Abstract
Glycylation is an uncommon posttranslational modification. It has been found that tubulin glycylation is essential for cell survival in Tetrahymena. Here we describe PGP1, a Tetrahymena gene encoding an Hsp70 homologue that is a novel glycylated protein. Pgp1p is a conserved glycoprotein that localizes within the lumen of the endoplasmic reticulum (ER). We demonstrate that PGP1 is essential for viability and present evidence that both glycosylation and ER retention are necessary but not sufficient for glycylation.
Glycylation and glutamylation are two unusual posttranslational modifications originally identified on tubulins. The two modifications are formally similar to each other, involving the addition of glycyl or glutamyl units to the γ-carboxyl groups of specific glutamate residues in the tubulin C-terminal tails (9, 28, 34). Glutamylation has also been found on nucleosome assembly proteins NAP1 and NAP2 (29). While this work was in progress, glycylation was reported to occur on a protein (g14-3-3) in Giardia duodenalis (21).
The exact function of glycylation is not clear. Mutagenic analyses indicate that tubulin glycylation is essential for cell survival in Tetrahymena thermophila and plays an important role in axonemal organization and in other cellular functions, such as microtubule severing during cytokinesis (33-35). Little is known about the enzymes that carry out glycylation. Glycylation, glutamylation, and tyrosination all involve the addition of an amino acid to a glutamate residue through the formation of an amide bond. Recent studies have shown that enzymes that glutamylate tubulins are encoded by tubulin tyrosine ligase-like (TTLL) genes (18) related to the gene for tubulin tyrosine ligase (TTL), an enzyme that tyrosinates tubulin at its C terminus (11). TTLL genes constitute a large superfamily of conserved eukaryotic proteins with a TTL homology domain. Humans have 14 predicted TTLL genes, Drosophila has 11, and Tetrahymena has an extraordinary 50. In addition, the TTLLs of diverse eukaryotes belong to several conserved subtypes, one or more of which may glycylate tubulins (18).
The diversity of TTLL genes, coupled with the finding that g14-3-3 is glycylated, raises the possibility that glycylation might be a more general protein posttranslational modification. Here we demonstrate that glycylation occurs on Pgp1p, a conserved, glycosylated, Hsp70-related, essential endoplasmic reticulum (ER) resident protein, and that the C-terminal ER retention signal KQTDL of Pgp1p is required for glycylation.
MATERIALS AND METHODS
Strains and culture conditions.
The strains used in this study are listed in Table 1. Tetrahymena cells were grown in 1× SPP medium (1% proteose peptone, 0.2% glucose, 0.1% yeast extract, 0.003% EDTA-ferric sodium salt) at 30°C (14). For conjugation, log-phase cells of different mating types were starved in 10 mM Tris (pH 7.5) for 16 to 24 h at 30°C and then mixed at a concentration of 2 × 105 cells per ml. Wild-type CU428 cells were heat shocked by being transferred from 30°C to 41°C for 0.5 and 1.0 h. Heat-shocked cells were then harvested for total RNA preparation. RNAs were also isolated from wild-type log-phase cells treated for 6 h in 1× SPP medium containing tunicamycin (TUN; Sigma) (0.25 and 0.5 μg/ml) or thapsigargin (THSP; Sigma) (1.25 and 2.5 μM).
TABLE 1.
Strains used in this studyc
| Strain | Mic genotype | Mac genotype | Mac phenotypea |
|---|---|---|---|
| B2086b | WT | WT | II |
| CU428b | mpr1-1/mpr1-1 | WT | mp-s, VIId |
| PGP1KO1 | pgp1::neo3/pgp1::neo3 | WT | pm-s |
| PGP1KO4 | pgp1::neo3/pgp1::neo3 | WT | pm-s |
| PGP1-HA | pgp1::neo3/pgp1::neo3 | PGP1-HA/pgp1::neo3 | pm-r |
| PGP1-MHA | pgp1::neo3/pgp1::neo3 | MTT1-PGP1-HA/pgp1::neo3 | pm-r |
| PGP1-MH2 | pgp1::neo3/pgp1::neo3 | MTT1-PGP1-ΔMC-HA/pgp1::neo3 | pm-r |
| PGP1-HA-ΔKQTDL | pgp1::neo3/pgp1::neo3 | PGP1-HA-ΔKQTDL/pgp1::neo3 | pm-r |
Including drug resistance and mating type.
Provided by P. Bruns (Cornell University, Ithaca, NY).
WT, wild type.
mp-s, 6-methylpurine sensitive.
Protein immunoprecipitation.
