Abstract
Saccharomyces cerevisiae Cdc42p functions as a GTPase molecular switch, activating multiple signaling pathways required to regulate cell cycle progression and the actin cytoskeleton. Regulatory proteins control its GTP binding and hydrolysis and its subcellular localization, ensuring that Cdc42p is appropriately activated and localized at sites of polarized growth during the cell cycle. One of these, the Rdi1p guanine nucleotide dissociation inhibitor, negatively regulates Cdc42p by extracting it from cellular membranes. In this study, the technique of bimolecular fluorescence complementation (BiFC) was used to study the dynamic in vivo interactions between Cdc42p and Rdi1p. The BiFC data indicated that Cdc42p and Rdi1p interacted in the cytoplasm and around the periphery of the cell at the plasma membrane and that this interaction was enhanced at sites of polarized cell growth during the cell cycle, i.e., incipient bud sites, tips and sides of small- and medium-sized buds, and the mother-bud neck region. In addition, a ring-like structure containing the Cdc42p-Rdi1p complex transiently appeared following release from G1-phase cell cycle arrest. A homology model of the Cdc42p-Rdi1p complex was used to introduce mutations that were predicted to affect complex formation. These mutations resulted in altered BiFC interactions, restricting the complex exclusively to either the plasma membrane or the cytoplasm. Data from these studies have facilitated the temporal and spatial modeling of Rdi1p-dependent extraction of Cdc42p from the plasma membrane during the cell cycle.
Highly conserved among eukaryotes, the Rho GTPase Cdc42 functions as a binary molecular switch capable of activating signaling pathways that regulate polarized cell growth, cell cycle progression, gene transcription, and vesicle trafficking (reviewed in references 16, 17, and 29). Rho GTPases cycle between active (GTP-bound) and inactive (GDP-bound) states in a spatially and temporally controlled manner, which is achieved through interactions with three classes of regulatory proteins: guanine nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs) (3).
Three mammalian RhoGDIs have been identified (33, 54, 55): RhoGDIα is ubiquitously expressed, while RhoGDIβ and RhoGDIγ exhibit tissue-specific expression. Saccharomyces cerevisiae has one RhoGDI, Rdi1p, which has 36% amino acid identity with human RhoGDIα (39). RhoGDIs have multiple functions within cells. They antagonize the action of GEFs by inhibiting GDP dissociation (54), and they antagonize GAPs by inhibiting GTP hydrolysis (7). In addition, RhoGDIs extract Rho GTPases from membranes (34), sequestering them in the cytoplasm (41). RhoGDIs contain two functional domains: an amino-terminal “regulatory arm” that interacts with the switch I and switch II domains of the Rho GTPase and a carboxyl-terminal “geranylgeranyl (GG)-binding domain” that interacts with the geranylgeranyl moiety bound to the carboxyl terminus of the Rho GTPase (26). Interactions between the geranylgeranyl moiety and the RhoGDI GG-binding domain are believed to facilitate the extraction of Rho GTPases from cellular membranes (26, 44).
In vitro kinetic studies utilizing fluorescence resonance energy transfer between human Cdc42 and RhoGDI suggested a two-step model for RhoGDI extraction of Cdc42 from membranes (40). This model was supported by the crystallographic structure of the human GDP-bound Cdc42-bovine RhoGDI complex (26). In the first step of this model, the carboxyl-terminal GG-binding domain of RhoGDI interacts with Cdc42. This interaction is also believed to guide the RhoGDI amino-terminal regulatory arm into contact with the Cdc42 switch I and switch II regions, thus preventing Cdc42 from interacting with GEFs, GAPs, and downstream effector proteins. In the second step, the Cdc42 geranylgeranyl moiety isomerizes from the membrane into the RhoGDI GG-binding domain, resulting in extraction from the membrane and shielding of the geranylgeranyl moiety from the aqueous environment (41). Extraction is facilitated by the interaction of the Cdc42 carboxyl-terminal polylysine region [Cdc42(K183-187)] with an acidic patch in the GG-binding domain of RhoGDI.
Current models (13, 41) suggest that the GDP-bound GTPase is complexed with RhoGDI in the cytoplasm. A signal triggers the translocation of the complex to a cellular membrane, where a displacement factor facilitates GTPase release from RhoGDI. The exchange of GDP for GTP is then catalyzed by GEFs, and the active, GTP-bound GTPase interacts with downstream effector proteins to activate various signaling pathways. Upon signal down-regulation, RhoGDIs extract either the GTP-bound GTPase or, after GAP-mediated GTP hydrolysis, the GDP-bound GTPase from the membrane. The cytoplasmic RhoGDI-GTPase complex may be a source of readily activatable GTPase, or it may be shuttled to other membranes within the cell (36). The RhoGDI-Rho GTPase complex can also be directly involved in signaling; for example, the RhoGDI-Rac complex activates NADPH oxidase (12), and the RhoGDIβ-Cdc42 complex activates phospholipase C-β2 (28).
S. cerevisiae Rdi1p is a nonessential protein that, when overexpressed, causes growth inhibition, presumably by extracting Cdc42p from membranes and sequestering it in the cytoplasm (39, 46). Rdi1p has been shown to coimmunoprecipitate with Cdc42p from cytoplasmic fractions (32). In addition, a green fluorescent protein (GFP)-tagged Rdi1p was previously shown to be a cytoplasmic protein that consistently localized only to the plasma membrane at the tips of small-sized buds and at the mother-bud neck region (46). Cdc42p was found in both soluble and particulate fractions (57), suggesting that it localized to both the cytoplasm and membranes. In addition, GFP-Cdc42p was previously shown to localize around the plasma and internal membranes and was observed to cluster at incipient bud sites, tips and sides of small- and medium-sized buds, and at the mother-bud neck region (45). Rdi1p and Cdc42p therefore colocalized in the cytoplasm, at the tips and sides of enlarging buds, and at the mother-bud neck region.
The new technique of bimolecular fluorescence complementation (BiFC) was utilized to determine if Rdi1p and Cdc42p actually interacted at sites where they colocalized and to study the dynamic localization of the Cdc42p-Rdi1p complex during the cell cycle. BiFC enables the visualization of protein-protein interactions in vivo (27, 31). In this approach, the two nonfluorescent halves of the GFP variant yellow fluorescent protein (YFP) or cyan fluorescent protein (CFP) are fused separately to two potential interacting proteins. The interaction of the two fusion proteins leads to the reconstitution of the GFP variant and, hence, fluorescence. BiFC has recently been used to successfully dissect interactions between cytokine receptors (20), cytochromes (42), transcription factors (25), and proteins involved in S. cerevisiae cytokinesis (2).
In this study, BiFC data indicated that Cdc42p and Rdi1p interacted in the cytoplasm and around the periphery of the cell at the plasma membrane. In addition, there were enhanced interactions at incipient bud sites, the tips and sides of small- and medium-sized buds, and the mother-bud neck region. These interactions were confirmed using time-lapse photomicroscopy and cell cycle synchrony, which also revealed the presence of a novel Cdc42p-Rdi1p complex in a transient ring-like structure following release from G1 arrest. Genetic analyses indicated that mutations predicted to affect Rdi1p regulatory-arm function [rdi1(D38A), rdi1(W44A), and cdc42(R66E)] restricted Cdc42p-Rdi1p BiFC interactions exclusively to the plasma membrane, as did the rdi1(P167K) mutation, which was predicted to interfere with the binding of the Cdc42p geranylgeranyl moiety. The cdc42(K183-187Q) and cdc42(C188S) mutations, which affect the localization of Cdc42p to the plasma membrane, restricted Cdc42p-Rdi1p interactions to the cytoplasm. These data have provided valuable insights into the mechanisms by which Rdi1p interacts with Cdc42p during the cell cycle and have highlighted the overall usefulness of BiFC as a technique to study in vivo protein-protein interactions.
