Abstract
The dynamics of cytoplasmic microtubules (MTs) is largely controlled by a protein complex at the MT plus end. In Schizosaccharomyces pombe and in filamentous fungi, MT plus end-associated proteins also determine growth directionality. We have characterized the Dis1/XMAP215 family protein AlpA from Aspergillus nidulans and show that it determines MT dynamics as well as hyphal morphology. Green fluorescent protein-tagged AlpA localized to MT-organizing centers (centrosomes) and to MT plus ends. The latter accumulation occurred independently of conventional kinesin or the Kip2-familiy kinesin KipA. alpA deletion strains were viable and only slightly temperature sensitive. Mitosis, nuclear migration, and nuclear positioning were not affected, but hyphae grew in curves rather than straight, which appeared to be an effect of reduced MT growth and dynamics.
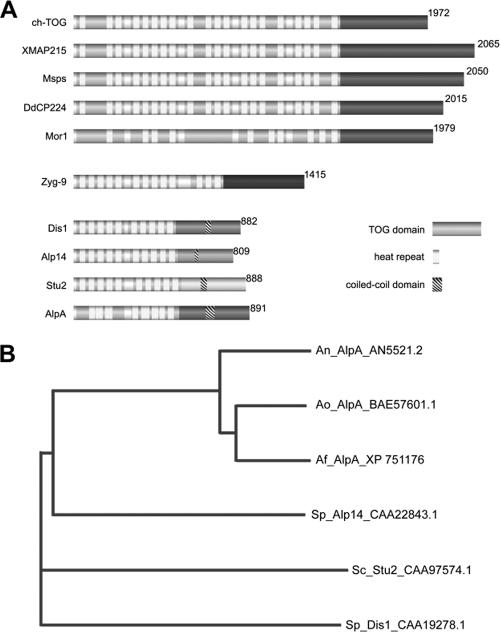
Microtubules (MTs) are hollow tubes which are generated from microtubule-organizing centers, and they perform multiple structural and dynamic functions in a cell. Although comprising an important part of the cell skeleton, MTs are very dynamic structures, which assemble at one end α,β-tubulin dimers, stop growth after some time, undergo a catastrophe event, and subsequently shrink. This dynamic instability is regulated by a number of different MT-associated proteins (MAPs), one of which was discovered in Xenopus and named XMAP215 (5). Similar proteins, which are meanwhile classified in the Dis1/XMAP215 family, exist in eukaryotes from yeast to plants and humans (17). Common to all of them is their association with MTs and the presence of TOG domains and HEAT repeats, which are responsible for interactions with many different associated proteins. One MAP can interact through its TOG domains and HEAT repeats with several other MAPs. The proteins were classified into three different groups (17). Members of the first group have four TOG domains, including one to five HEAT repeats within each of them, and a conserved C terminus. Human ch-TOG belongs to the first group together with Xenopus XMAP215, Drosophila (Msps), Dictyostelium (DdCP224), and Arabidopsis (MOR1) (Fig. 1). The second group has only one known member from Caenorhabditis elegans (ZYG-9). Members of the third group have only two TOG domains with several HEAT repeats and, in comparison to group one members, do not have a conserved C terminus. However, all of them harbor a coiled-coil region instead. XMAP215 proteins have a prominent MT-stabilizing function (12). Recently, it was shown nicely in Saccharomyces cerevisiae that the Dis1/XMAP215 protein Stu2 binds to tubulin heterodimers and associates to the MT plus end, where it appears to be responsible for the loading of α,β-tubulin dimers to the growing end (1). This activity may explain the Stu2 stabilization activity of MTs in living cells.
FIG. 1.
AlpA belongs to the third group of the Dis1/XMAP215 family. (A) Two TOG domains, eight HEAT repeats, and a coiled-coil region were identified, which are common to all class three members, including S. pombe Alp14 and Dis1, which is the Dis1/XMAP215 family-founding protein, and S. cerevisiae Stu2. Members of the first group include human ch-TOG, Xenopus XMAP215, Drosophila melanogaster Msps, D. discoideum DdCP224, and Arabidopsis thaliana MOR1. So far, there is only one known group two member, namely ZYG-9 of Caenorhabditis elegans. (B) Phylogenetic analysis of S. pombe Alp14 (Sp) homologues with S. cerevisiae (Sc), A. nidulans (An), A. fumigatus (Af), and A. oryzae (Ao). Accession numbers are indicated.
Besides the MT stabilization activity of Dis1/XMAP215 proteins, DdCP224, the Dictyostelium discoideum homologue, is involved in MT-cortex interactions. There is evidence that this contact is mediated by cortical dynein with which DdCP224 is able to physically interact (9).
In this paper, we have analyzed the function of the Dis1/XMAP215-like protein AlpA in Aspergillus nidulans. The protein localized at the spindle pole bodies (the fungal homologues of centromeres) and at MT plus ends. Interestingly, deletion of the gene was not lethal, although a drastic reduction of the MT array and MT dynamics was observed. Hyphae of an alpA deletion strain grew in curves, suggesting that AlpA is involved in the determination of growth directionality.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Supplemented minimal and complete media for A. nidulans were prepared as described previously, and standard strain construction procedures were as described by Hill and Käfer (10). A list of A. nidulans strains used in this study is given in Table 1. Standard laboratory Escherichia coli strains (XL1-Blue) were used. Plasmids are listed in Table 2.
