic conditions in defined liquid media with and without farnesola
| Condition | Temp (°C) | Cell density (OD600) | Mating efficiency (%) at:
|
||
|---|---|---|---|---|---|
| 1 day | 2 days | 3 days | |||
| No added farnesol | 25 | 0.05 | 1.9 | 2.2 | 1.9 |
| 25 | 0.15 | 1.4 | 1.6 | 64.2 | |
| 25 | 0.25 | 3.4 | 16.4 | 74.3 | |
| 37 | 0.05 | 14 | 22.5 | 76.4 | |
| 37 | 0.15 | 20.5 | 25.2 | 82.3 | |
| 37 | 0.25 | 19.5 | 46.1 | 84.5 | |
| 20 μM E,E-farnesol | 25 | 0.05 | <0.2 | 2.5 | 1.4 |
| 25 | 0.15 | 0.2 | 1.5 | 78.8 | |
| 25 | 0.25 | 2.6 | 5.5 | 68 | |
| 37 | 0.05 | 2.4 | 7 | 69.1 | |
| 37 | 0.15 | 33.1 | 18 | 76 | |
| 37 | 0.25 | 15.5 | 34 | 81.7 | |
A quantitative mating assay was used to determine mating efficiency at two temperatures and three initial cell densities (see Materials and Methods). Data are the averages of two mating experiments, one between strains 3740 (MTLα/α) and 3745 (MTLa/a) and the other between strains 3294 (MTLa/a) and 3315 (MTLα/α). Equivalent data were observed for cells grown at 25 and 37°C with 10 μM farnesol (not shown) and at 30°C with 0, 10, and 20 μM farnesol (not shown).
FIG. 2.
In vitro anaerobic mating in C. albicans. MTLa/a (strain 3745) cells were stained with rhodamine-ConA, and MTLα/α (strain 3740) cells were stained with FITC-ConA by the method of Lockhart et al. (16). The stained cells were mixed, incubated in anaerobic medium (6) for 72 h at 25°C, and analyzed by confocal microscopy. The left panel is the fluorescent image, whereas the right panel merges the phase-contrast and fluorescent images to visualize the unstained conjugation tube. Excitation and emission wavelengths were 543 and 560 nm for rhodamine-ConA and 488 and 505 to 525 nm for FITC-ConA. Bar, 2 μm.
FIG. 3.
(A) Growth curves for anaerobically grown opaque MTLa/a cells (strain 3745) and opaque MTLα/α cells (strain 3740) at 25°C and 37°C. Opaque MTLa/a and MTLα/α cells were inoculated separately in 10 ml of anaerobic GPP medium (6) supplemented with the auxotrophic requirements and incubated for 1 week at either 25°C or 37°C. The OD600 was monitored every 24 h. (B) Growth curves for aerobically grown opaque MTLa/a and MTLα/α cells at 25°C. Opaque cells were inoculated separately in 10 ml of aerobic GPP medium supplemented with the required amino acids. The OD600 was monitored every hour.
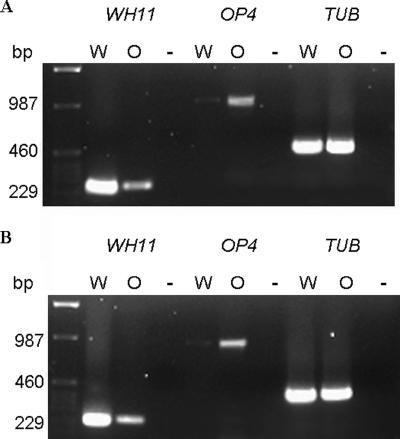
Critically, the opaque cells exhibit long-term stability anaerobically, even at 37°C. Confirmation that these cells are still opaque after 1 week at 37°C is provided by their having the larger size, elongated shape, and phloxin B staining expected of opaque cells (2, 30), and they can mate. After 2 weeks of anaerobic growth at 37°C, C. albicans 3740 cells were plated onto YPD supplemented with phloxine B (1) and incubated aerobically at 25°C. Over 40% of these colonies were opaque in appearance. Additionally, even after 1 week at 37°C, the anaerobic cells still have an opaque phase-specific gene expression pattern (2, 19, 30): the expression of opaque phase-specific OP4 (20) is elevated, while the expression of white phase-specific WH11 (31) is dramatically reduced (Fig. 4). Thus, the distinctive properties of anaerobically grown opaque cells might facilitate mating in locations, such as the colon, which are naturally anaerobic.
FIG. 4.