MATERIALS AND METHODS
DNA manipulations.
p415MET(YN-RDI1) and p416MET(CC-CDC42) were made by D-loop PCR utilizing primers (Invitrogen, Carlsbad, CA) that removed amino acids (aa) 155 to 238 from YFP in p415MET(YFP-RDI1) (46) and aa 1 to 154 from CFP in p416MET(CFP-CDC42) (45). The eight-Ala linker remained intact at the carboxyl termini of both constructs. p416MET(CC-CDC42) was converted to p416MET(YC-CDC42) by site-directed mutagenesis, changing Thr203 of CFP to Tyr. Subcellular localization of fluorescent protein-tagged wild-type and mutant proteins was performed with transformants containing either p415MET(GFP-Rdi1p) or p416MET(GFP-Cdc42p), except for p416MET(CFP-Cdc42-R66Ep). Primer sequences and cycling conditions are available upon request. Site-directed mutagenesis was performed in these plasmids and in pKT10(RDI1) (39) utilizing primers (Invitrogen) with the QuikChange kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions, with the exception of the creation of p416MET(CC-cdc42-C188S). This primer set (SigmaGenosys, St. Louis, MO) required an annealing temperature of 52°C. All mutations were verified by the Vermont Cancer Center DNA Sequencing Facility. p414MET(YN-RDI1) was made by releasing YN-RDI1 from p415MET(YN-RDI1) with SpeI and XhoI and inserting it into p414MET cut with SpeI and XhoI.
Reagents, media, and strains.
Growth media, maintenance of bacterial and yeast strains, and yeast transformations were described previously by Sambrook et al. (47) and Sherman et al. (51). Low-fluorescence (LF) yeast nitrogen base, as described previously by Sheff and Thorn (50), was used in all media for microscopy. The yeast strains used for microscopy, immunoblotting, and cell synchrony experiments were BJ5459 (MATa pep4::HIS3 prb1-Δ1.6R his3Δ200 lys2-801 trp1 ura3-52 can1 leu2Δ1) (30) and 1607-5D (MATα bar1 cln1 cln2 cln3 ura3 his2 ade1 arg4 trp1 leu2::LEU2::GAL1::CLN3) (8).
Fluorescence microscopy.
BJ5459 cells transformed with BiFC or GFP variant (for subcellular localization studies) plasmids were grown in the appropriate LF synthetic complete (SC) liquid medium to mid-log phase, collected by centrifugation, sonicated, and viewed using differential interference contrast optics. Fluorescence microscopy using an E400 Nikon microscope (Omega Optical, Brattleboro, VT) utilized an Omega XF100 optical filter cube for GFP-tagged proteins, a Chroma Cyan GFP V2 (catalog number 31044 V2) optical filter cube for CFP-tagged proteins, and a Chroma Yellow GFP (catalog number 41028) optical filter cube for both YFP-tagged proteins and BiFC constructs. Exposure times for BiFC constructs were empirically determined: 25 s for YN/CC partners and 8 s for YN/YC partners.
Immunoblot analysis.
Total protein was isolated from BJ5459 or 1607-5D cells expressing the BiFC constructs described above. A 1:5,000 dilution of polyclonal rabbit anti-GFP α-Av antibody (BD Biosciences, San Jose, CA) and a 1:8,000 dilution of goat anti-rabbit horseradish peroxidase (Sigma, St. Louis, MO) were used to detect YN, CC, and YC fusion proteins as previously described (53).
Homology modeling of Cdc42p and Rdi1p.
The SWISS-MODEL (http://swissmodel.expasy.org/workspace/) first-approach mode was utilized to build a homology model (22, 43, 49) of the Cdc42p-Rdi1p complex from the template structure (PDB accession number 1DOA) of the human GDP-bound Cdc42-bovine RhoGDI complex (26). The first three amino-terminal amino acids of Rdi1p were added to the returned structure using the build function of SwissPDB Viewer.
RESULTS
A Cdc42p-Rdi1p BiFC signal was observed in the cytoplasm and around the periphery of the cell at the plasma membrane.
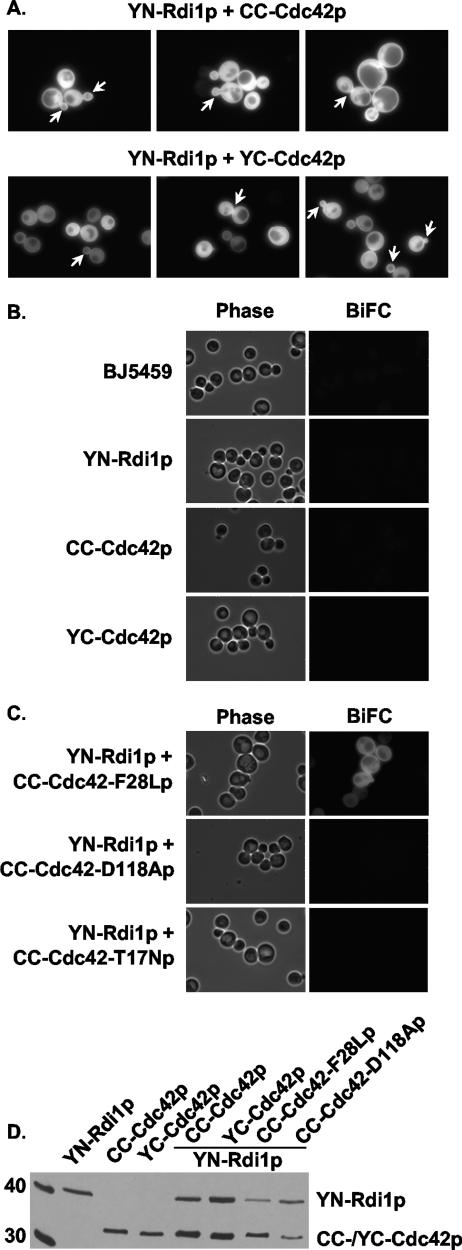
BiFC was used to determine if Cdc42p and Rdi1p physically interacted at sites where they have been shown to colocalize: in the cytoplasm, at the tips of small-sized buds, and at the mother-bud neck region (45, 46). The BiFC constructs were generated by fusing Rdi1p to the amino terminus of YFP (aa 1 to 154) (YN-Rdi1p) and fusing Cdc42p to the carboxyl terminus of either CFP or YFP (aa 155 to 238) (CC-Cdc42p or YC-Cdc42p). A Cdc42p-Rdi1p BiFC signal was observed in the cytoplasm and around the periphery of the cell at the plasma membrane (Fig. 1A). An enhanced BiFC signal was observed at incipient bud sites, the tips and sides of small- and medium-sized buds, and the mother-bud neck region (Fig. 1A). The same BiFC signal pattern was observed with YN-Rdi1p and both CC-Cdc42p and YC-Cdc42p (Fig. 1A).
FIG. 1.