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| GR5 | pyrG89 wA3 pyroA4 | 28 |
| GFP-NudA | pyrG89 wA2 pyroA4 ΔnudA::pyr4 alcA(p)::GFP::nudA::pyr4 | 6 |
| GFP-NudF | pyrG89 wA2 pyroA4 ΔnudF::pyr4 alcA(p)::GFP::nudF::pyr4 | 6 |
| RMS011 | pabaA1 yA2 ΔargB::trpCΔB trpC801 | 25 |
| SCE01 | SJW02 transformed with pCE05; wA3 ΔargB::trpCΔB pyroA4 alcA(p)::alpA::mRFP1 alcA(p)::GFP::tubA | This work |
| SCE05 | SRF200 transformed with pCE08; ΔargB::trpCΔB pyroA4 alcA(p)::GFP::alpA (single homologous integration) | This work |
| SCE10 | SRF200 transformed with pCE06; pyrG89 ΔargB::trpCΔB pyroA4 alcA(p)::alpA::GFP | This work |
| SCE12 | SRF200 transformed with pCE06, pJW18, and pPND1; alcA(p)::mRFP1::stuA alcA(p)::alpA::GFP alcA(p)::mRFP::kipBrigor | This work |
| SCE35 | SSK44 transformed with pCE06; pabaA1 wA3 ΔargB::trpCΔB ΔkipA::pyr4 alcA(p)::alpA::GFP (single integration) | This work |
| SCS13a/b | TNO2A3 transformed with pAT3; pyrG89 pyroA4 argB2 ΔnkuA::argB ΔalpA::pyr4 | This work |
| SDV69f | SNR1 transformed with pCE06; ΔkinA::pyr4 pyroA4 alcA(p)::GFP::alpA (single integration) | This work |
| SDV83b | SCS13 crossed with RMS011; pabaA1 yA2 pyrG89 ΔalpA::pyr4 | This work |
| SDV86 | SDV83b crossed with SJW02; pabaA1 yA2 pyrG89 alcA(p)::GFP::tubA ΔalpA::pyr4 | This work |
| SDV87 | SDV83b crossed with SSK92; pabaA1, yA2, pyrG89; alcA(p)::GFP::kipA ΔalpA::pyr4 | This work |
| SDV96 | TN02A3 transformed with pPND1 and pCE06; alcA(p)::mRFP1::kipBrigor alcA(p)::alpA::GFP | This work |
| SDV100 | SDV83 transformed with pGFP-NudA; ΔalpA alcA(p)::GFP::nudA (single integration) | This work |
| SDV101 | SDV83 transformed with pGFP-NudF; ΔalpA alcA(p)::GFP::nudF (single integration) | This work |
| SDV102 | SDV83 crossed to GFP-ClipA; ΔalpA alcA(p)::GFP::clipA | This work |
| SJW02 | wA3 pyroA4 alcA(p)::GFP::tubA ΔargB::trpCΔB | J. Warmbold, Marburg, Germany |
| AnKin26 | pyrG89 yA2 ΔargB::trpCΔB ΔkinA::pyr4 | 18 |
| SRF200 | pyrG89 ΔargB::trpCΔB pyroA4 | 11 |
| SSK44 | pabaA1 wA3 ΔargB::trpCΔB ΔkipA::pyr4 | 13 |
| SSK92 | wA3 pyroA4 alcA(p)::GFP::kipA | 13 |
| TNO2A3 | pyrG89 pyroA4 argB2 ΔnkuA::argB | 16 |
| SNR1 | ΔargB::trpCΔB pyroA4 ΔkinA::pyr4 | 18 |
| ΔclipA | pyrG89 wA3 ΔclipA::pyroA | 2 |
| SAD1c | SSK44 crossed with SCS13; wA3 pyroA4 ΔkipA::pyr4 ΔalpA::pyr4 | This work |
Only relevant genotypes are indicated. All strains carry the veA1 mutation.
TABLE 2.
Plasmids used in this study
| Plasmids | Constructiona | Source or reference |
|---|---|---|
| pAT1 | 1,000 bp upstream of alpA ORF (= LB) cloned into pCR2.1 | This work |
| pAT2 | 1,001 bp downstream of alpA ORF (= RB) cloned into pCR2.1 | This work |
| pAT3 | alpA-LB::N. crassa pyr-4::alpA-RB in pCS2.1 | This work |
| pCE05 | alcA::alpA::mRFP1 argB; gateway construct of pMT01 | 26; this work |
| pCE06 | alcA::alpA::GFP argB; gateway construct of pMT-sGFP | 26; this work |
| pCE08 | alcA(p)::GFP::alpA pyr4; 1 kb of alpA with AscI und PacI inserted into pMCB17apx | 27; this work |
| pCR2.1-TOPO | Cloning vector | Invitrogen |
| pCS1 | N. crassa pyr-4 selectable marker as NotI fragment in pUMA208 | This work |
| pGFP-NudA | alcA(p)::GFP::nudA::pyr4 | 6 |
| pGFP-NudF | alcA(p)::GFP::nudF::pyr4 | 6 |
| pPND1 | Rigor mutant of KipB for staining MTs, alcA(p)::mRFP1::kipBrigor | 21 |
| pMCB17apx | pMCB17 version for fusion of GFP to N termini of proteins of interest | 27 |
| pJW18 | Red nuclei, alcA(p)::mRFP1::stuA | J. Warmbold, Marburg, Germany |
ORF, open reading frame.
Light and fluorescence microscopy.