Cells (5 × 106) were centrifuged at 383 × g for 2 min at room temperature, and the supernatant was discarded. To cells and residual medium in approximately 1 ml total volume, 1 ml 2× NP-40 lysis buffer (300 mM NaCl, 2% NP-40, 10 mM Tris, pH 7.5) with protease inhibitors (Complete protease inhibitor cocktail; Roche) was added and gently mixed. Cells were left on ice for 30 min, with occasional mixing, and then pelleted for 10 min at 10,000 × g (4°C). One milliliter of lysate, containing soluble proteins, was transferred to a fresh 1.5-ml microcentrifuge tube and was precleared by the addition of 50 μl of normal rabbit serum and incubated on ice for 1 h. Fifty microliters of protein A-Sepharose beads (Pharmacia) was then added and incubated on ice for 30 min (4°C) with gentle mixing. The beads were removed from the lysate by centrifugation at 10,000 × g for 15 min (4°C).
To 1 ml of the precleared lysate, 5 μl of a rabbit antipolyglycine polyclonal antibody (anti-polyG; originally named R-polygly) raised against a Cys-(Gly)9 peptide (7) or an anti-hemagglutinin (anti-HA) antibody (16B12; Zymed) was added and incubated on ice for 1 h. Next, 50 μl of protein A-Sepharose beads was added, and the lysate was incubated for 1 h at 4°C. The beads were collected by centrifugation at 10,000 × g for 15 s and then washed three to five times with lysis buffer. To the washed beads, 50 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (10% β-mercaptoethanol, 20% glycerol, 4% SDS, 120 mM Tris-HCl, pH 6.8) was added. The mixture was heated to 85°C for 10 min, and the beads were removed by centrifugation at 13,000 × g. The supernatant was then analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
N-terminal protein and peptide sequencing.
Immunoprecipitated proteins separated by 10% SDS-PAGE were electroblotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp.) and stained with Coomassie blue. A band at approximately 135 kDa was excised from the membrane, and N-terminal protein and peptide sequencing was performed at the Baylor College of Medicine Protein Chemistry Core Facility (Houston, TX).
Isolation of PGP1.
The N-terminal sequence of the purified protein (Pgp1p) exactly matched residues 22 to 40 of a predicted protein encoded by a cDNA from a Tetrahymena expressed sequence tag cDNA library (12). The full-length sequence of PGP1 was obtained by 5′ and 3′ rapid amplification of cDNA ends (36). The predicted Pgp1p sequence is almost the same as the protein sequence predicted for open reading frame (ORF) 56.m00217 in the Tetrahymena genome database (http://db.ciliate.org/cgi-bin/locus.pl?locus=56.m00217), but there are differences in their C-terminal sequences. No ER retention signal (KQTDL) was found in the C terminus of ORF 56.m00217.
The primers used were as follows: NotI-Adapter, GAGGCGGCCGCAAATGAACACTGCGTTTGCGGG; NotI-Adapter-PCR, CCGCAAATGAACACTGCGTTTGC; T7-Asc-d(T), GTAATACGACTCACTATAGGGCGAATGGCGCGCCTTTTTTTTTTTTTTTTTTTT; PGP1 P7F, AGTAGAATTCGGTGGTATTAGAGTTCCAAAAATCC; PGP1 P8R, TGAACCGCGGATTCTTCCATTCTTTTTTGAATAGGC; and oligo(dT)17, TTTTTTTTTTTTTTTTT.
Germ line knockouts.
To make the PGP1 knockout construct, the neo3 cassette, which confers paromomycin resistance (pm-r) (30), was inserted between a 0.75-kb fragment of the PGP1 5′-flanking region and a 1.1-kb fragment of the PGP1 3′-flanking sequence by overlapping PCR to produce a 3.8-kb fragment (see Fig. 3A). CU428 and B2086 cells were mated and transformed with 3 μg of the 3.8-kb PCR fragment and 2.5 μg of carrier EcoRI-linearized pBlueScript SK(+) plasmid DNA, using the Dupont Biolistic PDS-1000/He particle delivery system (Bio-Rad), at 2.5 to 4 h postmixing (3). The pgp1::neo3 knockout transformants were selected with 90 μg/ml paromomycin sulfate (true potency; Sigma Chemical Co.). Using standard Tetrahymena genetic procedures (15), two homozygous knockout heterokaryon strains, PGP1KO1 and PGP1KO4, were obtained. These cell lines are homozygous for the PGP1 knockout in the germ line micronucleus and homozygous for wild-type PGP1 in the macronucleus and can mate with each other.