RT-PCR analysis of white phase- and opaque phase-specific gene expression for strain 3740 (MTLα/α) (A) and strain 3745 (MTLa/a) (B). White-phase cells (W) were grown aerobically at 37°C in GPP medium for 24 h, and opaque cells (O) were grown anaerobically at 37°C for 7 days. Total RNA was prepared from harvested cultures, and mRNA expression of a white phase-specific gene (WH11), an opaque phase-specific gene (OP4), and a control gene, β-tubulin (TUB), was performed using RT-PCR analysis as described in Materials and Methods. The negative control was no cDNA (−).
In vivo mating assay.
To test the prediction that C. albicans mating may occur in the large intestine, which is an anaerobic environment, we devised an in vivo mating assay in mice. The mice were provided with opaque C. albicans in their drinking water at a cell density of 105/ml. Twelve mice were tested: two were controls not given C. albicans, three were provided with C. albicans MTLa/a (3745) only, and three were provided with C. albicans α/α (3740) only. For the remaining four mice, the water bottles with MTLa/a or MTLα/α C. albicans strains were switched at 3-h intervals over a 24-h time span with cycles of MTLa/a followed by MTLα/α and then back to MTLa/a. In this way, the opposite mating types will encounter one another only in the mouse gastrointestinal tract. After an initial 24-h colonization period, the mice were returned to plain water only for another 3 days whereupon they were euthanized using carbon dioxide and the colon and cecum were removed by dissection. The 4-day time period was chosen on the basis of the time needed for anaerobic mating in vitro (Table 3). In each case, the weights of water consumed per mouse were equivalent, and all of the mice appeared healthy throughout. The colon contents were collected, dispersed, and spread on minimal SD agar plates. Under these minimal conditions, the individual a and α mating types should not grow, and as expected, no colonies were obtained from the no-C. albicans or single-MTL controls. However, prototrophic colonies were obtained from the mice colonized with MTLα/α and MTLa/a alternately. For the four mouse colons, each with four replicates, the numbers of colonies detected were 4 (3, 1, 0, and 0), 15 (6, 2, 3, and 4), 20 (5, 5, 4, and 6), and 0 (0, 0, 0, and 0) while for the four ceca. the CFU were 3 (1, 1, 1, and 0), 1 (0, 1, 0, 0), 3 (1, 1, 1, and 0) and 0 (0, 0, 0, and 0), respectively. The colonies were transferred onto a secondary SD agar plate to confirm that they were prototrophic for the genetic markers and onto a BiGGY agar plate (24) to confirm that they were C. albicans. Negative-control mice in this experiment, as well as experiments conducted for the past 3 years (21), did not show any C. albicans colonies. This absence suggests that the CF-1 mice used (Charles River Laboratories) were not initially colonized by C. albicans.
Genetic analysis of the in vivo mating progenies.
To confirm that the prototrophic colonies recovered were true mating products, we conducted a three-pronged analysis: cells were (i) stained with 4′,6′-diamidino-2-phenylindole (DAPI) to confirm the presence of a single nucleus, (ii) subjected to FACS analysis to analyze the DNA content, and (iii) analyzed by PCR to identify the specific hisG gene from Salmonella enterica serovar Typhimurium inserted into the 3740 parental strain (ura3Δ his1Δ arg5,6Δ).
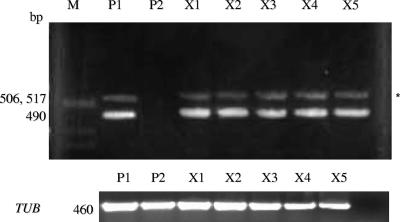
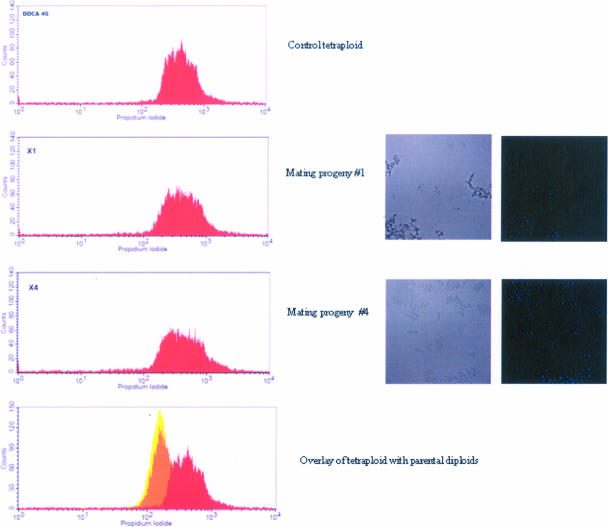
The construction of strain 3740 was explained by Negredo et al. (22). This hisG sequence is from S. enterica serovar Typhimurium; it is not normally found in C. albicans. Our PCR analysis detected the hisG sequence in strain 3740 but not in strain 3745 (Fig. 5) and in the tetraploid DDCA45 but not in the wild-type strain SC5314 (data not shown). Then, we randomly selected 10 of the 46 colonies from in vivo matings and used PCR to amplify this sequence. We recovered an S. enterica serovar Typhimurium hisG band of the expected size (490 bp) in all 10 mating progenies (Fig. 5). Staining with DAPI showed that they were all mononuclear. We also performed FACS analysis on the derived prototrophic strains. The prototrophic strains had twice the DNA content of the parent strains, which is consistent with them being tetraploid (Fig. 6). FACS analysis was performed on five randomly selected mating products, and all of them appeared to be tetraploid. Figure 6 shows two of these strains. These genetic experiments confirm that the prototrophic colonies recovered from the large intestine are indeed true mating products.