For live-cell imaging, cells were grown in glass-bottom dishes (World Precision Instruments, Berlin, Germany) in 4 ml of minimal medium containing either 2% glycerol (or ethanol) or 2% glucose as a carbon source. Medium was supplemented with pyridoxine, p-aminobenzoic acid, biotin, arginine, uracil, or uridine depending on the auxotrophy of the strains. Cells were incubated at room temperature for 1 to 2 days, and images were captured using an Axiophot microscope (Zeiss, Jena, Germany), a Plan-apochromatic 63× or 100× oil immersion objective lens, and an HBO50 Hg lamp. Alternatively, a Zeiss AxioImager Z1 with the latest AxioVision software (v 4.5) was used. Fluorescence was observed using standard Zeiss filter combinations no. 09 (fluorescein isothiocyanate, green fluorescent protein [GFP]) and no. 15 (DsRed). Images were collected and analyzed with a Hamamatsu Orca ER II camera system and the Wasabi software (version 1.2) or Zeiss Axiocam and AxioVision software. Time-lapse series were obtained with an automated Wasabi program that acquires series of images with 2- or 5-s intervals, 0.1- or 0.75-s exposure time, and about 100 exposures in a sequence. Image and video processing were done with the Wasabi software from Hamamatsu, Adobe Photoshop, ImageJ (NIH, Bethesda, MD), and virtual dub (http://www.virtualdub.org).
To determine SPK position, strains were grown on a microscope slide for 24 h at room temperature in MM containing 17% gelatin, and images were captured using differential interference contrast microscopy.
Molecular Techniques.
Standard DNA transformation procedures were used for A. nidulans (30) and E. coli (22). For PCR experiments, standard protocols were applied using a Biometra Personal Cycler (Biometra, Göttingen, Germany) for the reaction cycles. DNA sequencing was done commercially (MWG Biotech, Ebersberg, Germany). Total DNA was extracted from A. nidulans in the following way. Spores were inoculated in liquid minimal medium plus supplements and grown for 16 to 24 h at 37°C without shaking. Hyphal mats were harvested, dried with tissue paper, and ground in liquid nitrogen. The resulting powder was mixed with extraction buffer (50 mM EDTA, 0.2% sodium dodecyl sulfate) and incubated for 30 min to 2 h at 68°C in a water bath. Sodium dodecyl sulfate was removed from the suspension by addition of sodium acetate solution (8 M, pH 4.2) and centrifugation. From the supernatant, total DNA was precipitated with isopropanol, and the pellet was washed twice with 70% ethanol, air dried, resuspended in TE buffer, and stored at 4°C. Southern hybridizations were performed according to the DIG Application Manual for Filter Hybridization (Roche Applied Science, Technical Resources; Roche Diagnostics GmbH, Mannheim, Germany).
Deletion of alpA and construction of a ΔalpA/ΔkipA double mutant.
The alpA flanking regions were amplified by PCR using genomic DNA and the primers alpA_LB_fwd (5′-TCAAGGGCAGAGAGGGATGCAATC-3′) and alpA_LB_rev_Sfi (5′-CGGCCATCTAGGCCTGCGGAAGGTGGCGATG-3′) for the upstream region of alpA and alpA_RB_fwd_Sfi (5′-CGGCCTGAGTGGCCTGTACGGTCAACTTTAGG-3′) and alpA_RB_rev (5′-GAGTTCGCTAAGCTCCTCAGTGCCATC-3′) for the downstream region and cloned into pCR2.1-TOPO to generate pAT1 and pAT2, respectively (the Sfi restriction sites are underlined in the primer sequences). In a three-fragment ligation, the pyr4 gene from plasmid pCS1 was ligated between the two alpA-flanking regions, resulting in vector pAT3. The vector pAT3 was digested with restriction enzyme KpnI, and the linearized plasmid was transformed into the uracil/uridin-auxotrophic strain TNO2A3. Among six transformants, analyzed by PCR, five displayed homologous integration of the deletion cassette at the alpA locus. As primers for the indicative PCR, we used oligonucleotides derived from the pyr4 gene: pyr4-5′ (5′-GGTTGAGGAAGCAGTCGAGAGC-3′) and pyr4-3′ (5′-CTCGAGGACGAGCCGC-3′) and the alpA external primers alpA_5′-outside (5′-TACCCTAAGGTCACTACG-3′) and alpA_3′-outside (5′-AGATGGGTGTTCCTTACG-3′). Two of the ΔalpA strains (SCS13a and SCS13b) were also analyzed by Southern blotting (data not shown). In both strains, uracil/uridine prototrophy was linked to the alpA deletion, as shown by crossing them with uracil/uridin-auxotrophic alpA wild-type strains (data not shown).
To generate a ΔalpA/ΔkipA double mutant, we crossed the kipA deletion strain SSK44 with the deletion strain of alpA (SCS13). Heterokaryon formation was forced on MM, where none of the parent strains can grow alone. Progeny strains were screened by PCR and Southern blotting for the double deletion (data not shown).
Bioinformatics.
Protein sequences were aligned using vector NTI software (Invitrogen), MegAlign, and ClustalW software (http://www.embl-heidelberg.de). TOG domains and heat repeats of AlpA were identified using “REP” from the ExPASy database.
RESULTS
Identification of a Dis1/XMAP215 family protein in A. nidulans.