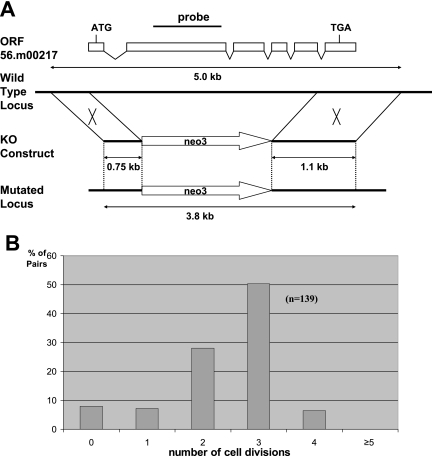
FIG. 3.
PGP1 is essential for vegetative growth. (A) Schematic drawing of the PGP1 locus and the knockout (KO) construct used to disrupt it. A pm-r marker (neo3) was inserted into the PGP1 gene, replacing most of the coding sequence. The knockout construct was introduced into the PGP1 locus by homologous recombination. The position of the probe used for hybridization is indicated. (B) Most PGP1 knockout cells died after two or three divisions. Two germ line homozygous PGP1 knockout heterokaryon strains (the macronucleus is wild type, and both copies of the PGP1 gene in the micronucleus are disrupted) were mated, and single pairs were isolated in drops of 1× SPP medium. The cells in each drop were counted before they died. The results were categorized as follows: 2 cells, no cell divisions; 3 or 4 cells, one division; 5 to 8 cells, two divisions; 9 to 16 cells, three divisions; 17 to 32 cells, four divisions; and more than 32 cells, more than five divisions.
Northern blot analysis.
Tetrahymena total RNA was prepared from mid-log-phase, starved, conjugating, heat-shocked, TUN-treated or THSP-treated cells by using Trizol (Invitrogen) according to the manufacturer's instructions. Thirty to 40 μg RNA was separated in 2.2 M formaldehyde-1.2% agarose gels, blotted, and hybridized (1). Hybridizations were done at 42°C in 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× SPED (0.1% Ficoll, 0.1% bovine serum albumin, 0.1% polyvinylpyrrolidone, 6 mM SDS, 2 mM sodium pyrophosphate, 2 mM EDTA), 1% SDS, and 100 μg/ml salmon sperm DNA.
The PGP1 probe was a PCR product amplified from Tetrahymena genomic DNA, using primers PGP1 P9 (TTCTCATCCTCTCTATTGAAGAAG) and PGP1 P10 (TCCGAGAGCAGTTGATTCATCTC). See Fig. 3A for the locations of the primers. As a control, a probe for the ribosomal RPL21 gene was PCR amplified from Tetrahymena genomic DNA, using primers L21 FW (AAGTTGGTTATCAACTGTTGCGTT) and L21 RV (CCCAGAAAGTTCCTGCTGCAT). Northern blots were stripped by boiling in 0.1% SDS for 10 to 15 min. All probes were radiolabeled by random priming. Northern blot data were quantified using Lab Work 4.0 image analysis software (UVP, Inc., Upland, CA).
Tagged and rescue constructs.
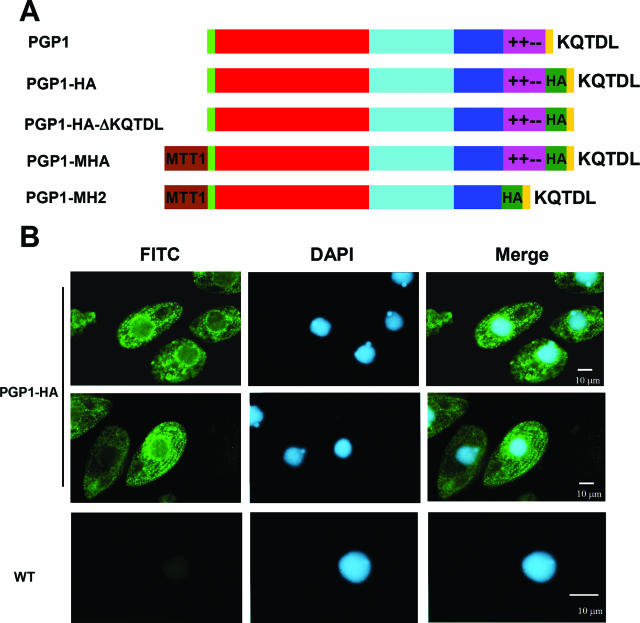
Wild-type, tagged, and mutated PGP1 constructs (Table 1; see Fig. 4A) were used to rescue mating heterokaryon strains PGP1KO1 and PGP1KO4 by biolistic transformation 24 h after they were mixed. All constructs were inserted into the endogenous PGP1 locus by homologous integration as diagrammed in Fig. 3A. The rescued progeny were selected with 90 μg/ml paromomycin in the presence of 1.0 μg/ml CdCl2, which induces the MTT1 promoter. To make the PGP1-HA construct, the HA epitope sequence (YPYDVPDYA) was inserted into the C-terminal region of Pgp1p before the sequence DGNQKQTDL. PGP1-MHA is a construct with the cadmium-inducible MTT1 promoter (30) inserted just before the start codon of an HA-tagged PGP1 gene. The PGP1-MH2 construct has the MTT1 promoter, with the mixed-charge region from K821 to K867 of Pgp1p deleted and the HA motif inserted into the C-terminal region. PGP1-HA-ΔKQTDL is the same as the PGP1-HA construct, but with the ER retention signal sequence KQTDL deleted.