BiFC interactions between Cdc42p and Rdi1p. (A) BJ5459 cells expressing YN-Rdi1p and either CC-Cdc42p or YC-Cdc42p were grown in low-fluorescent SC medium without Met to mid-log phase and observed by fluorescence microscopy. Arrows indicate an enhanced BiFC signal at the tips and sides of small- and medium-sized buds and at the mother-bud neck region. (B) BJ5459 cells expressing either YN-Rdi1p, CC-Cdc42p, or YC-Cdc42p alone were grown and observed as described above (A). (C) BJ5459 cells expressing YN-Rdi1p and either CC-Cdc42(F28Lp), CC-Cdc42(D118Ap), or CC-Cdc42(T17Np) were observed as described above (A). (D) Immunoblot analysis of Cdc42p and Rdi1p fusion proteins. Thirty micrograms of total protein from BJ5459 cells expressing the indicated proteins were resolved on 13% SDS-PAGE gels and immunoblotted with anti-GFP α-Av antibody. Left lane, size markers (in kDa).
To confirm that the observed BiFC signals depended solely on Cdc42p-Rdi1p interactions, wild-type and mutant YN-Rdi1p, YC-Cdc42p, and CC-Cdc42p were expressed in BJ5459 cells. Cells expressing wild-type YN-Rdi1p, CC-Cdc42p, or YC-Cdc42p alone did not exhibit fluorescence above the background of BJ5459 cells (Fig. 1B), verifying that the YN, YC, and CC fluorophore fragments fused to Rdi1p and Cdc42p were nonfluorescent. In addition, a BiFC signal was not observed when YN-Rdi1p was expressed with either CC-Cdc42-D118Ap or YC-Cdc42-T17Np, two cdc42 mutants that do not bind guanine nucleotide and therefore do not interact with RhoGDI (19, 56) (Fig. 1C). However, a BiFC signal with a wild-type localization pattern was observed when YN-Rdi1p was expressed with CC-Cdc42-F28Lp, a “fast-cycling” cdc42 mutant that can spontaneously exchange guanine nucleotide (37) and bind to RhoGDI (36) (Fig. 1C). The observation that single point mutations in Cdc42p known to abolish interactions with RhoGDI resulted in a lack of a BiFC signal indicated that specific interactions between Cdc42p and Rdi1p, and not nonspecific interactions between the YN and CC or YC fluorophore fragments, were responsible for the BiFC signals.
The absence of a BiFC signal could also be the result of the instability of the fusion proteins. Immunoblot analysis indicated that in cells that did not display a BiFC signal, YN-Rdi1p, CC-Cdc42p, and YC-Cdc42p were stably expressed at levels comparable to that observed in cells where they were coexpressed and exhibited a BiFC signal (Fig. 1D). CC-Cdc42-F28Lp and CC-Cdc42-D118Ap were also stably expressed at levels comparable to those of wild-type fusion proteins (Fig. 1D). Therefore, the absence of a BiFC signal was not the result of an instability of any of these proteins.
A BiFC signal was observed in a transient ring-like structure in G1 phase.
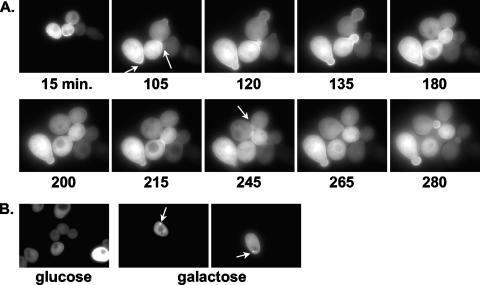
Time-lapse photomicroscopy together with cell cycle synchrony was utilized to monitor Cdc42p-Rdi1p interactions throughout the cell cycle. 1607-5D cells containing YN-Rdi1p and YC-Cdc42p were arrested in G1 phase by glucose-induced depletion of the G1 cyclin Cln3p in a cln1Δ cln2Δ background (8). Following release from G1 arrest, cells were monitored for ∼280 min either in liquid medium (data not shown) or on agar slides (n = 25) (Fig. 2A). As seen with the asynchronous population of cells, a Cdc42p-Rdi1p BiFC signal was observed in the cytoplasm and around the periphery of the cell at the plasma membrane throughout the cell cycle. In cells grown in liquid culture, an enhanced BiFC signal was observed at incipient bud sites ∼5 to 30 min following release (data not shown), whereas the appearance at incipient bud sites was slightly delayed until ∼60 to 110 min following release on agar slides (Fig. 2A). Enhanced BiFC signals were also observed in small- and medium-sized buds and at the mother-bud neck region (Fig. 2A). These results are consistent with the notion that an increase in Rdi1p at sites of polarized growth is required to remove clustered Cdc42p from membranes when polarized cell growth is no longer required (46).
FIG. 2.
Time-lapse photomicroscopy of Rdi1p-Cdc42p BiFC interactions during the cell cycle. 1607-5D cells expressing YN-Rdi1p and YC-Cdc42p were grown overnight in LF SC medium without Trp or Ura and with Met plus 2% galactose and 2% raffinose. Cells were collected by centrifugation, rinsed twice in double-distilled water, and resuspended in LF SC medium without Trp, Ura, or Met and with 2% glucose plus 2% raffinose for 3 h at 30°C (>90% cell cycle arrest as unbudded cells). Cells were collected and rinsed twice in double-distilled water, and 200 μl was resuspended in LF SC medium without Trp, Ura, or Met and with 2% galactose plus 2% raffinose to release cells from cell cycle arrest. Cells were viewed on 1% agar pads made with release medium. Numbers indicate times after release from the block. (A) Arrows represent enhanced BiFC signals at incipient bud sites, the periphery of small- and medium-sized buds, and the mother-bud neck region. (B) Cells were grown as described above (A). Left, G1-arrested cells (grown in glucose); right, cells released from arrest (grown in galactose). Within 5 min of release from G1 arrest, a ring-like BiFC structure appeared (arrows).
A BiFC signal was also observed in a ring-like structure following release from G1 arrest (Fig. 2B). Approximately 50% of cells (n = 100) released into galactose-containing liquid medium had this ring-like structure present within ∼5 min of release. Cells (n = 100) that were arrested in G1 phase by growth in glucose medium did not display this ring-like structure. This structure was transient in that it disappeared in all cells examined within ∼5 min (data not shown). This observation was interesting since BiFC complexes are irreversible in vitro (see Discussion). However, these data suggest that the Cdc42p-Rdi1p BiFC complex was reversible and dynamic in nature.
Homology modeling of the Cdc42p-Rdi1p complex facilitated a genetic analysis of Cdc42p-Rdi1p BiFC interactions.
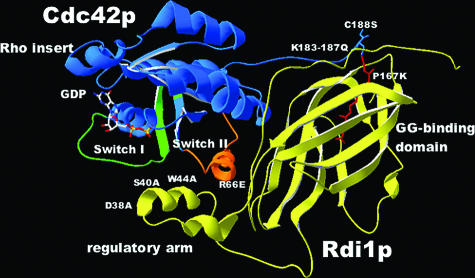
To facilitate the genetic analysis of Cdc42p-Rdi1p complex formation, a homology model of the S. cerevisiae Cdc42p-Rdi1p complex (Fig. 3) was generated based on the crystal structure of the human Cdc42-bovine GDI complex (26). Human Cdc42p and S. cerevisiae Cdc42p share 80.7% amino acid sequence identity, whereas the bovine GDI and Rdi1p are 38.6% identical. Automated homology modeling is believed to be reliable when the target and template have amino acid identity that is greater than 50% (1). Although bovine GDI-Rdi1p identity falls below this threshold, the homology model generated was sufficient for guiding site-directed mutagenesis studies, as the critical residues at the interface of human Cdc42p and RhoGDI were conserved. Therefore, mutations predicted to affect the functional interaction between the proteins were made in the Rdi1p regulatory arm (D38A, S40A, W44A, and Δ37-47) and GG-binding domain (P167K) and in the Cdc42p switch II domain (R66E) and geranylgeranylation site (C188S and K183-187Q) (Fig. 3 and Table 1).
