Abstract
The yeast Saccharomyces cerevisiae is able to use some biotin precursors for biotin biosynthesis. Insertion of a sulfur atom into desthiobiotin, the final step in the biosynthetic pathway, is catalyzed by biotin synthase (Bio2). This mitochondrial protein contains two iron-sulfur (Fe/S) clusters that catalyze the reaction and are thought to act as a sulfur donor. To identify new components of biotin metabolism, we performed a genetic screen and found that Isa2, a mitochondrial protein involved in the formation of Fe/S proteins, is necessary for the conversion of desthiobiotin to biotin. Depletion of Isa2 or the related Isa1, however, did not prevent the de novo synthesis of any of the two Fe/S centers of Bio2. In contrast, Fe/S cluster assembly on Bio2 strongly depended on the Isu1 and Isu2 proteins. Both isa mutants contained low levels of Bio2. This phenotype was also found in other mutants impaired in mitochondrial Fe/S protein assembly and in wild-type cells grown under iron limitation. Low Bio2 levels, however, did not cause the inability of isa mutants to utilize desthiobiotin, since this defect was not cured by overexpression of BIO2. Thus, the Isa proteins are crucial for the in vivo function of biotin synthase but not for the de novo synthesis of its Fe/S clusters. Our data demonstrate that the Isa proteins are essential for the catalytic activity of Bio2 in vivo.
Biotin is a water-soluble vitamin that functions as a prosthetic group of many carboxylases. It is covalently bound to lysine residues in highly conserved domains of the apoproteins. In Saccharomyces cerevisiae, the best-characterized biotin-containing enzymes are acetyl coenzyme A (acetyl-CoA) carboxylase, pyruvate carboxylase, and urea amidohydrolase. There are two isoforms of acetyl-CoA carboxylase in yeast. The cytosolic Acc1 protein appears to be the dominant form, which is necessary for the generation of malonyl-CoA for fatty acid biosynthesis and elongation (18). Moreover, the mitochondrial protein Hfa1 was recently shown to have acetyl-CoA carboxylase activity and to be essential for the generation of lipoate (21). There are also two isoforms of pyruvate carboxylase (Pyc1 and Pyc2 [56]), but in contrast to the situation in mammals, both are cytosolic enzymes in yeast (36, 60). Urea amidohydrolase, produced from the DUR1,2 gene, is essential for the utilization of urea as a nitrogen source and harbors two enzymatic activities: urea carboxylase, which depends on biotin, and allophanate hydrolase, which is biotin independent (57). Moreover, an additional biotin protein of yeast was recently identified to correspond to Arc1, a protein not acting as a carboxylase (30). The covalent linkage of biotin to its target proteins is catalyzed by biotin protein ligase which is encoded by the essential BPL1 gene (8). Since Acc1 is the only essential substrate of Bpl1, it appears that the essential function of Bpl1 lies in the covalent modification of Acc1 with biotin.
Most strains of S. cerevisiae are classified as biotin auxotrophs (32). However, the cells are proficient to carry out the last three steps of biotin biosynthesis and generate biotin from the precursors 7-keto-8-aminopelargonic acid (KAPA), 7,8-diaminopelargonic acid (DAPA), and desthiobiotin (also called dethiobiotin) making use of the enzymes encoded by BIO3 (DAPA aminotransferase), BIO4 (desthiobiotin synthase), and BIO2 (biotin synthase). Efficient utilization of KAPA and DAPA requires BIO5, which encodes a plasma membrane permease for both substrates (45). The plasma membrane biotin transporter Vht1 was recently identified in S. cerevisiae (54) and Schizosaccharomyces pombe (53) where it was also found to mediate the uptake of desthiobiotin. Active biotin uptake is not essential for viability, and vht1Δ mutants are fully complemented by adding high concentrations of biotin to the growth medium (53, 54).
The last step in biotin biosynthesis is the insertion of a sulfur atom for two hydrogen atoms at the saturated C-6 and C-9 positions of desthiobiotin, thus creating the thiophane ring of biotin. This reaction is performed by biotin synthase, which is localized to mitochondria. Yeast biotin synthase is related (>40% sequence identity) to the Escherichia coli BioB protein, the structure of which has recently been solved and shown to contain a 2Fe/2S and a 4Fe/4S cluster (2). Bio2 carries out a highly complex radical reaction that in vitro requires the presence of a reducing agent (dithiothreitol), flavodoxin, S-adenosylmethionine (SAM), and NADPH (2, 11). Although the sulfur donor for this reaction is still under debate, current evidence strongly implicates the 2Fe/2S cluster of biotin synthase as the source of biotin sulfur (2). Thus, biotin synthase may behave like a reactant which requires the repair of the 2Fe/2S cofactor for subsequent reaction cycles (25). The mitochondrial localization of Bio2 necessitates that all reactants required in vivo have to be present in mitochondria. Although the mitochondrial transporter for SAM was recently identified and found to be necessary for biotin synthesis (38), most of the other transport proteins are still elusive. For example, the protein required for desthiobiotin uptake across the mitochondrial inner membrane is still unknown. Moreover, biotin produced by Bio2 has to be exported to the cytosol where the majority of the biotinylated proteins are found (30).
The goal of the present study was to identify genes that are necessary for the utilization of desthiobiotin by S. cerevisiae cells. Besides BIO2, we found that biotin biosynthesis from desthiobiotin requires the ISA2 gene. The encoded mitochondrial matrix protein Isa2 and the related Isa1 are components of the iron-sulfur cluster (ISC) assembly machinery that catalyzes the synthesis of iron-sulfur (Fe/S) clusters and their insertion into Fe/S proteins. The ISC assembly machinery is essential for the biosynthesis of mitochondrial Fe/S proteins, and it is also required for the maturation of cytosolic and nuclear Fe/S proteins (35). Most of its members, including the Isa proteins, are related to components encoded by the isc operon of bacteria (27), and the presence of ISC homologs in virtually all eukaryotic species suggests that this biosynthetic process is highly conserved. Despite this high degree of conservation, the precise role of the Isa proteins in cellular Fe/S cluster assembly remains unknown. Here, we performed a detailed analysis of the role of Isa2 in biotin biosynthesis in S. cerevisiae.
MATERIALS AND METHODS
Yeast strains and cell growth.
The following wild-type strains of Saccharomyces cerevisiae were used: W303-1A (MATa ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112), W303-1B (MATα ura3-1 ade2-1 trp1-1 his3-11,15 leu2-3,112), and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). The following strains were derived from W303-1A: Gal-ISA2 (isa2Δ and isa1Δ/isa2Δ) (44), Gal-ISU1/isu2Δ (14), Gal-YAH1 (33), Gal-SSQ1 (39), Gal-ATM1 (31), isa1Δ (29), yfh1Δ (12), Gal-BPL1 (46), Gal-BPL1/bio2Δ (Gal-BPL1 bio2Δ::natNT2 [23; this work]), and Gal-ISA1 (pISA1::GAL1-10-HIS3 [this work]). In strain Gal-ISA1, the upstream region of ISA1 was exchanged for the galactose-inducible GAL1-10 promoter by PCR-mediated gene replacement. The functionality of various BIO2 constructs was tested below (see Fig. 3A) using bio2Δ::kanMX4 cells (BY4742 YGR286CΔ::kanMX4) (EUROSCARF, Frankfurt/Main, Germany). Cells were grown in rich (YP [1% yeast extract, 2% Bacto Peptone]) medium and minimal (synthetic complete [SC]) medium or minimal medium lacking added iron chloride. We refer to this as iron-poor medium because it contains traces of iron from laboratory glassware, distilled water, and various chemicals. Carbon sources were added as required (52). Biotin-free medium was prepared as described previously (53) and supplemented with biotin, desthiobiotin, or KAPA to obtain the desired concentrations. Desthiobiotin, which is chemically synthesized and certified to be free of biotin, was obtained from IBA (Göttingen, Germany).
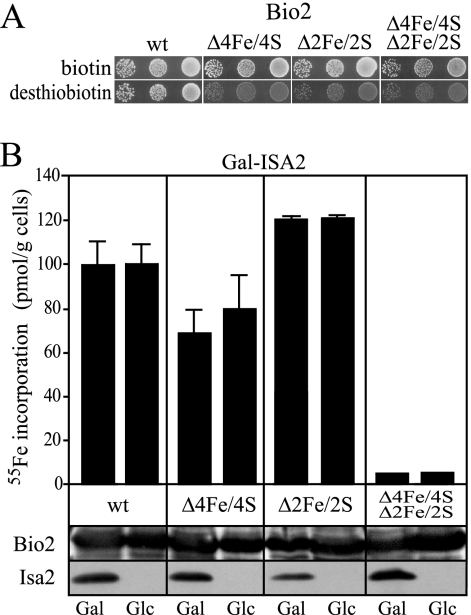
FIG. 3.