FIG. 5.
PCR detection of hisG in the mating progenies. PCR was done on 10 randomly selected prototrophic colonies recovered from the large intestine. The cells were restreaked onto YPD plates and grown for 2 days at 25°C whereupon PCR was done on individual colonies. The hisG gene from S. enterica serovar Typhimurium was recovered in all 10 prototrophs tested (the first five [X1 to X5] are shown). P1, 3740 parent; P2, 3745 parent; X, progeny. For a control, the tubulin (TUB) gene was used. M, molecular size markers.
FIG. 6.
FACS analysis of the DNA content of mating progenies and parental strains. Progeny cells derived from the in vivo mating experiment are mononuclear and have twice the DNA of control diploid cells. The control tetraploid (DDCA45) was made by aerobic mating of 3740 × 3745 (Table 1). Bar, 10 μm.
DISCUSSION
The white and opaque phases of C. albicans are dramatically distinct: they differ with regard to colony morphology, cell morphology, cell wall architecture, sterol content, antigenicity, accessibility to vital dyes, susceptibility to antifungal agents, sensitivity to neutrophils and oxidants, sugar assimilation, gene expression, and virulence in a mouse model (30). We have now extended this list: first, aerobically, white cells of C. albicans show exceptional tolerance for farnesol, while opaque cells are highly sensitive to farnesol (Fig. 1), and second, white cells of C. albicans excrete farnesol during aerobic growth, but opaque cells do not. As a corollary, aerobic mating is eliminated in the presence of farnesol (Table 2), probably because of the sensitivity of opaque cells to farnesol (Fig. 1).
How do the findings of this paper pertain to the evolution of mating in C. albicans? Farnesol does not block anaerobic mating because the 3-day mating efficiencies were effectively identical in the presence and absence of 20 μM farnesol (Table 3). However, in the presence of farnesol, the mating-competent opaque cells are stable only under anaerobic conditions. Anaerobically, opaque cells remain stable at 37°C for at least 1 week (Fig. 4). Thus, aerobic farnesol production may help restrict opaque cells to anaerobic conditions, thereby directing mating to anaerobic sites at mammalian body temperatures. In their comprehensive review on mating in C. albicans, Bennett and Johnson (2) suggested that the white-opaque switch (29, 30) had evolved to allow C. albicans to direct mating to specific environmental niches in the host. Our findings support this prediction.
With regard to where anaerobic mating would occur, the most obvious location is the gastrointestinal tract of the mammalian host. The GI tract is a natural environment for C. albicans (15) and one in which C. albicans associates with cell surfaces (15). We have now shown that the GI tract is permissive for mating. Thus, the relevance of anaerobic mating may focus on how C. albicans survives as a commensal in mammalian hosts rather than on invasion. This idea is consistent with the suggestion of Magee and Magee (18) that mating may facilitate the commensal lifestyle of this fungus.
As far as we know, the GI tract is the only naturally occurring anaerobic environment in mammals. However, C. albicans may create its own anaerobic environment in the interior of biofilms. These biofilms could be either C. albicans only or polymicrobial bacterial fungal biofilms, possibly associated with oral candidiasis (9). A similar scenario, e.g., kidney-associated biofilms, could explain the findings of Hull et al. (11). They found that that mating progenies were obtained from mouse kidneys following tail vein injection of the appropriate opposite mating type, a delivery route whereby it is not obvious how C. albicans cells could encounter prolonged anaerobic conditions. Alternatively, there may be another set of mating conditions still awaiting discovery.