FIG. 3.
SWISS-MODEL-generated structure of an S. cerevisiae Cdc42p (blue)-Rdi1p (yellow) complex. Highlighted are the switch I (lime green) and switch II (orange) regions of Cdc42p and the geranylgeranyl moiety (red). Mutated residues are shown in white.
TABLE 1.
Summary of BiFC interactionsa
| Protein | Interface or function affected | Localizationb | BiFC signalb |
|---|---|---|---|
| Cdc42p | Wild type | CPIc | CP |
| Cdc42(D118Ap) | Nucleotide binding | CPI | — |
| Cdc42(T17Np) | Nucleotide binding | CPI | — |
| Cdc42(F28Lp) | Nucleotide binding | CPI | CP |
| Cdc42-R66Ep | GG binding, switch II | CPI | P |
| Cdc42(K183-187Qp) | Acidic patch/polylysine | C | C |
| Cdc42(K183-187Q,R66Ep) | See above | NT | — (90%), CP (10%) |
| Cdc42(C188Sp) | Geranylgeranylation | Cc | C |
| Rdi1p | Wild type | CPd | CP |
| Rdi1(D38Ap) | Regulatory arm/switch I | NT | P |
| Rdi1(S40Ap) | Regulatory arm/switch I | NT | CP |
| Rdi1(W44Ap) | Regulatory arm/switch I | CP | P |
| Rdi1(Δ37-47p) | Regulatory arm/switch I, II | CP | P |
| Rdi1(P167Kp) | GG binding | CP | P |
| Cdc42(C188Sp) + Rdi1(P167Kp) | See above | C, CP | C |
Unless otherwise stated, all Cdc42p mutations were tested for BiFC interactions with wild-type Rdi1p, and all Rdi1p mutations were tested for BiFC interactions with wild-type Cdc42p.
Subcellular localization of GFP-tagged proteins and BiFC signals were categorized as either cytoplasm (C), plasma membrane (P), or internal membranes (I). NT, not tested; —, no BiFC signal was observed.
Assayed in reference 45.
Assayed in reference 46. Plasma membrane localization (P) of wild-type and mutant Rdi1p occurred only at tips and sides of small-sized buds and the mother-bud neck region.
Mutations in the Rdi1p regulatory arm and GG-binding domain and the Cdc42p switch II domain did not block Cdc42p-Rdi1p complex formation.
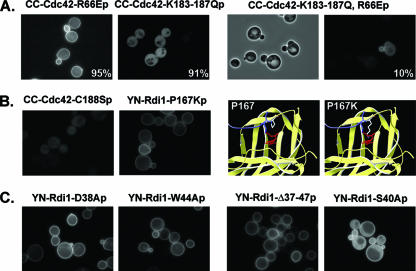
Previous studies showed that plasma membrane localization of GFP-Cdc42p depended on both geranylgeranylation of a carboxyl-terminal Cys residue and the polylysine region (KKSKK187) next to the Cys residue (9, 45). It is believed that a carboxyl-terminal “acidic patch” within the RhoGDI GG-binding domain can compete with negatively charged membrane phospholipids for binding to the positively charged polylysine region, thereby facilitating GDI-dependent extraction of Cdc42p from membranes (26). Changing the four Lys residues to Gln (K183-187Q) resulted in a BiFC signal between YN-Rdi1p and CC-Cdc42(K183-187Qp), indicating that altering the Lys residues did not block complex formation, but the BiFC signal was observed only in the cytoplasm (Fig. 4A). As GFP-Cdc42(K183-187Qp) localized to the cytoplasm and internal membranes but not to the plasma membrane (45) (Table 1), this result suggested that Cdc42(K183-187Qp) could interact with Rdi1p in the cytoplasm but that the Cdc42(K183-187Qp)-Rdi1p complex could not be targeted to the plasma membrane.
FIG. 4.
Analysis of mutations affecting the switch I/switch II regulatory-arm interactions and geranylgeranyl binding. BiFC interactions were observed as described in the legend of Fig. 1. (A) BJ5459 cells expressing YN-Rdi1p and either CC-Cdc42(R66Ep), CC-Cdc42(K183-187Qp), or CC-Cdc42(K183-187Q,R66Ep). For cells expressing YN-Rdi1p and CC-Cdc42(K183-187Q,R66Ep), both differential interference contrast (left) and BiFC fluorescence (right) photomicrographs are shown to highlight the observation that only ∼10% of cells exhibit a BiFC signal. The percentages of cells displaying a predominantly membrane [Cdc42(R66Ep)] or cytoplasmic [Cdc42(K183-187Qp)] BiFC signal are indicated. (B) BJ5459 cells expressing YN-Rdi1p plus CC-Cdc42(C188Sp) (left) or YN-Rdi1(P167Kp) plus CC-Cdc42p (right). Far right, predicted structural effect of the P167K mutation. P and mutant K residues are shown in white; the geranylgeranyl moiety is shown in red. (C) BJ5459 cells expressing YC-Cdc42p and either YN-Rdi1(D38Ap), YN-Rdi1(W44Ap), YN-Rdi1(Δ37-47p), or YN-Rdi1(S40Ap).
The proximity of the Cdc42 switch II Arg66 residue with amino acids in both the RhoGDI regulatory arm and the external face of the GG-binding domain (Fig. 3) suggested that this amino acid played an important role in the stability of the Cdc42p-Rdi1p complex (13). A BiFC signal was observed between YN-Rdi1p and CC-Cdc42(R66Ep), indicating that the R66E mutation does not block complex formation; but in this case, the BiFC signal was observed predominantly at the plasma membrane (Fig. 4A). Since GFP-Cdc42(R66Ep) localized normally to the cytoplasm and to plasma and internal membranes (Table 1), these results suggested either that Cdc42(R66Ep) could interact only with Rdi1p at the plasma membrane and not in the cytoplasm or that Rdi1p could not extract Cdc42(R66Ep) from the plasma membrane.
To address these possibilities, the cdc42(R66E) (plasma membrane BiFC) and cdc42(K183-187Q) (cytoplasmic BiFC) mutations were combined. If the cdc42(R66E) mutation prevented the formation of a stable complex with Rdi1p in the cytoplasm, then the double mutation would result in a complete loss of interactions with Rdi1p. A BiFC signal with wild-type localization was observed in only 10% of cells that expressed YN-Rdi1p and CC-Cdc42(K183-187Q,R66Ep) (Fig. 4A), suggesting that the R66E mutation does interfere with the formation of a Cdc42p-Rdi1p complex in the cytoplasm. However, this result does not rule out the possibility that Rdi1p cannot effectively extract Cdc42(R66Ep) from the plasma membrane (see below).