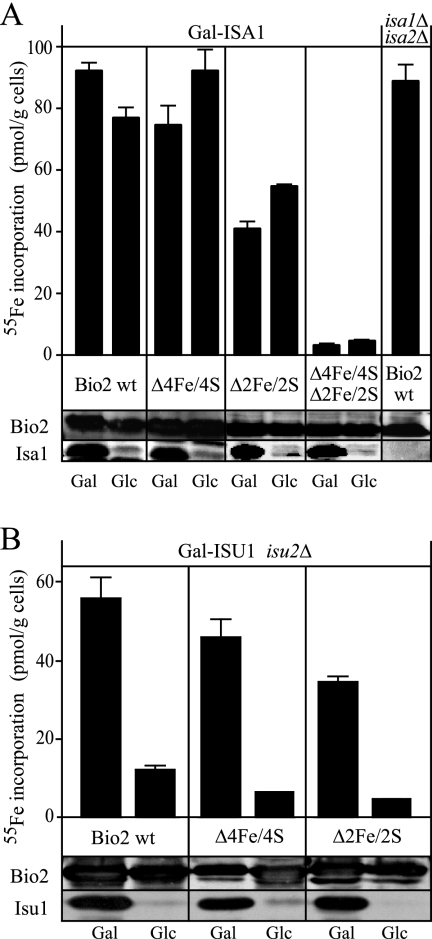
Fe/S cluster formation on Bio2 does not require Isa2 in vivo. (A) S. cerevisiae bio2Δ mutants (BY4742 bio2Δ::kanMX) were transformed with plasmids to overproduce wild-type (wt) Bio2, the Bio2 Δ4Fe/4S variant lacking the 4Fe/4S cluster, the Bio2 Δ2Fe/2S variant lacking the 2Fe/2S cluster, or a Bio2 variant lacking both clusters. Dilutions of the cells were spotted on media containing 0.2 μg liter−1 of either biotin or desthiobiotin, and growth was recorded after 3 days at 30°C. (B) Gal-ISA2 cells overproducing wild-type Bio2 and mutant Bio2 variants as in panel A were incubated in iron-poor minimal medium supplemented with galactose (Gal) or glucose (Glc) in order to induce or repress ISA2, respectively. Cells were incubated with 10 μCi 55Fe for 2 h, and Bio2 was precipitated from cell lysates with a polyclonal serum against Bio2. The amount of radioactivity that coprecipitated with Bio2 was quantified by liquid scintillation counting. Error bars represent the standard deviations. Western blot analysis of the cell lysates for Bio2 and Isa2 is shown below the bar graph.
Mutagenesis of yeast cells.
Yeast strain W303-1A was grown in yeast extract-peptone-dextrose, washed in water, and treated with 2% ethylmethane sulfonate for 2 h at 30°C. The cells were plated on SC medium containing 2 μg/liter biotin. Colonies appearing after 2 days at 30°C were replica plated to SC medium containing either 0.2 μg/ml biotin or 0.2 μg/liter desthiobiotin. Colonies forming on the biotin-containing plates but absent from the desthiobiotin-containing plates were purified and backcrossed to W303-1B. Wild-type and mutant spores from the resulting diploid were used in most experiments.
β-Galactosidase assays.
β-Galactosidase assays were performed according to published protocols (28) with minor modifications. Ten OD600 (optical density at 600 nm) units of logarithmically growing yeast cells were washed and broken with glass beads in 250 μl of 50 mM Na phosphate buffer (pH 7.0) containing 4 mM phenylmethylsulfonyl fluoride using a BIO101 FastPrep instrument. After the addition of 250 μl of 50 mM Na phosphate buffer (pH 7.0), the extract was centrifuged for 5 min in a microcentrifuge, and the protein concentration of the supernatant was determined by the Bradford assay (5). In parallel, the β-galactosidase activity of the supernatant was determined with ortho-nitrophenyl-β-d-galactoside (ONPG) at 37°C as described previously (28). Activities are reported in micromoles of ONPG hydrolyzed per milligram of protein per minute.
Plasmids.
To investigate iron binding to Isa2 and Bio2 in vivo, the wild-type genes and the respective mutant alleles of ISA2 and BIO2 were inserted into the 2μm plasmid p426-GPD downstream of the strong constitutive TDH3 promoter or into the low-copy plasmid p416-MET25 where expression is driven from the promoter of MET25 (13). The BIO2 alleles encoding proteins with defects in Fe/S cluster coordination were generated by site-directed mutagenesis using PCR. Replacement of three cysteine residues (C99, C103, and C106) by serine generated the Δ4Fe/4S mutant. Replacement of cysteine 143 by Ala created the Δ2Fe/2S mutant. Both mutations were combined to obtain a double mutant lacking both Fe/S clusters.
The BIO2-GFP plasmid was generated by ligation of a PCR product containing 1,000 bp of the BIO2 promoter and the entire BIO2 open reading frame into the URA3-CEN plasmid YCplac33-GFP (37). This plasmid complements the growth defect of bio2Δ mutants on desthiobiotin-containing plates and, as assessed by fluorescence microscopy, targets green fluorescent protein (GFP) to mitochondria (M. J. Gerl and J. Stolz, unpublished results). The same promoter fragment was fused to a full-length lacZ gene in a centromeric vector to give the reporter plasmid used in Fig. 5B (46). The yeast genomic library in the CEN-TRP1 vector pRS200 was described before (16) and obtained from the American Type Culture Collection (ATCC 77164).
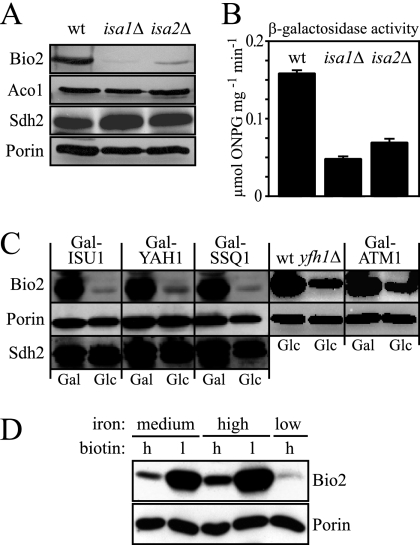
FIG. 5.
Expression of BIO2 is low in cells with defects in Fe/S protein maturation and under iron-limiting conditions. (A) Mitochondria were isolated from wild-type (wt), isa1Δ, and isa2Δ cells, and the abundance of biotin synthase (Bio2), aconitase (Aco1), the iron-sulfur protein subunit of succinate dehydrogenase (Sdh2), and the mitochondrial porin (Por1) were determined by Western blotting. (B) β-Galactosidase activities were determined for wild-type, isa1Δ, and isa2Δ cells expressing the lacZ gene under control of the BIO2 promoter. The assays were performed in triplicate on soluble cell extracts, and the activities are reported as micromoles of ortho-nitrophenyl-β-d-galactoside consumed per milligram of protein per minute. Error bars represent the standard deviations. (C) Gal-ISU1/Δisu2, Gal-YAH1, Gal-SSQ1, Gal-ATM1, and yfh1Δ cells were grown in minimal medium supplemented with galactose (Gal) or glucose (Glc), and the expression levels of Bio2, porin, and the Fe/S protein subunit of succinate dehydrogenase (Sdh2) were analyzed by Western blotting of whole-cell extracts. (D) W303-1A wild-type cells were grown in standard SC medium (containing 0.74 μM FeCl3 [medium iron]), high-iron medium [SC medium plus 300 μM Fe(NH4)2(SO4)2], or low-iron medium (SC medium plus 100 μM of the iron chelator 4,7-diphenyl-1,10-phenanthrolinedisulfonate). In addition, medium contained either the standard amount (2 μg liter−1 [lanes labeled h for high]) or a reduced amount of biotin (0.02 μg liter−1 [lanes labeled l for low]). Whole-cell extracts were analyzed by Western blotting using polyclonal sera directed against Bio2 or the mitochondrial outer membrane protein porin. Poor growth prevented the analysis of cells grown in low-biotin/low-iron media.
Miscellaneous methods.
In vivo labeling of yeast cells with radioactive 55FeCl3 (ICN) and measurement of 55Fe incorporation into Fe/S proteins by precipitation with specific sera and scintillation counting was carried out as described previously (31, 39). Growth assays on agar plates were performed according to published methods (55). Biotinylated proteins were detected in whole-cell extracts using streptavidin-coupled peroxidase (strep-PO) as described previously (46). To remove strep-PO from desthiobiotinylated proteins, Western blots were incubated for 16 h in TBST (Tris-buffered saline with 0.05% Tween 20) buffer containing 2 mM desthiobiotin and 1% bovine serum albumin. The following published methods were used: manipulation of DNA (50), transformation of yeast cells (15), preparation of yeast mitochondria (10), and immunological techniques (17).