Acknowledgments
We thank Danielle Shea for help with the FACS analysis and Julie Stone and Sadaf Khan for assistance with RNA extraction.
This work was supported in part by grants from CIHR (MOP 42516), and National Science Foundation (MCB-0110999), the University of Nebraska Tobacco Settlement Biomedical Research Enhancement Fund, the John C. and Nettie V. David Memorial Trust Fund, and the Farnesol and Candida albicans Research Fund, University of Nebraska Foundation.
This is NRC publication 47511.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Anderson, J., and D. R. Soll. 1987. Unique phenotype of opaque cells in the white-opaque transition in Candida albicans. J. Bacteriol. 169:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. J., and A. D. Johnson. 2005. Mating in Candida albicans and the search for a sexual cycle. Annu. Rev. Microbiol. 59:233-255. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, R. J., M. G. Miller, P. R. Chua, M. E. Maxon, and A. D. Johnson. 2005. Nuclear fusion occurs during mating in Candida albicans and is dependent on the KAR3 gene. Mol. Microbiol. 55:1046-1059. [DOI] [PubMed] [Google Scholar]
- 4.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, DC.
- 5.Dignard, D., and M. Whiteway. 2006. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot. Cell 5:192-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumitru, R., J. M. Hornby, and K. W. Nickerson. 2004. Defined anaerobic growth medium for studying Candida albicans: basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 48:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girishkumar, H., A. M. Yousuf, J. Chivate, and E. Geisler. 1999. Experience with invasive Candida infections. Postgrad. Med. J. 75:151-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, C. B., G. Cheng, J. Chandra, P. Mukherjee, M. A. Ghannoum, and L. L. Hoyer. 2004. RT-PCR detection of Candida albicans ALS gene expression in the reconstituted human epithelium (RHE) model of oral candidiasis and in model biofilms. Microbiology 150:267-275. [DOI] [PubMed] [Google Scholar]
- 10.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 12.Kakar, S. N., R. M. Partridge, and P. T. Magee. 1983. A genetic analysis of Candida albicans: isolation of a wide variety of auxotrophs and demonstration of linkage and complementation. Genetics 104:241-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kucsera, J., K. Yarita, and K. Takeo. 2000. Simple detection method for distinguishing dead and living yeast colonies. J. Microbiol. Methods 41:19-21. [DOI] [PubMed] [Google Scholar]
- 14.Kulkarni, R. K., and K. W. Nickerson. 1981. Nutritional control of dimorphism in Ceratocystis ulmi. Exp. Mycol. 5:148-154. [Google Scholar]
- 15.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. Cell biology of mating in Candida albicans. Eukaryot. Cell 2:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 18.Magee, P. T., and B. B. Magee. 2004. Through a glass opaquely: the biological significance of mating in Candida albicans. Curr. Opin. Microbiol. 7:661-665. [DOI] [PubMed] [Google Scholar]
- 19.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 20.Morrow, B., T. Srikantha, J. Anderson, and D. R. Soll. 1993. Coordinate regulation of two opaque-specific genes during white-opaque switching in Candida albicans. Infect. Immun. 61:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Navarathna, D. H. M. L. P., J. M. Hornby, N. Hoerrmann, A. M. Parkhurst, G. E. Duhamel, and K. W. Nickerson. 2005. Enhanced pathogenicity of Candida albicans pre-treated with subinhibitory concentrations of fluconazole in a mouse model of disseminated candidiasis. J. Antimicrob. Chemother. 56:1156-1159. [DOI] [PubMed] [Google Scholar]
- 22.Negredo, A., L. Monteoliva, C. Gil, J. Pla, and C. Nombela. 1997. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 143:297-302. [DOI] [PubMed] [Google Scholar]
- 23.Nickerson, K. W., A. L. Atkin, and J. M. Hornby. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72:3805-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nickerson, W. J. 1953. Reduction of inorganic substances by yeasts. I. Extracellular reduction of sulfite by species of Candida. J. Infect. Dis. 93:43-56. [DOI] [PubMed] [Google Scholar]
- 25.Ramage, G., S. P. Saville, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rikkerink, E. H. A., B. B. Magee, and P. T. Magee. 1988. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J. Bacteriol. 170:895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., p. 9.87-9.88. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 28.Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350:3-41. [DOI] [PubMed] [Google Scholar]
- 29.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soll, D. R. 1997. Gene regulation during high-frequency switching in Candida albicans. Microbiology 143:279-288. [DOI] [PubMed] [Google Scholar]
- 31.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]