To characterize the role of the MT plus end complex for polarized growth, we searched the A. nidulans database with the Schizosaccharomyces pombe Alp14 protein sequence (4) (http://www.broad.mit.edu). The putative homologue AlpA (An5521.2) is a 96.4-kDa protein comprised of 891 amino acid residues. The open reading frame is disrupted by three short introns, 70 bp, 72 bp, and 72 bp in size. The intron-exon borders were confirmed by reverse transcription-PCR of small cDNAs, subsequent sequencing, and comparison with the sequence of genomic DNA. Protein analysis revealed eight HEAT repeats embedded in two TOG domains at the N terminus and a coiled-coil region at the C terminus (Fig. 1A). According to Ohkura et al. (17), MAPs of the Dis1/XMAP215 family are grouped into three classes (Fig. 1A). Sequence comparison of A. nidulans AlpA, which belongs to group three of the Dis1/TOG family, with related proteins from S. pombe showed a similarity of 30% to Alp14 and 23% to Dis1 (Fig. 1B). In contrast to S. pombe, where two proteins of this family exist, a protein with higher similarity to Dis1 was not found in A. nidulans. Sequence similarities of putative Alp14 homologues in Aspergillus oryzae (Ao_AlpA) and Aspergillus fumigatus (Af_AlpA) displayed similarities of 44.8% and 43.6% to Alp14 but of 84.2% and 84.5% to A. nidulans AlpA and 89.1% to each other. A Dis1 homologue was also not identified in the latter two Aspergillus species.
AlpA localizes to MT plus ends during mitosis and in interphase.
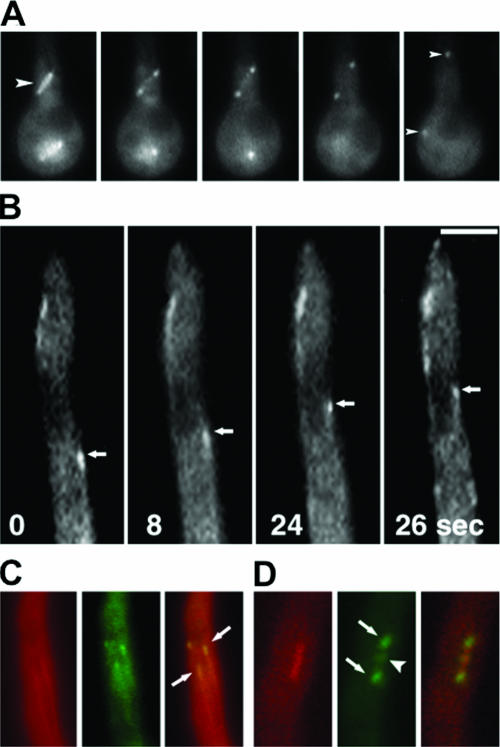
To analyze the function of alpA in A. nidulans, we studied the subcellular localization of the protein. We fused the alpA gene at the 3′ or 5′ end with GFP (pCE06, pCE08) or mRFP1 (pCE05) and transformed it into strain TN02A3 (SDV96) or SJW02 (SCE01). MTs were labeled in green (GFP) or red (mRFP1). The alpA construct was expressed under the control of the alcA promoter, with glycerol as a carbon source. Glycerol leads to derepression of the promoter but not induction, unlike ethanol (3). The expression levels under these conditions are quite low, and the problem of mislocalization of fusion proteins is minimized. Several transformant strains were analyzed in vivo, and identical results were obtained. AlpA localization and behavior were identical in C- and N-terminally fused GFP constructs (SCE10, SCE05). In general, the AlpA-GFP and AlpA-mRFP1 signal intensities were very low, which sometimes made a high-resolution analysis difficult. Figure 2A shows a mitotic spindle decorated with associated GFP-AlpA. During early mitosis, the complete spindle was covered with GFP-AlpA. As the spindle elongated, GFP-AlpA was distributed exclusively to the spindle poles (Fig. 2A). At some stage of mitosis, presumably the early metaphase, GFP-AlpA was detected in the middle of the spindle, suggesting association with the plus ends of the spindle MTs contacting the kinetochors (Fig. 2D). In interphase cells, GFP-AlpA localized to MTs as well, notably to the MT plus ends (Fig. 2B, C), and followed MT growth as comet-like structures (see Movie S01 in the supplemental material). This was similar to kinesin KipA, dynein heavy chain NudA, and NudF localization (29).
FIG. 2.
AlpA localization during mitosis and in interphase. (A) During mitosis, GFP-AlpA was distributed along short spindles (arrowhead in the first frame). As the spindle elongated GFP-AlpA was redistributed to the spindle poles (arrowheads in the last frame). Frames are shown in 2-min intervals. The strain was SCE05. (B) GFP-AlpA movement can be seen as comet-like structures, indicating the association with the MT plus ends (see Movie S01 in the supplemental material) (C, D) MTs were visualized by decoration with a red-labeled kinesin rigor mutant protein (mRFP1-KipBrigor) during interphase (C) and mitosis (D) (strain SDV96). The arrows point to a GFP-AlpA signal at MT plus ends (C) and to the spindle pole bodies (D). The arrow head in panel D points to the center of the spindle, where the protein could be associated with the kinetochores. Bars, 3 μm (A) and 2 μm (B to D).
alpA deletion strains show defects in polarized growth.