RESULTS
S. cerevisiae mutants with defects in the utilization of desthiobiotin.
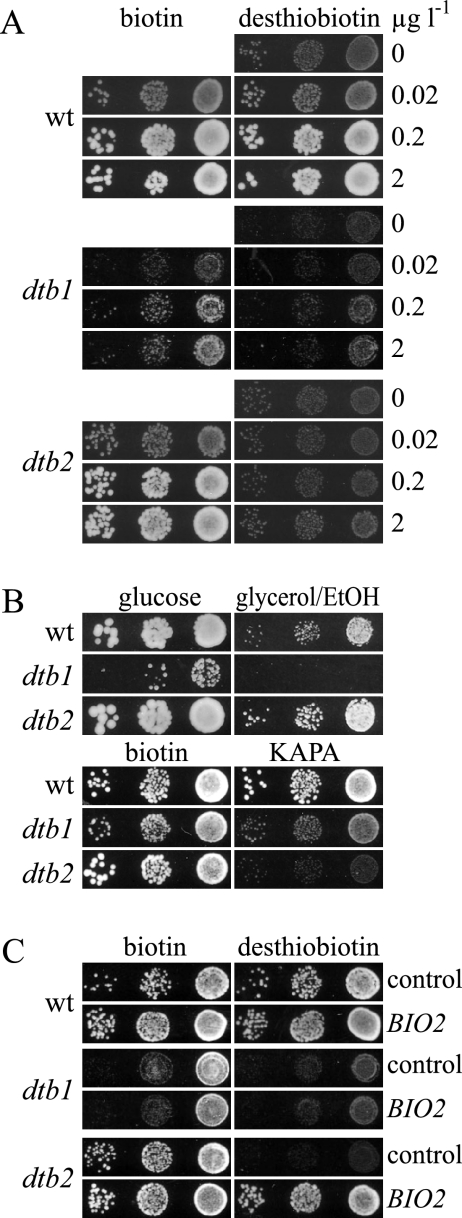
To identify genes that are essential for the utilization of desthiobiotin, we performed a genetic screen in which S. cerevisiae cells were treated with the mutagen ethylmethane sulfonate and replica plated on either biotin- or desthiobiotin-containing plates. Two mutants (dtb1 and dtb2) with specific growth defects on desthiobiotin plates were isolated from the pool of 15,000 surviving cells (Fig. 1A). In addition to growth defects on desthiobiotin, dtb1 cells grew much slower than wild-type cells in the presence of biotin. The dtb1 cells were also unable to use glycerol or ethanol as a carbon source and produced only small colonies on glucose-containing complete medium (Fig. 1B), indicating a defect in respiration (petite phenotype). The dtb2 mutants behaved like wild-type cells and were able to utilize glycerol/ethanol. We also performed growth assays on plates containing the biotin precursor KAPA (Fig. 1B). Whereas wild-type cells were competent to use KAPA, dtb1 mutants showed reduced growth on KAPA and dtb2 mutants were even more affected. All above-described phenotypes of the dtb1 mutants cosegregated in two successive crossings, indicating that they resulted from a single mutation. As expected from the different growth phenotypes, a diploid strain generated by mating of the dtb1 and dtb2 mutants showed full complementation and possessed normal growth on desthiobiotin-containing media (data not shown). Thus, dtb1 and dtb2 define different complementation groups.
FIG. 1.
Phenotypic characterization of the dtb1 and dtb2 biotin biosynthetic mutants. (A) S. cerevisiae wild-type (wt) cells (W303-1A) or the isogenic dtb1 and dtb2 mutants were grown on plates containing 2 μg liter−1 biotin, washed, diluted, and plated on SD plates containing the indicated concentrations of biotin or desthiobiotin. Growth was recorded after 2 days at 30°C. (Β and C) Growth of the same strains used in panel A was assayed on plates containing 0.8 nM (equivalent to 0.2 μg liter−1) biotin, 0.8 nM desthiobiotin, or 0.8 nM KAPA. The plates in panel B contained YP supplemented with either 2% glucose or 3% glycerol and 3% ethanol (EtOH). Growth on YP plates was recorded after 3 days at 30°C, whereas the other plates were scanned after 2 days. In panel C, complementation tests of the dtb1 and dtb2 mutants were performed with the BIO2 plasmid YCplac33-BIO2-GFP or with an empty YCplac33 control.
An obvious candidate for a gene conferring the Dtb phenotype is BIO2, the structural gene encoding biotin synthase. We tested whether the dtb1 or dtb2 cells carried a mutation in BIO2 by transformation with a centromeric plasmid expressing a functional Bio2-GFP fusion protein (see Materials and Methods). Expression of BIO2-GFP did not correct the growth defect of the dtb1 mutant but did correct that of dtb2 cells (Fig. 2C). Crossing of a bio2Δ strain with the dtb2 mutant resulted in a diploid that was unable to utilize desthiobiotin as a biotin source (data not shown). The bio2 allele from dtb2 cells contained a point mutation that substituted Gln189 for a stop codon, truncating the Bio2 protein to about half of its original size. We therefore conclude that the dtb2 cells carry a null mutation in BIO2 (designated bio2-1) and this causes their failure to convert desthiobiotin to biotin.
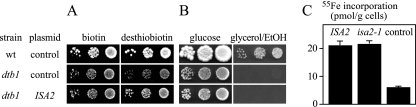
FIG. 2.
Cloning of ISA2 and characterization of Isa2 from the dtb1 mutant. The wild-type (wt) strain, dtb1 mutants overexpressing ISA2 from vector p426-GPD, or dtb1 mutants harboring an empty p426-GPD control plasmid were grown in liquid culture containing 2 μg liter−1 biotin. The cells were washed and plated on SD plates containing 0.2 μg liter−1 biotin or desthiobiotin (A) or YP plates containing either glucose or a mixture of 3% glycerol and 3% ethanol (EtOH) (Β). Growth was recorded after 2 days (A) or 3 days (B) at 30°C. (C) Wild-type yeast cells harboring an empty p426-GPD control plasmid or overproducing Isa2 or Isa2 G91D (encoded by the isa2-1 allele) from the same plasmid were cultivated in iron-poor minimal medium supplemented with glucose. Cells were labeled with 10 μCi 55Fe for 2 h, cell lysates were prepared, and Isa2 was precipitated with a specific antiserum. The amount of radioactivity precipitating with Isa2 was quantified by liquid scintillation counting. A wild-type strain containing the empty plasmid served as a control. Error bars represent the standard deviations.
dtb1 carries a mutation in ISA2.
Next, we addressed the gene defective in the dtb1 mutant. We initially tried, without much success, to complement the growth defect of the dtb1 mutant on glycerol-containing plates (Fig. 1B) by transformation with single-copy or multicopy genomic libraries. When we used the same libraries to complement the desthiobiotin defect of the dtb1 mutant on glucose, however, transformations were immediately successful and produced 30 colonies that were capable of using desthiobiotin as a biotin source. Plasmids isolated from these cells contained almost identical fragments from chromosome XVI. The smallest plasmid contained a truncated version of ROX1 and the genes UBA3, ISA2, HOS1, SPE3, and MED1. Isa2 was previously identified as a mitochondrial protein involved in the formation of Fe/S clusters (3), and biotin synthase carries two Fe/S clusters that are required for its activity (2, 19). Complementation of a dtb1 mutant with a plasmid expressing ISA2 from a strong promoter restored the growth of the dtb1 mutant on desthiobiotin-containing plates (Fig. 2A), indicating that ISA2 was responsible for complementation. Expression of ISA2, however, did not correct the growth defect of the dtb1 mutant back to wild-type levels. ISA2 transformants still displayed growth retardation relative to wild-type cells on biotin-containing minimal (Fig. 2A) or complete medium (Fig. 2B) containing glucose as a carbon source, and they were not able to grow on containing glycerol and ethanol plates (Fig. 2B). It is known that mutation of ISA2 leads to a loss of mitochondrial DNA, a defect that cannot be genetically complemented (26, 44). This secondary defect of isa2 mutants explains our inability to clone the gene defective in dtb1 by complementation of the petite phenotype. A diploid strain generated by crossing the dtb1 mutant with a isa2Δ strain was indistinguishable from its two haploid parents, had growth defects on desthiobiotin, and was unable to grow on glycerol/ethanol plates (data not shown). The ISA2 gene from dtb1 cells carried a point mutation that changed the codon for Gly91 to Asp (G91D). Gly91 is part of a conserved sequence motif present in Isa2. This motif (88-GCHGFQY-94) is called ISAI, and Gly91 within this motif is highly conserved in organisms ranging from bacteria to humans (26, 29, 44). These results indicate that dtb1 carries a mutated ISA2 allele that encodes a nonfunctional protein.
S. cerevisiae contains two homologous ISA genes which are not functionally redundant, as yeasts deleted for either ISA1 or ISA2 show virtually identical phenotypes that are not exacerbated by deletion of both genes (26, 44). We therefore analyzed the growth behavior of an isa1Δ strain and found that, similar to the dtb1 mutant and to the isa2Δ strain, it was unable to utilize desthiobiotin (data not shown). Thus, both Isa proteins appear to be necessary for biotin biosynthesis.