Geranylgeranylation of the Rho GTPase Rac is essential for high-affinity binding to RhoGDI (35), and geranylgeranylation-defective Cdc42(C188Sp) does not localize to the plasma membrane (45, 57). Therefore, it was predicted that CC-Cdc42(C188Sp) and YN-Rdi1p would have little or no interactions and, if observed, would be localized to the cytoplasm. In fact, an attenuated BiFC signal between Cdc42(C188Sp) and Rdi1p was observed only in the cytoplasm (Fig. 4B), indicating that the geranylgeranylation of Cdc42p was important but not essential for interactions with Rdi1p in the cytoplasm.
The Cdc42p-Rdi1p complex model was examined to determine if an amino acid(s) at the opening of the Rdi1p carboxyl-terminal GG-binding domain could be mutated such that the insertion of the geranylgeranyl moiety would be prevented. Mutation of Pro167 to Lys was predicted to provide both steric and charge hindrances to the insertion of the large 20-carbon hydrophobic geranylgeranyl moiety (Fig. 4B). Interestingly, a BiFC signal between YN-Rdi1(P167Kp) and YC-Cdc42p was observed (Fig. 4B), indicating that the Rdi1(P167Kp) mutant protein could still interact with Cdc42p. However, the BiFC signal was observed only at the plasma membrane, suggesting that Rdi1(P167Kp) could not extract Cdc42p from membranes, possibly because it could not bind the geranylgeranyl moiety (see below and Discussion). In addition, a predominantly cytoplasmic BiFC signal was observed between YN-Rdi1(P167Kp) and YC-Cdc42(C188Sp) (data not shown), indicating that Rdi1(P167Kp) could bind to Cdc42p lacking a geranylgeranyl moiety.
The Asp45 and Ser47 residues in the bovine RhoGDI regulatory arm interact with the Thr35 and Val36 residues, respectively, in the Cdc42 switch I domain (13, 26). Also, the Tyr51 regulatory-arm residue interacts with conserved Tyr64, Leu67, and Leu70 hydrophobic residues in Cdc42 (26). The equivalent residues in Rdi1p (Asp38, Ser40, and Trp44) were mutated to Ala residues. A BiFC signal was observed in cells expressing either YN-Rdi1(D38Ap), YN-Rdi1(W44Ap), or YN-Rdi1(S40Ap) together with YC-Cdc42p (Fig. 4C), indicating that these mutations do not interfere with Cdc42p binding. A BiFC signal was also observed between YN-Rdi1(D38Ap) and CC-Cdc42(R66Ep) (data not shown). Although the Cdc42p-Rdi1(S40Ap) BiFC signal showed a wild-type localization pattern, the Cdc42p-Rdi1(D38Ap), Cdc42p-Rdi1(W44Ap), and Cdc42(R66Ep)-Rdi1(D38Ap) BiFC signals were observed only at the plasma membrane (Fig. 4C and data not shown), suggesting that mutant Rdi1p may not be able to extract Cdc42p from membranes (see below).
Deletion of the amino-terminal 59 amino acids of bovine RhoGDI, which includes the regulatory arm, did not interfere with binding to Cdc42 but did block membrane extraction of Cdc42 (21). Therefore, regulatory-arm amino acids 37 to 47 of Rdi1p, which encompass the αC helix that lies in proximity to the switch I/II domain of Cdc42p, were deleted to assess their function in complex formation. Interestingly, a BiFC signal between YN-Rdi1(Δ37-47p) and YC-Cdc42p was observed (Fig. 4C), again indicating that the deletion of the Rdi1p regulatory arm did not interfere with Cdc42p binding, but the BiFC signal was observed only at the plasma membrane, suggesting that mutant Rdi1(Δ37-47p) could not extract Cdc42p from the membrane.
GFP-Cdc42p release from membranes can be stimulated by expression of Rdi1p regulatory-arm mutants.
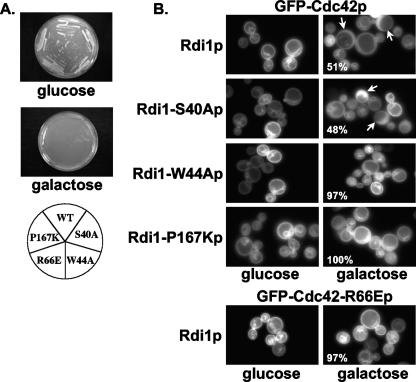
Overexpression of wild-type Rdi1p resulted in two cellular phenotypes: inhibition of cell growth and a loss of GFP-Cdc42p membrane localization (32, 39, 46). Therefore, these two phenotypes were used in assays to examine whether the effects of mutations that restricted Cdc42p-Rdi1p BiFC interactions to the plasma membrane [cdc42(R66E), rdi1(W44A), and rdi1(P167K)] were due to an inability of Rdi1p to extract Cdc42p from membranes.
In the growth inhibition assay, cells expressing wild-type Rdi1p or mutant Rdi1(W44Ap), Rdi1(P167Kp), or Rdi1(S40Ap) (a control that showed wild-type BiFC signal patterns) under the control of a galactose-inducible promoter could grow on repression medium (glucose) but could not grow on derepression medium (galactose) (Fig. 5A). This result indicated that the three rdi1 mutants had retained the ability to cause growth arrest. However, in the membrane extraction assay, cells expressing Rdi1(S40Ap) showed predominately cytosolic GFP-Cdc42p localization, whereas cells expressing Rdi1(W44Ap) or Rdi1(P167Kp) displayed predominantly plasma membrane GFP-Cdc42p localization (Fig. 5B, right panels). Taken together, these data suggested that Rdi1(S40Ap) had retained the ability to extract Cdc42p from membranes, thereby causing growth arrest, but Rdi1(W44Ap) and Rdi1(P167Kp) had lost that ability. Therefore, the nature of the growth arrest seen with Rdi1(W44Ap) and Rdi1(P167Kp) was unclear but may be due to the generation of dominant-negative complexes at the membrane, as Cdc42p-Rdi1(W44Ap) and Cdc42p-Rdi1(P167Kp) BiFC signals were restricted to the plasma membrane.
FIG. 5.
Growth inhibition (A) and membrane extraction (B) assays with Cdc42p and Rdi1p mutants. (A) BJ5459 cells expressing either wild-type Rdi1p, Rdi1(S40Ap), Rdi1(W44Ap), Rdi1(P167Kp), or Cdc42(R66Ep) were grown on repression (glucose) and derepression (galactose) media. (B) Top, BJ5459 cells expressing wild-type GFP-Cdc42p and either wild-type Rdi1p or the indicated mutant Rdi1p were grown in LF SC medium without Ura, Leu, or Met and with 2% raffinose and either 2% glucose (left) or 2% galactose (right) for 6 h and then observed by fluorescence microscopy. Arrows indicate cells with predominantly cytoplasmic GFP-Cdc42p localization. The percentages of cells [n = 100 for Rdi1(P167Kp); n = 300 for Rdi1(S40Ap) and Rdi1(W44Ap)] with predominantly membrane GFP-Cdc42p localization are indicated. Bottom, BJ5459 cells expressing GFP-Cdc42(R66Ep) and wild-type Rdi1p were grown and observed as described above.