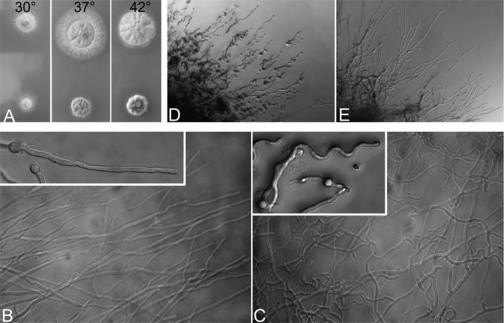
The alpA gene was deleted by homologous recombination where the alpA ORF was replaced by the Neurospora crassa pyr4 gene. We used A. nidulans strain TNO2A3, which has a very high frequency of homologous integration (16). Homologous single integration was verified by Southern blot and PCR analysis in 3 of 7 tested strains (data not shown). Compared to the wild type, alpA deletion strains showed reduced colony size and compact growth, especially at higher temperatures (Fig. 3A). Although the conidiospore number was slightly reduced in alpA deletion strains, the morphology of conidiophores was indistinguishable from that of the wild type. Interestingly, and in contrast to the situation in S. cerevisiae, deletion strains were viable. This was surprising because we found only one Alp14 similar protein in the A. nidulans genome in comparison to two in S. pombe, where Dis1 can substitute for Alp14 (Fig. 1). Hyphae of the alpA deletion strain did not show any difference with regard to nuclear distribution or septation, but hyphal morphology was changed. While wild-type hyphae grow relatively straight, the alpA deletion strain produced curly or curved hyphae (Fig. 3B, C), which were similar to hyphae of a kipA deletion strain (13). In addition to the curved growth phenotype, we noticed an increased number of branches in older hyphae. To show that the observed phenotypes were due to the deletion event, we constructed a plasmid where about 1 kb of the 5′ end of the alpA gene was fused to GFP and under the control of the inducible alcA promoter. The construct was integrated at the alpA locus (confirmed by Southern blotting), resulting in a full-length, GFP-fused version under the control of the alcA promoter (strain SCE05). The strain was used in the localization experiments described above. Under repressing conditions (glucose), SCE05 showed the knockout-like curved growth (Fig. 3D), whereas under inducing conditions (ethanol), wild-type-like growth was restored (Fig. 3E). This result proved that the GFP-tagged protein version was fully functional.
FIG. 3.
Phenotype of an alpA deletion strain. (A) In comparison to a control strain (top, RMS011), the ΔalpA strain (bottom, SDV83b) grew slower and colonies were more compact. While wild-type hyphae grew straight (B), hyphae of the alpA deletion strain showed a curved growth phenotype (C). (D) In a strain having the only functional copy of alpA under the control of the inducible alcA promoter (SCE05), curved growth was observed under repressing conditions (glucose), but wild-type hyphal morphology was restored when grown under inducing conditions (ethanol) (E).
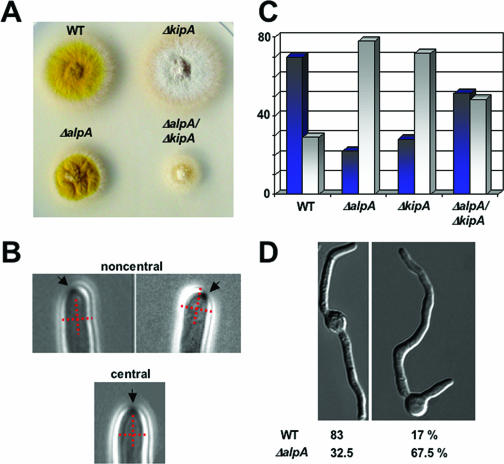
The curved growth phenotype in the alpA mutant resembled that of a kipA deletion strain (13). Therefore, we asked whether the lack of both genes would result in a similar or a different phenotype than that of the single mutations. The double mutant showed a more severe phenotype than the individual mutations. Hyphae appeared even more curly and similar to the alpA mutant, with more branches in older hyphae. Colonies were much smaller than the colonies of the parent strains, indicating an additive effect of ΔkipA and ΔalpA (Fig. 4A).
FIG. 4.
Localization of the Spitzenkörper in hyphae and germination pattern. The Spitzenkörper was observed in growing hyphae as described previously (19). (A) Colonies of the wild-type (RMS011) (WT), ΔalpA (SDV83), ΔkipA (SSK44), and ΔalpA ΔkipA (SAD1c) strains on an agar plate after 3 days of growth at 37°C. (B) Representative hyphae with a Spitzenkörper in the center of the cell or noncentral. To indicate the position of the organelle, we introduced a cross into the hypha. (C) Quantification of the location of the Spitzenkörper in the strains listed for panel A. Dark blue columns represent hyphae with the Spitzenkörper in the center and gray columns the ones where the Spitzenkörper was noncentral. Between 50 and 64 hyphae were analyzed for each strain. (D) Quantification of the germination pattern of conidiospores as displayed in the pictures. Wild type (RMS011), n = 200; alpA mutant (SDV83), n = 268.
Because the growth direction of hyphae depends on the localization of the Spitzenkörper in the apex, we analyzed the position of this organelle in the wild type and compared it to the one in the alpA, kipA, and alpA kipA double deletion strains. Whereas in the wild type, the Spitzenkörper was found in the center of the hyphae in 70% and noncentral in 30% of the cases (n = 50), in the alpA deletion strain, only 22% showed the central position and 78% the noncentral one (n = 50). In comparison, in the kipA deletion strain, the percentages were 28% (central) and 72% (noncentral) (n = 50), and in the alpA kipA double deletion strain, the percentages were 52% (central) and 48% (noncentral) (n = 64) (Fig. 4B, C). It was surprising that the number of central and noncentral positioning of the Spitzenkörper was almost even in the double mutant strain. In addition, we noticed that in 18% of the cases two Spitzenkörper were observed in the hyphal tip. In comparison, this number was only 5% in the wild type, 10% in the alpA mutant, and 5% in the kipA mutant. If there were two Spitzenkörper in the apex, we counted them as one event of noncentral organelles in the quantification shown in Fig. 4A.
To test whether AlpA might play a role in the initiation of polarized growth, we analyzed the germination pattern of conidiospores (Fig. 4D). Wild-type conidiospores produce a second germ tube after the first germ tube has reached a certain length, and this second hypha emerges from a place opposite the first hypha. In contrast, the alpA deletion strain produced the second germ tube normally in angles smaller than 180° from the first hypha (Fig. 4D). This germination pattern resembled the one from the kipA mutant strain (13).
AlpA determines cytoplasmic MT dynamics.