Isa2 from dtb1 mutants is not defective in iron binding.
Several members of the Isa/IscA family, including Isa1 from S. pombe have been shown to bind iron in vitro (40, 62). Recent crystallographic studies have provided evidence that iron binding is likely mediated by the cysteine-containing ISAII motif present at the C termini of all known Isa proteins (3, 9). In S. cerevisiae, mutations of any of the conserved cysteine residues result in loss of function of Isa1 in vivo, suggesting that bound iron is necessary for the function of the Isa proteins (29). We therefore tested whether Isa2 (G91D) showed a reduced affinity for iron. To this end, wild-type yeast cells were transformed with multicopy plasmids that contained either wild-type ISA2 or the isa2-1 allele under control of the strong TDH3 promoter. Cells were grown in iron-poor minimal medium supplemented with glucose and labeled with 55Fe in vivo. Extracts were prepared, and Isa2 was isolated by precipitation with a specific serum, followed by scintillation counting (Fig. 2C). Similar amounts of iron precipitated with wild-type Isa2 and Isa2 carrying the G91D substitution. Iron binding was specific, as much lower levels of radioactivity were precipitated from cells that did not overexpress Isa2. In summary, this experiment demonstrates that the G91D substitution does not affect iron binding of Isa2 and therefore provides no explanation why isa2 mutants are defective in synthesizing biotin from desthiobiotin.
Isa1 and Isa2 are not required for de novo Fe/S cluster assembly on biotin synthase.
IscA and SufA, two bacterial orthologs of the Isa proteins, have been shown to promote Fe/S cluster assembly on bacterial ferredoxins or biotin synthase in vitro (40, 42, 61, 62). We therefore analyzed whether the defect of the dtb1 mutant was the result of a defective maturation of Bio2 in vivo. To this end, we generated two versions of Bio2 in which the ligands of the 4Fe/4S or the 2Fe/2S clusters were replaced. These experiments were facilitated by the recent determination of the structure of E. coli biotin synthase (2), which identified the residues that coordinate the 4Fe/4S and 2Fe/2S clusters, all of which are conserved in the S. cerevisiae protein. To construct Bio2 Δ2Fe/2S, we replaced residue C143 with alanine. The Bio2 Δ4Fe/4S protein resulted from substitution of three closely spaced cysteine residues (C99, C103 and C106) each by serine. In addition, these mutations were combined to create a version of Bio2 lacking both Fe/S cofactors. The corresponding open reading frames were inserted into the yeast vector p426-GPD under the control of the strong constitutive TDH3 promoter and tested for functionality by transformation into a bio2Δ strain (Fig. 3A). Whereas the cells producing the wild-type Bio2 possessed full growth on plates containing desthiobiotin, expression of the Fe/S cluster mutants showed only background levels of growth. This finding was similar to the results of a mutational analysis of the E. coli BioB protein (19) and indicated that the Bio2 proteins harboring cysteine replacements were not functional in the conversion of desthiobiotin to biotin.
For the analysis of Fe/S cluster assembly, these BIO2-containing plasmids were introduced into the conditional yeast strain Gal-ISA2. This strain carries the endogenous ISA2 gene under the control of the GAL1-10 promoter, which is induced upon growth in the presence of galactose and repressed by glucose. The use of this strain was necessary, since yeast mutants with defective mitochondrial DNA, such as isa2Δ or isa2-1, possess pleiotropic phenotypes that frequently interfere with the analysis of Fe/S cluster maturation in vivo (26, 29). Prior to the analysis, Gal-ISA2 cells were grown for 4 days on solid minimal medium supplemented with glucose, and as judged by Western blotting, Isa2 levels were dramatically reduced by this treatment (Fig. 3B). For the analysis of Fe/S cluster formation, these cells were grown in iron-poor medium supplemented with glucose or galactose for 16 h, labeled with 55Fe in vivo, and harvested, and a cell lysate was prepared. Bio2 was isolated by precipitation with a polyclonal serum raised against Bio2, and the amount of 55Fe present in the precipitate was determined. High levels of 55Fe were precipitated from Gal-ISA2 cells overexpressing wild-type Bio2 (Fig. 3B). These were specific, as much lower levels of radioactivity were precipitated in strains producing the Bio2 Δ4Fe/4S Δ2Fe/2S protein. This residual level of radioactivity likely corresponds to wild-type Bio2 produced from the endogenous BIO2 locus. Strikingly, iron binding to wild-type Bio2 was similar in Gal-ISA2 cells grown under permissive (Gal) or repressive (Glc) conditions. In the case of Bio2 Δ4Fe/4S, the net amount of iron incorporation was slightly lower than for the wild-type version and slightly increased in cells depleted for Isa2. Similarly, iron binding of Bio2 Δ2Fe/2S was unchanged upon depletion of Isa2. Since the levels of Bio2 were similar in all extracts analyzed (Fig. 3B), these data strongly suggest that Isa2 is not required for the formation of the Fe/S clusters of Bio2 in vivo.
On the basis of in vitro studies, the Isa proteins have been proposed to function as Fe/S scaffolding proteins that may work in parallel with the Isu proteins, the major Fe/S scaffolding proteins in yeast mitochondria (14, 40, 43, 62). To further characterize the requirements of the maturation of the Fe/S cofactors of Bio2, we carried out in vivo 55Fe labeling experiments of Bio2 with the Gal-ISA1 and Gal-ISU1/Δisu2 strains (14). Under permissive conditions, significant amounts of 55Fe were precipitated by Bio2-specific sera from all cells, regardless of the overexpression of either wild-type Bio2 or the Δ4Fe/4S and Δ2Fe/2S versions (Fig. 4A). The net amount of iron incorporated into wild-type Bio2 remained almost unchanged in Gal-ISA1 cells depleted for Isa1 (Fig. 4A). In the Δ4Fe/4S and Δ2Fe/2S mutants, iron incorporation increased slightly in Gal-ISA1 cells cultivated under repressive conditions, where Isa1 was almost absent (Fig. 4A). Thus, our analyses with both conditional Gal-ISA strains demonstrated that the Isa proteins play no role in de novo Fe/S cluster formation of Bio2. As a further control, we analyzed 55Fe binding to Bio2 in a isa1Δ/isa2Δ double mutant overexpressing the wild-type allele of BIO2. The amount of 55Fe isolated with Bio2 in this strain was comparable to that observed with Gal-ISA1 or Gal-ISA2 cells grown under inducing conditions. An additional observation of the experiments presented in Fig. 3B and Fig. 4A was that the amount of iron bound to the cluster mutants of Bio2 was variable. This fact might result from the different sensitivities of the 2Fe/2S and 4Fe/4S clusters towards contact with air and towards reducing conditions (25), as well as from a possible partial retention of the 2Fe/2S cluster after removal of only one of its cysteine ligands (19). Nevertheless, all forms of Bio2 tested here did not significantly differ in their iron content depending on the presence or absence of the Isa proteins. Thus, our findings demonstrate that neither Isa1 nor Isa2 is involved in the net formation of the Fe/S clusters of yeast biotin synthase in vivo.
FIG. 4.
Fe/S cluster formation on Bio2 requires Isu1 in vivo. Gal-ISA1 and isa1Δ isa2Δ double mutants (A) or Gal-ISU1/Δisu2 cells (B) overproducing wild-type (wt) Bio2 or the Bio2 variants described in the legend to Fig. 3 were incubated in iron-poor minimal medium supplemented with galactose (Gal) or glucose (Glc) in order to induce or repress ISA1 or ISU1. Iron incorporation into Bio2 was analyzed as described in the legend to Fig. 3. Cell lysates were probed for Bio2, Isu1, and Isa1 by Western blotting.
A different result was obtained when we compared iron incorporation into wild-type Bio2, Bio2 Δ4Fe/4S, and Bio2 Δ2Fe/2S in Gal-ISU1/Δisu2 cells (Fig. 4B). We observed that the single Fe/S cluster mutants of Bio2 contained high levels of iron when ISU1 was expressed. Growth in glucose, however, caused a strong reduction in the levels of Isu1 and strongly reduced the incorporation of 55Fe into all forms of Bio2 (Fig. 4B). As judged by Western blotting, the levels of Bio2 were similar in all conditions. Taken together, these data indicate that the net synthesis of both Fe/S clusters of Bio2 requires the Isu proteins but not the Isa proteins in vivo. Moreover, the inability of the isa2-1 mutant to convert desthiobiotin to biotin is not caused by a defect in the de novo assembly of the two Fe/S clusters of Bio2.
Expression of BIO2 is reduced in isa mutants.