In the membrane extraction assay, the expression of Rdi1p resulted in predominantly cytosolic localization of wild-type GFP-Cdc42p but predominantly plasma membrane localization of GFP-Cdc42(R66Ep) (Fig. 5B). This result suggested that Rdi1p could not extract mutant Cdc42(R66Ep) from the plasma membrane, possibly generating a dominant-negative complex at the membrane, as cells expressing wild-type Rdi1p with mutant Cdc42(R66Ep) could not grow on derepression medium (galactose) (Fig. 5A). These data, taken together with the BiFC data, indicated that the restriction of the Cdc42(R66Ep)-Rdi1p BiFC signal to the plasma membrane was most likely due to the inability of Rdi1p to extract Cdc42(R66Ep) from membranes.
DISCUSSION
During the S. cerevisiae cell cycle, Cdc42p is required for actin rearrangements that occur at sites of polarized cell growth. Consistent with this cell cycle role, functional GFP-Cdc42p localized around the periphery of the cell at the plasma membrane and was observed to cluster at polarized growth sites, including incipient bud sites, tips and sides of small- and medium-sized buds, and the mother-bud neck region (45). Cdc42p was also observed in the cytoplasm (57), and, hence, a dynamic equilibrium must exist between cytoplasmic and membrane-bound pools. However, the mechanisms by which Cdc42p is trafficked from the cytoplasm to the plasma membrane and sites of polarized cell growth and subsequently extracted from membranes to cytoplasmic pools later in the cell cycle have remained elusive (45).
It is clear that mammalian RhoGDI proteins play an important role in the trafficking and extraction of Rho GTPase from membranes. As S. cerevisiae rdi1Δ cells are viable (39, 46) and GFP-Cdc42p localized and clustered normally in rdi1Δ cells (T. Richman and D. I. Johnson, unpublished results), the ability of Rdi1p to traffic and/or remove Cdc42p from the plasma membrane is not essential for cell growth. Although Cdc42p could localize to the plasma membrane independent of Rdi1p, Rdi1p was able to extract GFP-Cdc42p from the plasma membrane (46). Consistent with this function, GFP-tagged Rdi1p localized to the plasma membrane at the tips of small-sized buds and at the mother-bud neck region as well as to the cytoplasm (46).
The observed Cdc42p-Rdi1p BiFC interaction patterns described herein were consistent with the localization of the individual GFP-tagged proteins with three notable exceptions: (i) BiFC interactions between Cdc42p and Rdi1p were evident around the entire periphery of the cell at the plasma membrane instead of just at sites of polarized growth, (ii) enhanced BiFC signals were observed at incipient bud sites where GFP-Rdi1p did not consistently localize (46), and (iii) the proteins interacted around the tips and sides of large-sized buds (Fig. 1A). There are several possible explanations for these differences that will be explored in future studies: (i) the BiFC technique was able to capture a rapid transient event in which the proteins interacted around the entire periphery of the cell and at incipient bud sites; (ii) the reconstituted fluorophore irreversibly conjoined the fusion proteins, and they diffused away from sites of polarized growth but could not be extracted from membranes in large-sized buds; and/or (iii) the expression of the proteins overwhelmed a potential Rdi1p displacement factor, thereby causing the complex to remain membrane associated. Although it has been proposed that cytoplasmic RhoGDI shuttles Rho GTPases to other internal membranes within the cell (13), a Cdc42p-Rdi1p BiFC signal was not observed at internal membranes, even though GFP-Cdc42p has been localized to these membranes. The cytoplasmic Cdc42p-Rdi1p complex, therefore, may not be shuttling Cdc42p to internal membranes but may just be serving as a source of readily activatable GTPase (41).
Time-lapse photomicroscopy of G1-synchronized cells indicated that Cdc42p interacted with Rdi1p in the cytoplasm and around the periphery of the cell during a G1 arrest and subsequently throughout the cell cycle, indicating that a Cdc42p-Rdi1p complex does not need to be specifically targeted to the plasma membrane following Start. Surprisingly, within 5 min of release from the cyclin-depletion-induced cell cycle arrest, a Cdc42p-Rdi1p complex appeared in a ring-like structure, and this complex disappeared within ∼5 min. The rapid disappearance of the ring-like BiFC structure was presumably due to the disassembly of the Cdc42p-Rdi1p complex. Whereas the reassembly of GFP from two nonfluorescent halves is fundamentally irreversible in vitro, with the half-life of the dissociation of the halves being ∼10 years without a denaturing agent (38), the disassembly of other BiFC complexes has been previously observed in vivo (10, 23).
There are several mechanisms that may account for the dissociation of the Cdc42p-Rdi1p ring-like structure following release from G1 arrest: phosphorylation of the RhoGDI, RhoGDI displacement factors, and/or phospholipid-induced dissociation (13). Mammalian p21-activated kinase phosphorylated RhoGDIα on two Ser residues, which decreased the affinity of RhoGDIα for Rac1 (11). RhoGDI protein displacement factors are members of the ezrin, radixin, and moesin (ERM) superfamily that link actin filaments to plasma membrane proteins (4, 15, 48). Although ERM domain-containing proteins have not been identified in S. cerevisiae (4), it is likely that additional proteins mediate interactions between Cdc42p and Rdi1p. This supposition is supported by the observation that the deletion of the Cdc42p Rho insert domain, a 13-amino-acid domain (aa 122 to 134) not directly involved in protein-protein interactions with Rdi1p (Fig. 3), prevented Rdi1p-dependent membrane extraction of Cdc42p (46). Phospholipids have been shown to disrupt a cytosolic Rac-RhoGDI complex (6) and have been linked to increasing CD44's affinity for ERM (24), presumably resulting in the loss of Rac inhibition and the delivery of Rho GTPases to membrane signaling complexes. Studies are currently under way in the laboratory to address these three possibilities.
Cdc42-RhoGDI binding has been proposed to occur in two steps (14). Initially, amino acids in the RhoGDI regulatory arm interact with amino acids in the Cdc42 switch I/II domains, thereby detecting the nucleotide status of the GTPase. This is followed by an interaction between amino acids on the external face of the GG-binding domain and amino acids in the switch II domain, which positions the complex at the membrane for extraction. Surprisingly, neither mutations in D38A, S40A, or W44A or a deletion of Δ37-47, the Rdi1p regulatory arm, nor mutations in the GG-binding domain (P167K) alone could block the formation of a Cdc42p-Rdi1p complex (Fig. 4 and Table 1). These results indicated that neither domain was necessary for binding Cdc42p and that both domains most likely act cooperatively to form a Cdc42p-Rdi1p complex. However, the W44A mutation did affect the ability of Rdi1p to extract Cdc42p from the plasma membrane, suggesting that regulatory-arm interactions play a role in positioning Rdi1p in an extraction-competent conformation.
Mutations predicted to affect geranylgeranyl binding resulted in a shift of the Cdc42p-Rdi1p complex to either the cytoplasm [cdc42(K183-187Q) and cdc42(C188S)] or the plasma membrane [rdi1(P167K)]. The persistence of the Cdc42(K183-187Qp)-Rdi1p and Cdc42(C188Sp)-Rdi1p complexes in the cytoplasm was most likely due to the inability of mutant Cdc42p to target to the membrane (9, 45). The inability of Rdi1(P167Kp) to extract GFP-Cdc42p from the plasma membrane (Fig. 5) is the most likely explanation for the persistence of the Cdc42p-Rdi1(P167Kp) complex at the plasma membrane. Previous studies showed that the RhoGDI Ile177 residue (located at the base of the GG-binding pocket) was required for high-affinity geranylgeranyl binding and RhoGDI-dependent extraction of Cdc42 from membranes (26, 44). Mutation of this residue to Asp resulted in a dominant-negative protein that formed a complex with Cdc42 at the plasma membrane (52). It is likely that the Cdc42p-Rdi1(P167Kp) complex is analogous, since Rdi1(P167Kp) should not be able to bind the geranylgeranyl moiety, and it formed a dominant-negative Cdc42p complex restricted to the plasma membrane.