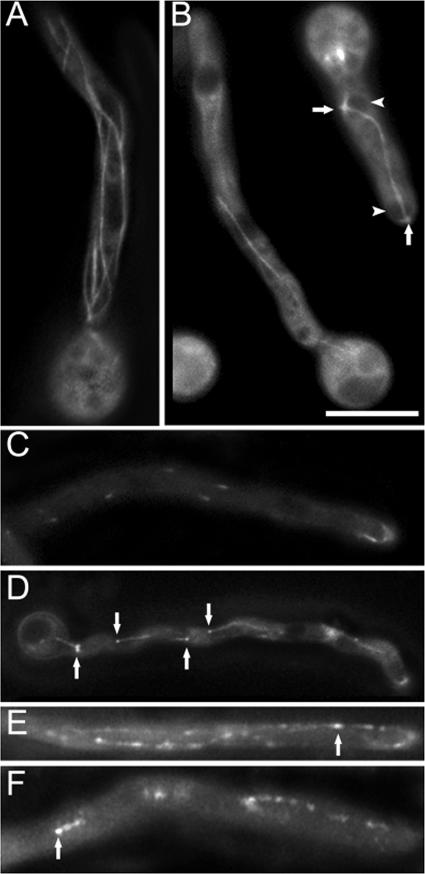
To further unravel the function of alpA in A. nidulans, we studied the effect of the alpA deletion on the MT cytoskeleton. MTs were visualized in the alpA deletion strain by GFP staining (6) (strain SDV86). Compared to he wild type (SJW02), the number of MTs was reduced in the ΔalpA strain. Basically, only one thick MT bundle (according to Veith et al. [27]), connecting adjacent nuclei, was visible, in addition to some shorter MTs emerging from the nuclear spindle pole bodies, while in the wild-type strain several single and bundled MTs were present (Fig. 5A, B). In addition, the normally highly dynamic MTs appeared more stable and less dynamic. Whereas wild-type MTs polymerize at a rate of 14 μm per min (6), the extension rate in the alpA mutant was only 6 μm per minute. It has to be noted that growth of MTs only occurred occasionally. Most MTs did not elongate nor shrink. After MTs have reached the hyphal tip, they normally disassemble (MT catastrophe) within 20 s (see Movie S02 in the supplemental material) (13). In the ΔalpA background, fewer MTs reached the tip (4 in 5 min, compared to 20 in the wild type [36 hyphae analyzed]), and disassembly did not occur within minutes (see Movie S03 in the supplemental material). In addition, the number of emerging MTs in the mutant was reduced by 85% (25 hyphae analyzed). The mitotic spindle and mitosis itself were indistinguishable from that of the wild type (see Movie S04 in the supplemental material).
FIG. 5.
alpA affects MTs and MT plus end-localized proteins. (A) Several MTs and bundles thereof stained with GFP are obvious in the wild type (SJW02). (B) The number and dynamics (see Movies S02 and S03 in the supplemental material) of MTs in an alpA deletion strain (SDV86) were reduced compared to the wild type. Arrowheads point to nuclei and arrows to spindle pole bodies. (C) While GFP-KipA localized to MT plus ends and moved as comets in the wild type (SSK92) (see Movie S05 in the supplemental material), (D) GFP-KipA decorated short fragments of MTs in the ΔalpA background (SDV87) (see Movie S06 in the supplemental material). Fragments of MTs were also GFP decorated in ΔalpA strains with GFP-NudA (E) and GFP-NudF (F) fusion proteins (strains SDV100 and SDV101). Arrows in panels D to F point to MT plus ends as determined by the growth at this end. Bars in panel B, 5 μm (A, C, and D), 6 μm (B), and 4 μm (E and F).
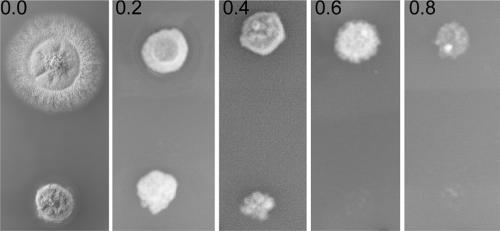
To analyze whether AlpA influences the stability of MTs, we tested the sensitivity of an alpA deletion strain (SDV83b) toward the microtubule-destabilizing agent benomyl. Whereas the wild type was able to produce colonies up to a concentration of 0.8 μg/ml, the alpA mutant was unable to grow at concentrations higher than 0.6 μg/ml (Fig. 6). This result suggests that AlpA stabilizes MTs in A. nidulans.
FIG. 6.
Benomyl sensitivity of the wild-type (upper row of colonies) and alpA deletion (lower row) strains. Benomyl was added in concentrations from 0 to 0.8 μg/ml, and colonies were grown for 2 days at 37°C.
Interdependence of AlpA with other MT plus end-associated proteins.
The fact that AlpA localized to the MT plus end in interphase cells raised the question of how it reaches the destination and whether this localization depends on the presence or activity of other MT plus end-associated proteins. To this end, we studied interactions between AlpA and the kinesins KinA and KipA, the dynein pathway components NudA and NudF, and the Clip170 homologue ClipA.
We analyzed AlpA MT plus end localization in ΔkipA and ΔkinA mutant backgrounds (strains SCE35 and SDV69f). The situation for GFP-AlpA in the ΔkipA and ΔkinA backgrounds was wild-type-like (not shown). Both KipA and KinA have been shown to be involved in MT plus end accumulation of ClipA and NudA, respectively (2, 31), but neither of those two kinesins was responsible for AlpA plus end localization. These results are in agreement with recent findings in S. cerevisiae, where Al-Bassam et al. (1) showed for the AlpA homologue Stu2 that it localizes to MT plus ends independently of any motor protein. Localization was dependent on the second TOG domain of Stu2, whereas the first TOG domain promotes the addition of α,β-tubulin dimers to the growing MT end.