The biotin synthesis defect of the isa mutants might be due to a reduced expression of Bio2 to levels that may be too low to support growth in the presence of desthiobiotin. To directly test this hypothesis, we performed Western blotting experiments with isa1Δ and isa2Δ cells. The amount of Bio2 was in fact severely reduced in mitochondria from both isa mutants compared to the wild type (Fig. 5A). In contrast, the abundance of the mitochondrial Fe/S protein aconitase (Aco1) or the Fe/S protein subunit of succinate dehydrogenase (Sdh2) appeared unchanged in both isa mutants, as were the levels of the mitochondrial outer membrane protein porin. This strongly suggests that the low levels of Bio2 in both isa mutants were specific for Bio2 and not generally observed for other Fe/S proteins.
We next transformed wild-type, isa1Δ, and isa2Δ cells with a reporter plasmid in which the promoter of the BIO2 gene was fused to lacZ. This reporter construct allowed us to measure the activity of the BIO2 promoter and to eliminate possible effects of protein or RNA stability that might have influenced the amount of Bio2 detected in Western blots. Figure 5B shows that lacZ activities in the isa1Δ strain amounted to only 30% of the wild-type control, whereas the isa2Δ strain possessed 43% of the wild-type activity. Thus, the reporter assays qualitatively agree with the results of Western blotting (Fig. 5B) and demonstrate that both isa mutants have reduced amounts of Bio2 because of reduced transcription of the BIO2 gene.
The expression of some of the genes of the biotin biosynthetic pathway is influenced by iron availability (1, 48, 51). In yeast, defects in cellular Fe/S cluster biogenesis frequently display a deregulated iron homeostasis, which is one of the factors associated with the induction of iron uptake genes that are under the control of the iron-sensing transcription factor AFT1 and the accumulation of iron within the mitochondria (49). In order to find an explanation for the low abundance of Bio2 in cells deleted for ISA1 or ISA2, we tested whether similar low levels of Bio2 could be observed in other mutants with defects in cellular Fe/S protein assembly. To this end, the conditional yeast mutants Gal-ISU1/Δisu2, Gal-YAH1, and Gal-SSQ1 were grown under inducing and repressing conditions, and the levels of Bio2 were analyzed (Fig. 5C). The abundance of Bio2 strongly decreased in all of these mutants under repressive conditions, whereas the levels of the Fe/S protein subunit of succinate dehydrogenase (Sdh2) or porin were similar in all samples analyzed. Low levels of Bio2 were also observed in a yfh1Δ strain lacking the yeast homolog of frataxin. These data suggest that various defects in the mitochondrial ISC assembly machinery lead to a decreased expression of Bio2. Low levels of Bio2 were also found in Gal-ATM1 cells, which are defective only in the maturation of extramitochondrial Fe/S proteins (Fig. 5C). Together, these observations indicate that the low Bio2 levels are the result of a deregulated gene expression in response to defects in cellular Fe/S protein maturation.
In order to verify that the abundance of Bio2 is directly influenced by the environmental iron concentration in our strain background, wild-type yeast cells were cultivated in low-, intermediate-, and high-iron media, and these treatments were combined with variation in the biotin concentration, which was either high (2 μg liter−1 as in standard SC medium) or low (0.02 μg liter−1). When whole-cell extracts from these cells were analyzed by Western blotting, we found that Bio2 levels were highest in cells from high-iron media and were hardly detectable in cells grown in low iron (Fig. 5D). Moreover, as observed before (46), Bio2 showed an inverse correlation with the biotin content and was more abundant in cells from low-biotin media. The mitochondrial outer membrane protein porin, in contrast, had a similar abundance in all samples, indicating that the changes in protein levels described above are specific for Bio2. Taken together, these results suggest that the drastically reduced cellular levels of Bio2 in ISC mutants may result from the iron-deplete status prevailing in these cells. Functional defects of the Isa proteins and other ISC components thus indirectly affect biotin synthesis from desthiobiotin via low cytosolic iron levels causing decreased expression of the BIO2 gene.
Biotin synthase is not functional in isa mutants.
If the observed low cellular levels of Bio2 alone were responsible for the inability of the isa mutants to utilize desthiobiotin, this phenotype should be cured by constitutive expression of BIO2. In order to test this possibility, a isa1Δ/isa2Δ double mutant that expressed BIO2 from either a high-copy (p426-GPD) or a low-copy vector (p416-MET25) was depleted of biotin for 40 h and either spotted on minimal medium plates supplemented with desthiobiotin (20 μg/liter) or diluted into biotin-free medium in the absence or presence of moderate (20 μg/liter) or high concentrations (100 μg/liter) of desthiobiotin. Neither of the two BIO2 plasmids rescued the growth defect of the isa1Δ/isa2Δ double mutant on media containing only desthiobiotin (Fig. 6A and B). In contrast, the yfh1Δ strain displayed a slow but robust growth in desthiobiotin-containing media, despite having a reduced amount of Bio2 (Fig. 5C) and defects in Fe/S protein maturation (12). This indicates that reduced levels of Bio2 alone are not sufficient to explain the inability of the isa mutants to thrive on desthiobiotin. In addition, wild-type cells, bio2-1, yfh1Δ, and isa1Δ isa2Δ mutants failed to grow in the absence of biotin but recovered in biotin-containing medium, indicating that cells were still viable after cultivation in biotin-free medium for 40 h (Fig. 6B). Taken together, these data strongly suggest that Bio2 is not functional in cells lacking the Isa proteins, despite the fact that iron can be efficiently incorporated into both Fe/S clusters of Bio2.
FIG. 6.
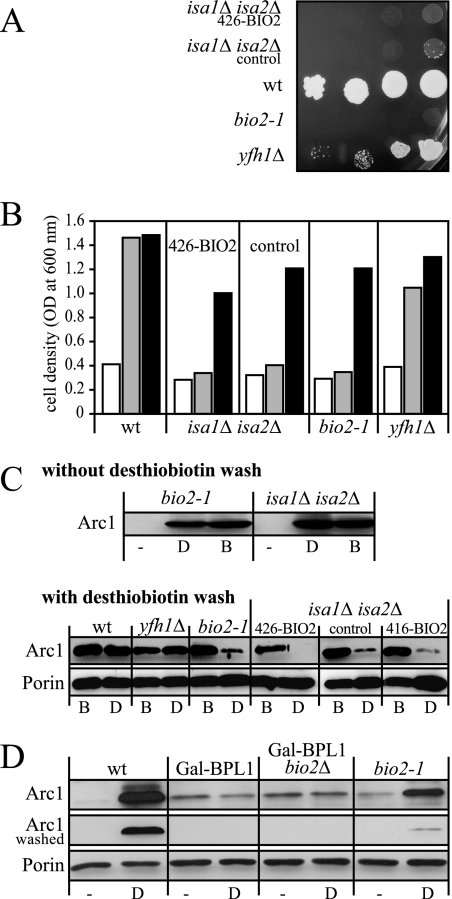
Biotin synthase is inactive in isaΔ cells. S. cerevisiae wild-type (wt) cells, a isa1Δ isa2Δ double mutant expressing BIO2 from the low-copy plasmid p416-MET25 (416-BIO2) or the high-copy vector p426-GPD (426-BIO2), isa1Δ isa2Δ cells carrying the empty p416-MET25 plasmid (control), bio2-1, or yfh1Δ cells were grown for 40 h in liquid minimal SD medium lacking biotin. (A) Serial dilutions of yeast strains were spotted on SD plates containing 20 μg liter−1 desthiobiotin and incubated for 3 days at 30°C. (B) Equal numbers of cells were diluted in biotin-free SD medium without further supplementation (white bars) or supplemented with 20 μg liter−1 desthiobiotin (gray bars) or with 2 μg liter−1 biotin (black bars). Cell growth was recorded by measuring the optical density (OD) at 600 nm after incubation for 24 h at 30°C. (C) The cells grown in biotin (B) or desthiobiotin (D) used in Fig. 6B were harvested, cell lysates were prepared, and Arc1 was detected by streptavidin linked to peroxidase (strep-PO) in Western blots. The blots were developed immediately after decoration with strep-PO (top blot) or after a subsequent incubation with 2 mM desthiobiotin for 16 h to assess whether strep-PO can be removed (bottom blot). Reduced signal intensities after washing indicate that the strep-PO reactivity resulted from a reversible interaction, likely with desthiobiotinylated proteins on the blot. The extracts were also stained for porin, which served as a loading control. (D) The addition of desthiobiotin to Arc1 requires the activity of biotin-protein ligase. The yeast cells indicated above the lanes were grown for 48 h in glucose-containing minimal medium to decrease the expression of BPL1 in strains containing the Gal-BPL1 allele. The media used contained no biotin source. Cells were shifted for 16 h to the same medium (−) or to medium containing 20 μg liter−1 desthiobiotin (D). Protein extracts of the cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Arc1 was detected by strep-PO in Western blots. The blots were developed either immediately (top blot) or after washing with 2 mM desthiobiotin for 16 h to remove reversibly bound strep-PO (middle blot). A stain for porin (bottom blot) served as a loading control.