BiFC data indicated that mutant Cdc42(R66Ep) could interact with Rdi1p, in contrast to previous studies of mammalian cells (18, 36), but the membrane extraction and growth inhibition assays indicated that Rdi1p could not extract Cdc42(R66Ep) from the plasma membrane (Fig. 5), which agrees with analysis of the mammalian Cdc42(R66E) mutant protein. This inability to be extracted from membranes may be due to the role of the Cdc42 R66 residue in binding to the external face of the Rdi1p GG-binding domain, leading to the proper positioning of the Cdc42p-Rdi1p complex at the membrane (14).
Taken together, these data suggest a model for dynamic Cdc42p-Rdi1p interactions throughout the cell cycle. Rdi1p is likely to play an important, although not essential, role in trafficking Cdc42p to and from the plasma membrane at sites of polarized cell growth during the cell cycle. Rdi1p likely functions in the extraction of Cdc42p from membranes at these sites upon the completion of polarized cell growth following the apical-isotropic switch in the S/G2 phase. Rdi1p does not seem to restrict Cdc42p to polarized growth sites in S. cerevisiae, in contrast to other organisms such as Arabidopsis, in which a RhoGDI is required to spatially restrict ROP GTPases to sites of polarized growth (5). These results also suggested that the Rdi1p amino-terminal regulatory arm and the carboxy-terminal GG-binding domain act cooperatively to bind and extract Cdc42p from membranes, an observation that may shed new light on the proposed two-step model for Rho GTPase-RhoGDI binding. Additional BiFC and genetic analyses should provide important new insights into the mechanisms by which RhoGDIs bind and extract Rho GTPases from cellular membranes.
Acknowledgments
We thank Mariana Matraijt for anti-GFP antibodies, Matt Hogg for assistance with the homology modeling, Gary Ward and members of the Johnson laboratory for helpful discussions and critical comments on the manuscript, and the Vermont Cancer Center DNA Sequencing Facility for analysis of site-directed mutations.
This project was supported in part by National Science Foundation grant MCB-110138, a University of Vermont College of Medicine New Research Initiative grant, and USDA-Hatch grant VT-H01308. K.C.C. was supported in part by NCI Cancer Biology training grant T32-CAO9286-19 and Vermont Genetics Network grant P20 RR16462 from the INBRE Program of the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH).
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or NIH.
Footnotes
Published ahead of print on 12 January 2007.
REFERENCES
- 1.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 2.Blondel, M., S. Bach, S. Bamps, J. Dobbelaere, P. Wiget, C. Longaretti, Y. Barral, L. Meijer, and M. Peter. 2005. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast. EMBO J. 24:1440-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boguski, M. S., and F. McCormick. 1993. Proteins regulating Ras and its relatives. Nature 366:643-654. [DOI] [PubMed] [Google Scholar]
- 4.Bretscher, A., K. Edwards, and R. G. Fehon. 2002. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3:586-599. [DOI] [PubMed] [Google Scholar]
- 5.Carol, R. J., S. Takeda, P. Linstead, M. C. Durrant, H. Kakesova, P. Derbyshire, S. Drea, V. Zarsky, and L. Dolan. 2005. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438:1013-1016. [DOI] [PubMed] [Google Scholar]
- 6.Chuang, T.-H., B. P. Bohl, and G. M. Bokoch. 1993. Biologically active lipids are regulators of RacGDI complexation. J. Biol. Chem. 268:26206-26211. [PubMed] [Google Scholar]
- 7.Chuang, T.-H., X. Xu, U. G. Knaus, M. J. Hart, and G. M. Bokoch. 1993. GDP dissociation inhibitor prevents intrinsic and GTPase activating protein-stimulated GTP hydrolysis by the Rac GTP-binding protein. J. Biol. Chem. 268:775-778. [PubMed] [Google Scholar]
- 8.Cross, F. R. 1990. Cell cycle arrest caused by CLN gene deficiency in Saccharomyces cerevisiae resembles START-I arrest and is independent of the mating-pheromone signalling pathway. Mol. Cell. Biol. 10:6482-6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, C. R., T. J. Richman, S. B. Deliduka, J. O. Blaisdell, C. C. Collins, and D. I. Johnson. 1998. Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J. Biol. Chem. 273:849-858. [DOI] [PubMed] [Google Scholar]
- 10.Demidov, V. V., N. V. Dokholyan, C. Witte-Hoffmann, P. Chalasani, H. W. Yiu, F. Ding, Y. Yu, C. R. Cantor, and N. E. Broude. 2006. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proc. Natl. Acad. Sci. USA 103:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DerMardirossian, C., A. Schnelzer, and G. M. Bokoch. 2004. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol. Cell 15:117-127. [DOI] [PubMed] [Google Scholar]
- 12.Di-Poi, N., J. Faure, S. Grizot, G. Molnar, E. Pick, and M. C. Dagher. 2001. Mechanism of NADPH oxidase activation by the Rac/Rho-GDI complex. Biochemistry 40:10014-10022. [DOI] [PubMed] [Google Scholar]
- 13.Dovas, A., and J. R. Couchman. 2005. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dransart, E., A. Morin, J. Cherfils, and B. Olofsson. 2005. Uncoupling of inhibitory and shuttling functions of Rho GDP dissociation inhibitors. J. Biol. Chem. 280:4674-4683. [DOI] [PubMed] [Google Scholar]
- 15.Dransart, E., B. Olofsson, and J. Cherfils. 2005. RhoGDIs revisited: novel roles in Rho regulation. Traffic 6:957-966. [DOI] [PubMed] [Google Scholar]
- 16.Etienne-Manneville, S. 2004. Cdc42—the centre of polarity. J. Cell Sci. 117:1291-1300. [DOI] [PubMed] [Google Scholar]
- 17.Etienne-Manneville, S., and A. Hall. 2002. Rho GTPases in cell biology. Nature 420:629-635. [DOI] [PubMed] [Google Scholar]
- 18.Gibson, R. M., P. N. Gandhi, X. F. Tong, J. Miyoshi, Y. Takai, M. Konieczkowski, J. R. Sedor, and A. L. Wilson-Delfosse. 2004. An activating mutant of Cdc42 that fails to interact with Rho GDP-dissociation inhibitor localizes to the plasma membrane and mediates actin reorganization. Exp. Cell Res. 301:211-222. [DOI] [PubMed] [Google Scholar]
- 19.Gibson, R. M., and A. L. Wilson-Delfosse. 2001. RhoGDI-binding-defective mutant of Cdc42Hs targets to membranes and activates filopodia formation but does not cycle with the cytosol of mammalian cells. Biochem. J. 359:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giese, B., C. Roderburg, M. Sommerauer, S. B. Wortmann, S. Metz, P. C. Heinrich, and G. Muller-Newen. 2005. Dimerization of the cytokine receptors gp130 and LIFR analysed in single cells. J. Cell Sci. 118:5129-5140. [DOI] [PubMed] [Google Scholar]