To analyze the role of AlpA at the MT plus end and in polarized growth, we sought to determine whether AlpA is required for the recruitment of other proteins, such as the kinesin-like protein KipA, ClipA, the dynein motor NudA, or one of its regulators (NudF) to this place. Therefore, we constructed alpA deletion strains in which KipA or ClipA were labeled with GFP. Normally, both proteins accumulate at the MT plus end and hitchhike with the growing MT end. The visible movement of the KipA- or ClipA-GFP spots were described as comets (see Movie S05 in the supplemental material) (2, 13). KipA movement in an alpA deletion strain was reduced, and GFP-KipA partly decorated cytoplasmic MTs behind the plus end instead of moving with the MT plus end (Fig. 5C, D; see also Movie 06 in the supplemental material). An accumulation of the GFP fusion protein was still visible at the MT plus end. Because MTs did not extend as fast as in the wild type (see above), KipA-GFP comets were not observed. Similar results were obtained for ClipA (our own results and L. Zhuang and X. Xiang [Bethesda, MD], personal communication), dynein (NudA), and its regulator NudF. In strains with fusion proteins of GFP-NudA and GFP-NudF in a ΔalpA background (strains SDV100 and SDV101), MTs were similarly GFP decorated (Fig. 5E, F). However, in comparison to KipA, longer stretches of MTs were decorated with either NudA or NudF. Further experiments should address the question of whether the slight differences in localization are of functional importance or due to, e.g., different protein amounts of NudA, NudF, and KipA. The fact that the localization of components of the dynein pathway appears to be affected in alpA mutants does not cause nuclear distribution defects (see above) suggests that even in the absence of AlpA sufficient amounts of, e.g., dynein reach their normal place in the cell and serve the wild-type function.
DISCUSSION
In this paper, we characterized the Dis1/XMAP215 family protein AlpA from A. nidulans and found that it is associated with the MT plus end during mitosis and in interphase. AlpA plays a role in controlling MT dynamics and is important for the determination of growth polarity. Whereas the mechanism of MT stabilization was recently shown in S. cerevisiae (1), a role in polarized growth has not been described before. Polarized growth of filamentous fungi depends on the continuous delivery of secretory vesicles (7, 20). These vesicles provide new membranes and deliver, e.g., enzymes for cell wall biosynthesis. Because the vesicles are generated some distance away from the growing tip, they need to be transported long distances. It is assumed that MTs and conventional kinesin provide the basis for this long-distance transportation (18, 23). The first destination of the vesicles is an organelle close to the apex named the vesicle supply center or Spitzenkörper (8). The location of this organelle determines growth direction. For the last few micrometers between the Spitzenkörper and the cell membrane, fungi probably employ the actin cytoskeleton and its associated motors. According to this model, MTs contribute to polarized growth as tracks for the transportation of vesicles. Surprisingly, deletion of alpA seems not to affect long-distance vesicle transportation and accumulation of the vesicles in the Spitzenkörper significantly, despite the dramatic effects on MT organization. This result suggests that only few MTs are sufficient for efficient vesicle transportation. This is in agreement with observations that the growth rate of hyphae does not change during mitosis, although most of the cytoplasmic MTs are degraded during nuclear division (19). Another explanation for the observed growth in the alpA deletion strain could be the vesicle transport activity of the actin cytoskeleton. However, the fact that mutations in tubulin-encoding genes or in MT-dependent motor protein encoding-genes affect hyphal extension highly suggests an important role of MTs in polarized growth (18, 23). Although the Spitzenkörper was not obviously reduced in size in the alpA mutant, the position appears to be dependent on AlpA, as it was shown before for KipA (Fig. 5). An open question remains, however, why the number of centrally localized Spitzenkörpers increased again in the alpA kipA double mutant. Perhaps this is linked to the observation that the number of hyphae with two Spitzenkörpers in the apex was increased.
Results with S. pombe and A. nidulans suggest a second role for MTs in the determination of growth direction, and this feature is obviously affected in alpA deletion strains (13-15). According to the model of S. pombe, so called cell-end factors are transported towards the MT plus end and hitchhike with the growing MTs towards the cell cortex. A cell end factor is, for instance, the membrane-associated protein Mod5, which was suggested to act as an anchor for Tea1 and Tea4. The latter protein in turn binds the formin For3, which catalyzes actin polymerization (15, 24). Although we were not able to identify a Mod5 homologue in A. nidulans or other aspergilli yet, the presence of the kinesin KipA (Tea2) and TeaA (Tea1) as well as a curved growth phenotype upon deletion of either of them (13) (results for TeaA are unpublished) suggests at least partial conservation of the mechanism. If this is the case and if deletion of alpA caused a phenotype similar to that of deletion of kipA, the question is how AlpA is involved in polarity determination. It was shown in D. discoideum that DdCpd224 interacts with cortical dynein and thereby could mediate the contact between MT plus ends and the cortex (9). In A. nidulans, the situation could be similar, and a missing cortical contact could lead to the curved hyphal growth. However, it has to be noted that dynein mutants of A. nidulans do not display the same hyphal growth phenotype. In addition, dynein-mediated MT-cortical interactions are required for nuclear migration and nuclear positioning (27). Both phenomena were not affected in alpA mutants. Therefore, it seems likely that the lack of AlpA drastically reduces MT dynamics and that this leads to a reduction of specific cell end marker delivery. One of the key challenges is therefore to identify such cell end marker proteins in filamentous fungi. The fact that the alpA kipA double mutant displayed a more severe phenotype with regard to hyphal extension in comparison to the strains with only one mutation suggests that the two genes also act in different pathways.