To further explore the defects of the isa mutants, we isolated cell extracts and detected biotinylated proteins in Western blots using streptavidin coupled to peroxidase. Biotin is covalently attached to six yeast proteins. Of these, the Arc1 protein is a convenient marker of the biotin status because the abundance of biotinyl-Arc1 strongly depends on biotin availability (46). Thus, when desthiobiotin is provided as a biotin source, the amount of biotinyl-Arc1 reflects the biotin biosynthetic activity of the cell and can serve as a marker for Bio2 activity, which is difficult to directly assay in vitro.
As expected, biotin modifications on Arc1 were strongly reduced in cells cultivated without biotin or desthiobiotin (Fig. 6C). In contrast, Arc1 was readily bound by streptavidin when the cells were cultivated in the presence of biotin or desthiobiotin, even when isa1Δ/isa2Δ double mutants or the bio2-1 mutant that did not contain a functional Bio2 protein were used. Since streptavidin can bind to both desthiobiotin and biotin (20), a possible explanation of these surprising results is that yeast biotin ligase attached desthiobiotin instead of biotin to Arc1. To distinguish between biotinylated and desthiobiotinylated Arc1, we made use of the fact that the interaction of desthiobiotin with streptavidin is reversible, whereas its interaction with biotin is stable (20). The streptavidin-decorated blot was washed with desthiobiotin, and the detection procedure was repeated (Fig. 6C). We found that washing with desthiobiotin strongly reduced the signal for Arc1 in extracts from bio2-1 cells grown in the presence of desthiobiotin (Fig. 6C). A similar loss of streptavidin binding to Arc1 was observed in Δisa1/Δisa2 cells cultivated in the presence of desthiobiotin, regardless of whether these cells overexpressed BIO2 or not (Fig. 6C). In contrast, in wild-type and yfh1Δ cells that were able to utilize desthiobiotin, streptavidin could not be removed from Arc1 by washing with desthiobiotin, indicating that these cells contained predominantly biotinylated Arc1.
To further substantiate that the modification of Arc1 causing the reversible binding of streptavidin was desthiobiotin, the dependence of this modification on the activity of biotin-protein ligase, Bpl1, was analyzed. To this end, we used Gal-BPL1 cells in which the activity of BPL1 can be experimentally controlled (46) and additionally deleted BIO2 in these cells to create strain Gal-BPL1/bio2Δ. We then depleted Bpl1 in glucose-containing media in these cells and simultaneously depleted the cells of biotin. In contrast to wild-type and bio2-1 cells, the addition of desthiobiotin to glucose-grown Gal-BPL1 or Gal-BPL1/bio2Δ cells did not result in an increased binding of streptavidin to Arc1 (Fig. 6D). This indicates that the activity of Bpl1 is required for the modification of Arc1, which causes streptavidin binding. The fact that streptavidin binding to Arc1 in bio2 mutants can be reversed by washing with desthiobiotin and that it requires the activity of Bpl1 makes it likely that Arc1 is desthiobiotinylated when a functional form of Bio2 is missing. Thus, wild-type cells that are able to grow on desthiobiotin-containing plates contained a high proportion of biotinylated proteins. In contrast, cells with defects in biotin synthesis, such as bio2 mutants or isa1Δ isa2Δ double mutants, possessed mostly desthiobiotinylated proteins that lack catalytic activity.
In conclusion, our data demonstrate that the failure of the isa mutants to grow in the presence of desthiobiotin is confined to the last step of biotin synthesis, the insertion of sulfur into desthiobiotin, which is catalyzed by biotin synthase. The fact that the Fe/S clusters of Bio2 were assembled at normal rates in the absence of the Isa proteins indicates that the Bio2 holoprotein is not functional when the Isa proteins are absent. Together, these findings strongly suggest that the Isa proteins play a direct role in the catalytic cycle of biotin biosynthesis. They might be involved in regenerating the 2Fe/2S cluster of biotin synthase in vivo. The mechanism of this reaction is at present not clear, but it would minimally require the replacement of a single sulfur atom that is lost from Bio2 for each molecule of biotin formed.
DISCUSSION
We have isolated the ISA2 gene in a genetic screen for S. cerevisiae mutants with defects in the conversion of desthiobiotin to biotin. This sulfur insertion reaction is the last step of biotin biosynthesis and is catalyzed by biotin synthase (Bio2), a Fe/S protein from the family of radical SAM enzymes (11, 34). Isa2 is a member of the mitochondrial Fe/S cluster assembly system and itself is an iron-containing protein (35). The amount of iron bound to the G91D form of Isa2 isolated in the screen was similar to that of the wild-type protein, demonstrating that net iron binding was unaffected by the mutation. To address the function of Isa2 in biotin biosynthesis, we investigated whether the defect of isa2 mutants was caused by a defect in the maturation of the Fe/S clusters of Bio2. Surprisingly, our results demonstrated that neither Isa2 nor the paralogous Isa1 protein was required for the net de novo synthesis of any of the two Fe/S clusters of Bio2 in vivo. Studies carried out in vitro have shown that S. pombe Isa1 and the bacterial IscA or SufA proteins assist Fe/S cluster formation on biotin synthase or 2Fe/2S ferredoxins, in that they may harbor a mobile Fe/S cofactor that can be transferred to apoproteins in vitro (4, 40, 43, 62). Bacterial IscA was found to be transiently associated with biotin synthase during Fe/S cluster transfer from IscA to the apoprotein, suggesting a specific catalytic role in this process (43). On the basis of the results of these studies, it has been suggested that the IscA proteins perform a role as scaffold proteins similar to the IscU proteins. However, our experiments fail to demonstrate that the eukaryotic Isa proteins are required for the net synthesis of the Fe/S clusters of Bio2 in vivo.
Evidence for a role of the Isa proteins in Fe/S cluster biogenesis in vivo was mainly provided by studies carried out with S. cerevisiae cells. The two ISA genes are not required for yeast cell viability, and deletion of either gene is associated with identical, mild phenotypes (26, 29, 44). Although these and similar genetic studies of the iscA gene in E. coli have demonstrated the fundamental importance of this class of proteins in Fe/S cluster formation, their precise function has remained unclear. Our study on biotin synthase shows for the first time that the Isa proteins are not generally required for the de novo synthesis of all cellular Fe/S clusters, despite the fact that the Isa proteins are widely distributed and well conserved. In fact, more-detailed analyses in yeast have shown that they are required only for a small subset of Fe/S proteins in vivo (U. Mühlenhoff, unpublished data). In contrast, all S. cerevisiae Fe/S proteins analyzed so far, including Bio2, require the Isu proteins for net Fe/S cluster assembly (14, 39). The Isu proteins perform a central role as scaffolds for Fe/S cluster assembly in yeast mitochondria. They are essential for viability and show a remarkably high degree of sequence conservation.
Despite the fact that the Isa proteins are not required for the net synthesis of any of the two Fe/S centers of biotin synthase, the failure of the isa mutants to utilize desthiobiotin was confined to the last step of biotin biosynthesis, which is catalyzed by Bio2. This reaction involves SAM, desthiobiotin, a sulfur source, and a reductant (11, 24). The isa mutants are not impaired in desthiobiotin uptake, as was evident from the fact that their biotin-dependent enzymes were desthiobiotinylated in the absence of biotin. In addition, these cells must be capable of producing SAM, as this is an essential metabolite (58). Sulfur is most likely derived from cysteine and transferred via one of the two Fe/S clusters of biotin synthase (59). This step most likely involves the cysteine desulfurase Nfs1. Since NFS1 is essential in S. cerevisiae, we can rule out the possibility that this protein is defective in the isa mutants (31). The physiological reductant used in yeast mitochondria is unknown, but it likely involves ferredoxin (Yah1), which is also essential for viability (33). In conclusion, it seems unlikely that the failure of the isa mutants to produce biotin from desthiobiotin results from a missing substrate rather than from a loss of functional biotin synthase.
The precise catalytic mechanism of biotin synthase is unresolved. There is a general agreement that the 4Fe/4S cluster of the active form of biotin synthase mediates the electron transfer to SAM, while the 2Fe/2S cluster is considered to be the sulfur donor (11, 22, 24, 34, 41, 59). On the basis of experiments in vitro, biotin synthase has been proposed to function as a reactant that is capable of performing only a single sulfur insertion per reaction cycle (11). From the perspective of a living cell, this represents a considerable waste of resources and there is increasing evidence that biotin synthase behaves as an enzyme in vivo which is able to perform multiple rounds of sulfur insertions (6). This requires that the catalytic 2Fe/2S cluster of Bio2 must be regenerated after each round of catalysis. On the basis of our experimental results, we suggest that the Isa proteins may function in this regeneration step. This is an attractive idea, as it explains the phenotype of the isa mutants and is consistent with the observation that the Isa proteins are dispensable for the net synthesis of the Fe/S centers of Bio2. Alternatively, Bio2 may be inhibited by an unknown substance that accumulates in mitochondria when Fe/S cluster maturation is defective. The latter scenario seems unlikely, as cells deleted for the Fe/S assembly protein Yfh1 are capable of synthesizing biotin in vivo. Experimental proof that will allow us to distinguish between these possibilities will require further work with biotin synthase in vitro.