- 21.Gosser, Y. Q., T. K. Nomanbhoy, B. Aghazadeh, D. Manor, C. Combs, R. A. Cerione, and M. K. Rosen. 1997. C-terminal binding domain of rho GDP-dissociation inhibitor directs N-terminal inhibitory peptide to GTPases. Nature 387:814-819. [DOI] [PubMed] [Google Scholar]
- 22.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 23.Guo, Y., M. Rebecchi, and S. Scarlata. 2005. Phospholipase Cβ2 binds to and inhibits phospholipase Cδ1. J. Biol. Chem. 280:1438-1447. [DOI] [PubMed] [Google Scholar]
- 24.Hirao, M., N. Sato, T. Kondo, S. Yonemura, M. Monden, T. Sasaki, Y. Takai, S. Tsukita, and S. Tsukita. 1996. Regulation mechanism of ERM (exrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J. Cell Biol. 135:37-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoff, B., and U. Kuck. 2005. Use of bimolecular fluorescence complementation to demonstrate transcription factor interaction in nuclei of living cells from the filamentous fungus Acremonium chrysogenum. Curr. Genet. 47:132-138. [DOI] [PubMed] [Google Scholar]
- 26.Hoffman, G. R., N. Nassar, and R. A. Cerione. 2000. Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100:345-356. [DOI] [PubMed] [Google Scholar]
- 27.Hu, C.-D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 28.Illenberger, D., F. Schwald, D. Pimmer, W. Binder, G. Maier, A. Dietrich, and P. Gierschik. 1998. Stimulation of phospholipase C-b(2) by the rho GTPases Cdc42Hs and Rac1. EMBO J. 17:6241-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194:428-453. [DOI] [PubMed] [Google Scholar]
- 31.Kerppola, T. K. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7:449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch, G., K. Tanaka, T. Masuda, W. Yamochi, H. Nonaka, and Y. Takai. 1997. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene 15:417-422. [DOI] [PubMed] [Google Scholar]
- 33.Leffers, H., M. S. Nielsen, A. H. Andersen, B. Honore, P. Madsen, J. Vandekerckhove, and J. E. Celis. 1993. Identification of two human rho GDP dissociation inhibitor proteins whose overexpression leads to disruption of the actin cytoskeleton. Exp. Cell Res. 209:165-174. [DOI] [PubMed] [Google Scholar]
- 34.Leonard, D., M. J. Hart, J. V. Platko, A. Eva, W. Henzel, T. Evans, and R. A. Cerione. 1992. The identification and characterization of a GDP-dissociation inhibitor (GDI) for the CDC42Hs protein. J. Biol. Chem. 267:22860-22868. [PubMed] [Google Scholar]
- 35.Lian, L. Y., I. Barsukov, A. P. Golovanov, D. I. Hawkins, R. Badii, K. H. Sze, N. H. Keep, G. M. Bokoch, and G. C. K. Roberts. 2000. Mapping the binding site for the GTP-binding protein Rac-1 on its inhibitor RhoGDI-1. Structure 8:47-55. [DOI] [PubMed] [Google Scholar]
- 36.Lin, Q., R. N. Fuji, W. N. Yang, and R. A. Cerione. 2003. RhoGDI is required for Cdc42-mediated cellular transformation. Curr. Biol. 13:1469-1479. [DOI] [PubMed] [Google Scholar]
- 37.Lin, R., S. Bagrodia, R. Cerione, and D. Manor. 1997. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7:794-797. [DOI] [PubMed] [Google Scholar]
- 38.Magliery, T. J., C. G. Wilson, W. Pan, D. Mishler, I. Ghosh, A. D. Hamilton, and L. Regan. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146-157. [DOI] [PubMed] [Google Scholar]
- 39.Masuda, T., K. Tanaka, H. Nonaka, W. Yamochi, A. Maeda, and Y. Takai. 1994. Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. J. Biol. Chem. 269:19713-19718. [PubMed] [Google Scholar]
- 40.Nomanbhoy, T. K., J. W. Erikson, and R. A. Cerione. 1999. Kinetics of Cdc42 membrane extraction by Rho-GDI monitored by real-time fluorescence resonance energy transfer. Biochemistry 38:1744-1750. [DOI] [PubMed] [Google Scholar]
- 41.Olofsson, B. 1999. Rho guanine dissociation inhibitors: Pivotal molecules in cellular signalling. Cell. Signal. 11:545-554. [DOI] [PubMed] [Google Scholar]
- 42.Ozalp, C., E. Szczesna-Skorupa, and B. Kemper. 2005. Bimolecular fluorescence complementation analysis of cytochrome p450 2c2, 2e1, and NADPH-cytochrome p450 reductase molecular interactions in living cells. Drug Metab. Dispos. 33:1382-1390. [DOI] [PubMed] [Google Scholar]
- 43.Peitsch, M. C. 1995. Protein modeling by e-mail. Bio/Technology 13:658-660. [Google Scholar]
- 44.Platko, J. V., D. A. Leonard, C. N. Adra, R. J. Shaw, R. A. Cerione, and B. Lim. 1995. A single residue can modify target-binding affinity and activity of the functional domain of the Rho-subfamily GDP dissociation inhibitors. Proc. Natl. Acad. Sci. USA 92:2974-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richman, T. J., M. M. Sawyer, and D. I. Johnson. 2002. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot. Cell 1:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richman, T. J., K. A. Toenjes, S. E. Morales, K. C. Cole, B. T. Wasserman, C. M. Taylor, J. A. Koster, M. F. Whelihan, and D. I. Johnson. 2004. Analysis of cell-cycle specific localization of the Rdi1p RhoGDI and the structural determinants required for Cdc42p membrane localization and clustering at sites of polarized growth. Curr. Genet. 45:339-349. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sasaki, T., and Y. Takai. 1998. The Rho small G protein family-Rho GDI system as a temporal and spatial determinant for cytoskeletal control. Biochem. Biophys. Res. Commun. 245:641-645. [DOI] [PubMed] [Google Scholar]
- 49.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 51.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics: laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Sun, J. J., and J. T. Barbieri. 2004. ExoS Rho GTPase-activating protein activity stimulates reorganization of the actin cytoskeleton through rho GTPase guanine nucleotide disassociation inhibitor. J. Biol. Chem. 279:42936-42944. [DOI] [PubMed] [Google Scholar]
- 53.Toenjes, K. A., M. M. Sawyer, and D. I. Johnson. 1999. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr. Biol. 9:1183-1186. [DOI] [PubMed] [Google Scholar]
- 54.Ueda, T., A. Kikuchi, N. Ohga, J. Yamamoto, and Y. Takai. 1990. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J. Biol. Chem. 265:9373-9380. [PubMed] [Google Scholar]
- 55.Zalcman, G., V. Closson, J. Camonis, N. Honoré, M. F. Rousseau-Merck, A. Tavitian, and B. Olofsson. 1996. RhoGDI-3 is a new GDP dissociation inhibitor (GDI): identification of a non-cytosolic GDI protein interacting with the small GTP-binding proteins RhoB and RhoG. J. Biol. Chem. 271:30366-30374. [DOI] [PubMed] [Google Scholar]
- 56.Ziman, M., J. M. O'Brien, L. A. Ouellette, W. R. Church, and D. I. Johnson. 1991. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11:3537-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziman, M., D. Preuss, J. Mulholland, J. M. O'Brien, D. Botstein, and D. I. Johnson. 1993. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell 4:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]