Supplementary Material
Acknowledgments
We thank X. Xiang (Bethesda, MD) for sending us several clipA strains.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG), the Fonds der Chemischen Industrie, the Max-Planck-Institute for Terrestrial Microbiology, and the special program “Lebensmittel und Gesundheit” of the Ministery of Baden Württemberg.
Footnotes
Published ahead of print on 19 January 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Al-Bassam, J., M. van Breugel, S. C. Harrison, and A. Hyman. 2006. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172:1009-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Efimov, V., J. Zhang, and X. Xiang. 2006. CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol. Biol. Cell 17:2021-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felenbok, B., M. Flipphi, and I. Nikolaev. 2001. Ethanol catabolism in Aspergillus nidulans: a model system for studying gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 69:149-204. [DOI] [PubMed] [Google Scholar]
- 4.Galagan, J. E., S. E. Calvo, C. Cuomo, L.-J. Ma, J. R. Wortman, S. Batzoglou, S.-I. Lee, M. Bastürkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. d'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Peñalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 5.Gard, D. L., and M. W. Kirschner. 1987. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 105:2203-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han, G., B. Liu, J. Zhang, W. Zuo, N. R. Morris, and X. Xiang. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biol. 11:19-24. [DOI] [PubMed] [Google Scholar]
- 7.Harris, S. D., and M. Momany. 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41:391-400. [DOI] [PubMed] [Google Scholar]
- 8.Harris, S. D., N. D. Read, R. W. Roberson, B. Shaw, S. Seiler, M. Plamann, and M. Momany. 2005. Polarisome meets Spitzenkörper: microscopy, genetics, and genomics converge. Eukaryot. Cell 4:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hestermann, A., and R. Gräf. 2004. The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol. 5:24-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill, T. W., and E. Käfer. 2001. Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions. Fungal Genet. Newsl. 48:20-21. [Google Scholar]
- 11.Karos, M., and R. Fischer. 1999. Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol. Genet. Genomics 260:510-521. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita, K., B. Habermann, and A. A. Hyman. 2002. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12:267-273. [DOI] [PubMed] [Google Scholar]
- 13.Konzack, S., P. Rischitor, C. Enke, and R. Fischer. 2005. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell 16:497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, S. G., and F. Chang. 2003. Cell polarity: a new mod(e) of anchoring. Curr. Biol. 13:R711-730. [DOI] [PubMed] [Google Scholar]
- 15.Martin, S. G., W. H. McDonald, J. R. Yates III, and F. Chang. 2005. Tea4p links microtubule plus ends with the formin For3p in the establishment of cell polarity. Dev. Cell 8:479-491. [DOI] [PubMed] [Google Scholar]
- 16.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohkura, H., M. A. Garcia, and T. Toda. 2001. Dis1/TOG universal microtubule adaptors—one MAP for all? J. Cell Sci. 114:3805-3812. [DOI] [PubMed] [Google Scholar]
- 18.Requena, N., C. Alberti-Segui, E. Winzenburg, C. Horn, M. Schliwa, P. Philippsen, R. Liese, and R. Fischer. 2001. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 42:121-132. [DOI] [PubMed] [Google Scholar]
- 19.Riquelme, M., R. Fischer, and S. Bartnicki-Garcia. 2003. Apical growth and mitosis are independent processes in Aspergillus nidulans. Protoplasma 222:211-215. [DOI] [PubMed] [Google Scholar]
- 20.Riquelme, M., C. G. Reynaga-Peña, G. Gierz, and S. Bartnicki-García. 1998. What determines growth direction in fungal hyphae? Fungal Genet. Biol. 24:101-109. [DOI] [PubMed] [Google Scholar]
- 21.Rischitor, P., S. Konzack, and R. Fischer. 2004. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot. Cell 3:632-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russel. 1999. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 23.Seiler, S., F. E. Nargang, G. Steinberg, and M. Schliwa. 1997. Kinesin is essential for cell morphogenesis and polarized secretion in Neurospora crassa. EMBO J. 16:3025-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snaith, H. A., and K. E. Sawin. 2003. Fission yeast mod5p regulates polarized growth through anchoring of tea1p at cell tips. Nature 423:647-651. [DOI] [PubMed] [Google Scholar]
- 25.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171. [DOI] [PubMed] [Google Scholar]
- 26.Toews, M. W., J. Warmbold, S. Konzack, P. E. Rischitor, D. Veith, K. Vienken, C. Vinuesa, H. Wei, and R. Fischer. 2004. Establishment of mRFP1 as fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in Escherichia coli (GATEWAY). Curr. Genet. 45:383-389. [DOI] [PubMed] [Google Scholar]
- 27.Veith, D., N. Scherr, V. P. Efimov, and R. Fischer. 2005. Role of the spindle-pole body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J. Cell Sci. 118:3705-3716. [DOI] [PubMed] [Google Scholar]
- 28.Waring, R. B., G. S. May, and N. R. Morris. 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin coding genes. Gene 79:119-130. [DOI] [PubMed] [Google Scholar]
- 29.Xiang, S., G. Han, D. A. Winkelmann, W. Zuo, and N. R. Morris. 2000. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 10:603-606. [DOI] [PubMed] [Google Scholar]
- 30.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J., S. Li, R. Fischer, and X. Xiang. 2003. The accumulation of cytoplasmic dynein and dynactin at microtubule plus-ends is kinesin dependent in Aspergillus nidulans. Mol. Biol. Cell 14:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.