Our experiments showed that isa mutants and other mutants with defects in the mitochondrial Fe/S cluster biogenesis system display reduced levels of Bio2. Although these low expression levels may contribute to a reduced biotin production, they alone are not sufficient to explain the failure of the isa mutants to synthesize biotin. On the one hand, cells lacking the yeast frataxin homolog Yfh1, similar to the isa mutants, displayed low levels of Bio2, but nevertheless, they were able to grow on medium containing desthiobiotin. On the other hand, overexpression of BIO2 from plasmids failed to rescue the growth defects of isa mutants, indicating that even when produced at high levels, biotin synthase is inactive.
In S. cerevisiae, biotin synthesis genes are regulated by iron availability. This iron-regulated expression is under transcriptional control, and BIO2, BIO3, and BIO4 are poorly expressed, while BIO5 and VHT1 are induced by iron limitation (51). The physiological meaning of these observations is not understood. In yeast, defects in cellular Fe/S maturation induce a deregulated iron homeostasis that results in a constitutive induction of iron uptake genes that are under the control of the Aft transcription factors and an accumulation of iron within mitochondria. Proper iron sensing by Aft1 requires the function of the mitochondrial ISC assembly and export systems (49). The constitutive activation of Aft1 in ISC mutants is thus reminiscent of iron-starved cells. The observed low expression of BIO2 in all ISC mutants tested may thus be a physiological consequence of impaired mitochondrial Fe/S cluster assembly and export systems, and the resulting phenotype resembles that of an iron-starved cell. Most likely, the expression of other iron-regulated biotin synthesis genes is also affected. Low transcription levels for BIO2 and elevated levels of BIO5 and VHT1 were found in yeast cells lacking the ABC transporter Atm1 or the mitochondrial ferredoxin Yah1 (A. Hausmann et al., unpublished data), and levels of BIO5 were also increased in cells deleted for the glutaredoxin gene GRX5 (1).
We find that proteins isolated from bio2 mutants contain a modification that interacts with streptavidin in a reversible fashion and requires biotin-protein ligase to be established. Based on these findings, this modification is desthiobiotin, which is transferred to apoproteins by biotin-protein ligase when biotin is unavailable. A similar transfer of desthiobiotin was also reported for the E. coli biotin-protein ligase encoded by birA in vitro; however, this reaction was much less effective than the transfer of biotin, and it required high concentrations of desthiobiotin (63). Transfer of desthiobiotin to apoproteins was not observed in E. coli in vivo (6), demonstrating that the yeast and bacterial enzymes differ in their substrate specificities. This shows that S. cerevisiae cells are unable to efficiently discriminate between biotin and desthiobiotin at the level of protein modification. We have shown for S. pombe and have evidence for S. cerevisiae that biotin and desthiobiotin are acquired via the biotin transporter Vht1 (53; Gerl and Stolz, unpublished). Thus, the activity of Bio2 is essential to prevent desthiobiotin from becoming incorporated into apoproteins, which would lead to the formation of holoproteins that lack enzymatic activity. This role of biotin synthase is further supported by the fact that there are several fungal and bacterial species that contain homologs of BIO2 despite the fact that they lack a complete biotin biosynthetic pathway (7, 47, 53).
In conclusion, this work establishes two surprising connections between the assembly of Fe/S clusters and the synthesis of biotin. (i) Impairment of Fe/S cluster biogenesis reduces the expression of biotin synthase. This appears to result from the iron-limited status that prevails in cells with defects in Fe/S protein maturation. (ii) Biotin synthase present in isa mutants is inactive, indicating that the Isa proteins have a direct role in the catalytic cycle of biotin synthase in vivo. This observation points toward the Isa proteins playing a role in the regeneration of the 2Fe/2S cluster of Bio2. This function will have to be substantiated by future biochemical work, which involves the reconstitution of the biotin synthase reaction cycle with purified components.
Acknowledgments
We thank Sabine Laberer, Roland Donhauser, Petra Reihl, and Stefan Ringlstetter for technical assistance and Claude Alban (Bayer Crop Science, Lyon, France) for the gift of KAPA.
This work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB593, SFB521, STO 434/2-1, and the Gottfried-Wilhelm-Leibniz program), Fonds der Chemischen Industrie, Deutsches Humangenomprojekt, and the Fritz-Thyssen-Stiftung.
Footnotes
Published ahead of print on 26 January 2007.
REFERENCES
- 1.Belli, G., M. M. Molina, J. Garcia-Martinez, J. E. Perez-Ortin, and E. Herrero. 2004. Saccharomyces cerevisiae glutaredoxin 5-deficient cells subjected to continuous oxidizing conditions are affected in the expression of specific sets of genes. J. Biol. Chem. 279:12386-12395. [DOI] [PubMed] [Google Scholar]
- 2.Berkovitch, F., Y. Nicolet, J. T. Wan, J. T. Jarrett, and C. L. Drennan. 2004. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303:76-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilder, P. W., H. Ding, and M. E. Newcomer. 2004. Crystal structure of the ancient, Fe-S scaffold IscA reveals a novel protein fold. Biochemistry 43:133-139. [DOI] [PubMed] [Google Scholar]
- 4.Bonomi, F., S. Iametti, D. Ta, and L. E. Vickery. 2005. Multiple turnover transfer of [2Fe2S] clusters by the iron-sulfur cluster assembly scaffold proteins IscU and IscA. J. Biol. Chem. 280:29513-29518. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Choi-Rhee, E., and J. E. Cronan. 2005. Biotin synthase is catalytic in vivo, but catalysis engenders destruction of the protein. Chem. Biol. 12:461-468. [DOI] [PubMed] [Google Scholar]
- 7.Cliften, P. F., L. W. Hillier, L. Fulton, T. Graves, T. Miner, W. R. Gish, R. H. Waterston, and M. Johnston. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11:1175-1186. [DOI] [PubMed] [Google Scholar]
- 8.Cronan, J. E., Jr., and J. C. Wallace. 1995. The gene encoding the biotin-apoprotein ligase of Saccharomyces cerevisiae. FEMS Microbiol. Lett. 130:221-229. [DOI] [PubMed] [Google Scholar]
- 9.Cupp-Vickery, J. R., J. J. Silberg, D. T. Ta, and L. E. Vickery. 2004. Crystal structure of IscA, an iron-sulfur cluster assembly protein from Escherichia coli. J. Mol. Biol. 338:127-137. [DOI] [PubMed] [Google Scholar]
- 10.Diekert, K., A. I. de Kroon, G. Kispal, and R. Lill. 2001. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65:37-51. [DOI] [PubMed] [Google Scholar]
- 11.Fontecave, M., S. Ollagnier-de-Choudens, and E. Mulliez. 2003. Biological radical sulfur insertion reactions. Chem. Rev. 103:2149-2166. [DOI] [PubMed] [Google Scholar]
- 12.Foury, F. 1999. Low iron concentration and aconitase deficiency in a yeast frataxin homologue deficient strain. FEBS Lett. 456:281-284. [DOI] [PubMed] [Google Scholar]
- 13.Funk, M., R. Niedenthal, D. Mumberg, K. Brinkmann, V. Ronicke, and T. Henkel. 2002. Vector systems for heterologous expression of proteins in Saccharomyces cerevisiae. Methods Enzymol. 350:248-257. [DOI] [PubMed] [Google Scholar]
- 14.Gerber, J., K. Neumann, C. Prohl, U. Mühlenhoff, and R. Lill. 2004. The yeast scaffold proteins Isu1 and Isu2 are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol. Cell. Biol. 24:4848-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Halbrook, J., and M. F. Hoekstra. 1994. Mutations in the Saccharomyces cerevisiae CDC1 gene affect double-strand-break-induced intrachromosomal recombination. Mol. Cell. Biol. 14:8037-8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Hasslacher, M., A. S. Ivessa, F. Paltauf, and S. D. Kohlwein. 1993. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 268:10946-10952. [PubMed] [Google Scholar]
- 19.Hewitson, K. S., S. Ollagnier-de Choudens, Y. Sanakis, N. M. Shaw, J. E. Baldwin, E. Munck, P. L. Roach, and M. Fontecave. 2002. The iron-sulfur center of biotin synthase: site-directed mutants. J. Biol. Inorg. Chem. 7:83-93. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch, J. D., L. Eslamizar, B. J. Filanoski, N. Malekzadeh, R. P. Haugland, and J. M. Beechem. 2002. Easily reversible desthiobiotin binding to streptavidin, avidin, and other biotin-binding proteins: uses for protein labeling, detection, and isolation. Anal. Biochem. 308:343-357. [DOI] [PubMed] [Google Scholar]
- 21.Hoja, U., S. Marthol, J. Hofmann, S. Stegner, R. Schulz, S. Meier, E. Greiner, and E. Schweizer. 2004. HFA1 encoding an organelle-specific acetyl-CoA carboxylase controls mitochondrial fatty acid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 279:21779-21786. [DOI] [PubMed] [Google Scholar]
- 22.Jameson, G. N., M. M. Cosper, H. L. Hernandez, M. K. Johnson, and B. H. Huynh. 2004. Role of the [2Fe-2S] cluster in recombinant Escherichia coli biotin synthase. Biochemistry 43:2022-2031. [DOI] [PubMed] [Google Scholar]
- 23.Janke, C., M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947-962. [DOI] [PubMed] [Google Scholar]
- 24.Jarrett, J. T. 2005. Biotin synthase: enzyme or reactant? Chem. Biol. 12:409-410. [DOI] [PubMed] [Google Scholar]
- 25.Jarrett, J. T. 2005. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch. Biochem. Biophys. 433:312-321. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, L. T., and V. C. Culotta. 2000. Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol. 20:3918-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Kaut, A., H. Lange, K. Diekert, G. Kispal, and R. Lill. 2000. Isa1 is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem. 275:15955-15961. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H. S., U. Hoja, J. Stolz, G. Sauer, and E. Schweizer. 2004. Identification of the tRNA-binding protein Arc1 as a novel target of in vivo biotinylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:42445-42452. [DOI] [PubMed] [Google Scholar]
- 31.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1 and Nfs1 are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koser, S. A. 1968. Vitamin requirements of bacteria and yeasts. Charles C Thomas, Springfield, IL.
- 33.Lange, H., A. Kaut, G. Kispal, and R. Lill. 2000. A mitochondrial ferredoxin is essential for biogenesis of cellular iron-sulfur proteins. Proc. Natl. Acad. Sci. USA 97:1050-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Layer, G., D. W. Heinz, D. Jahn, and W. D. Schubert. 2004. Structure and function of radical SAM enzymes. Curr. Opin. Chem. Biol. 8:468-476. [DOI] [PubMed] [Google Scholar]
- 35.Lill, R., and U. Mühlenhoff. 2005. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 30:133-141. [DOI] [PubMed] [Google Scholar]
- 36.Lim, F., M. Rohde, C. P. Morris, and J. C. Wallace. 1987. Pyruvate carboxylase in the yeast pyc mutant. Arch. Biochem. Biophys. 258:259-264. [DOI] [PubMed] [Google Scholar]
- 37.Malinska, K., J. Malinsky, M. Opekarova, and W. Tanner. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marobbio, C. M., G. Agrimi, F. M. Lasorsa, and F. Palmieri. 2003. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mühlenhoff, U., J. Gerber, N. Richhardt, and R. Lill. 2003. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1. EMBO J. 22:4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ollagnier-de Choudens, S., L. Nachin, Y. Sanakis, L. Loiseau, F. Barras, and M. Fontecave. 2003. SufA from Erwinia chrysanthemi. Characterization of a scaffold protein required for iron-sulfur cluster assembly. J. Biol. Chem. 278:17993-18001. [DOI] [PubMed] [Google Scholar]
- 41.Ollagnier-de Choudens, S., Y. Sanakis, K. S. Hewitson, P. Roach, E. Munck, and M. Fontecave. 2002. Reductive cleavage of S-adenosylmethionine by biotin synthase from Escherichia coli. J. Biol. Chem. 277:13449-13454. [DOI] [PubMed] [Google Scholar]
- 42.Ollagnier-de-Choudens, S., T. Mattioli, Y. Takahashi, and M. Fontecave. 2001. Iron-sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J. Biol. Chem. 276:22604-22607. [DOI] [PubMed] [Google Scholar]
- 43.Ollagnier-de-Choudens, S., Y. Sanakis, and M. Fontecave. 2004. SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg. Chem. 9:828-838. [DOI] [PubMed] [Google Scholar]
- 44.Pelzer, W., U. Mühlenhoff, K. Diekert, K. Siegmund, G. Kispal, and R. Lill. 2000. Mitochondrial Isa2 plays a crucial role in the maturation of cellular iron-sulfur proteins. FEBS Lett. 476:134-139. [DOI] [PubMed] [Google Scholar]
- 45.Phalip, V., I. Kuhn, Y. Lemoine, and J. M. Jeltsch. 1999. Characterization of the biotin biosynthesis pathway in Saccharomyces cerevisiae and evidence for a cluster containing BIO5, a novel gene involved in vitamer uptake. Gene 232:43-51. [DOI] [PubMed] [Google Scholar]
- 46.Pirner, H. M., and J. Stolz. 2006. Biotin sensing in Saccharomyces cerevisiae is mediated by a conserved DNA element and requires the activity of biotin-protein ligase. J. Biol. Chem. 281:12381-12389. [DOI] [PubMed] [Google Scholar]
- 47.Rodionov, D. A., and M. S. Gelfand. 2006. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol. Lett. 255:102-107. [DOI] [PubMed] [Google Scholar]
- 48.Rutherford, J. C., S. Jaron, and D. R. Winge. 2003. Aft1 and Aft2 mediate iron-responsive gene expression in yeast through related promoter elements. J. Biol. Chem. 278:27636-27643. [DOI] [PubMed] [Google Scholar]
- 49.Rutherford, J. C., L. Ojeda, J. Balk, U. Mühlenhoff, R. Lill, and D. R. Winge. 2005. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 280:10135-10140. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Shakoury-Elizeh, M., J. Tiedeman, J. Rashford, T. Ferea, J. Demeter, E. Garcia, R. Rolfes, P. O. Brown, D. Botstein, and C. C. Philpott. 2004. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell 15:1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 53.Stolz, J. 2003. Isolation and characterization of the plasma membrane biotin transporter from Schizosaccharomyces pombe. Yeast 20:221-231. [DOI] [PubMed] [Google Scholar]
- 54.Stolz, J., U. Hoja, S. Meier, N. Sauer, and E. Schweizer. 1999. Identification of the plasma membrane H+-biotin symporter of Saccharomyces cerevisiae by rescue of a fatty acid-auxotrophic mutant. J. Biol. Chem. 274:18741-18746. [DOI] [PubMed] [Google Scholar]
- 55.Stolz, J., and M. Vielreicher. 2003. Tpn1, the plasma membrane vitamin B6 transporter of Saccharomyces cerevisiae. J. Biol. Chem. 278:18990-18996. [DOI] [PubMed] [Google Scholar]
- 56.Stucka, R., S. Dequin, J. M. Salmon, and C. Gancedo. 1991. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol. Gen. Genet. 229:307-315. [DOI] [PubMed] [Google Scholar]
- 57.Sumrada, R. A., and T. G. Cooper. 1982. Urea carboxylase and allophanate hydrolase are components of a multifunctional protein in yeast. J. Biol. Chem. 257:9119-9127. [PubMed] [Google Scholar]
- 58.Thomas, D., A. Becker, and Y. Surdin-Kerjan. 2000. Reverse methionine biosynthesis from S-adenosylmethionine in eukaryotic cells. J. Biol. Chem. 275:40718-40724. [DOI] [PubMed] [Google Scholar]
- 59.Tse Sum Bui, B., T. A. Mattioli, D. Florentin, G. Bolbach, and A. Marquet. 2006. Escherichia coli biotin synthase produces selenobiotin. Further evidence of the involvement of the [2Fe-2S]2+ cluster in the sulfur insertion step. Biochemistry 45:3824-3834. [DOI] [PubMed] [Google Scholar]
- 60.Walker, M. E., D. L. Val, M. Rohde, R. J. Devenish, and J. C. Wallace. 1991. Yeast pyruvate carboxylase: identification of two genes encoding isoenzymes. Biochem. Biophys. Res. Commun. 176:1210-1217. [DOI] [PubMed] [Google Scholar]
- 61.Wollenberg, M., C. Berndt, E. Bill, J. D. Schwenn, and A. Seidler. 2003. A dimer of the FeS cluster biosynthesis protein IscA from cyanobacteria binds a [2Fe2S] cluster between two protomers and transfers it to [2Fe2S] and [4Fe4S] apo proteins. Eur. J. Biochem. 270:1662-1671. [DOI] [PubMed] [Google Scholar]
- 62.Wu, G., S. S. Mansy, C. Hemann, R. Hille, K. K. Surerus, and J. A. Cowan. 2002. Iron-sulfur cluster biosynthesis: characterization of Schizosaccharomyces pombe Isa1. J. Biol. Inorg. Chem. 7:526-532. [DOI] [PubMed] [Google Scholar]
- 63.Wu, S. C., and S. L. Wong. 2004. Development of an enzymatic method for site-specific incorporation of desthiobiotin to recombinant proteins in vitro. Anal. Biochem. 331:340-348. [DOI] [PubMed] [Google Scholar]