FIG. 4.
Localization of HA-tagged Pgp1p. (A) Construction of HA-tagged and mutated PGP1 constructs. All constructs were used to rescue the PGP1 homozygous knockout heterokaryon progeny. Colored regions are as follows: green, signal peptide; red, conserved ATP-binding domain; blue, a region of similarity found in the C terminus among members of the Grp170 family; purple, the mixed-charge region; and dark green, the HA tag. PGP1-MHA and PGP1-MH2 expression is under the control of the MTT1 promoter (brown). (B) Immunofluorescence localization of HA-tagged PGP1 in Tetrahymena. Log-phase PGP1-HA and CU428 wild-type cells were fixed and processed for indirect immunofluorescence staining. The PGP1-HAp construct was localized by anti-HA antibody, followed by secondary labeling with fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G antibodies; DNA was stained with DAPI.
Indirect immunofluorescence staining.
Cells were processed for immunofluorescence labeling as described previously (33), with slight modifications. Several hundred cells in 30 μl of 10 mM Tris, pH 7.5, were applied to poly-l-lysine-coated coverslips. An equal volume of 0.5% Triton X-100 in PHEM buffer [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 25 mM HEPES, 10 mM EGTA, 2 mM magnesium chloride] at pH 6.9 was added. After 2 min, 30 μl of 2% paraformaldehyde in PHEM buffer was added. The fixed cells were air dried, treated with blocking solution (3% bovine serum albumin, 10% normal goat serum, and 0.1% Tween 20 in phosphate-buffered saline [PBS]) for 1 h at room temperature, and incubated for 1 h at room temperature (or overnight at 4°C) in blocking solution with anti-HA primary antibody (16B12; Zymed) at a 1:100 dilution. The coverslips were washed three times for 5 min each with PBS containing 0.1% Tween 20 and then were incubated for 1 h at room temperature with goat anti-mouse-fluorescein isothiocyanate secondary antibody (Sigma) at a 1:500 dilution in blocking solution. After two washes in PBS with 0.1% Tween 20 and a third wash in the same buffer containing 100 ng/ml DAPI (4′,6′-diamidino-2-phenylindole) to stain DNA, the coverslips were mounted with 8 μl of antibleaching solution containing DABCO (1,4-diazobicyclo-[2,2,2]-octane; Sigma) dissolved at 100 mg/ml in 90% glycerol in PBS.
Protein electrophoresis and immunoblotting.
Cells (1 × 106) were centrifuged and resuspended in 1 ml of 10 mM Tris, pH 7.5. The cells were then centrifuged, and the pellets were mixed with 150 μl of 2× SDS sample buffer containing 125 mM Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, and 10% β-mercaptoethanol and boiled for 5 to 10 min. Total protein from 3 × 104 cells in each lane was separated by 7.5% SDS-PAGE. Gels were electroblotted onto Immobilon-P polyvinylidene difluoride membranes by using a semidry transfer unit (Bio-Rad Laboratories). The blots were blocked in 1× TBST buffer (100 mM Tris-HCl, pH 7.5, 0.9% NaCl, 0.1% Tween 20) containing 5% dried milk for 1 h at room temperature and incubated overnight at 4°C with monoclonal anti-HA antibody or rabbit anti-polyG at a 1:10,000 dilution. Anti-polyG is a polyclonal antibody directed against a Cys-(Gly)9 peptide. It is highly sensitive and specific for posttranslationally added glycyl residues, as indicated by the fact that it reacts with Tetrahymena α-tubulin when five of six glycylatable glutamates have been mutated to aspartates but not when all six sites have been mutated (J. Duan, J. Bowen, and M. A. Gorovsky, unpublished observations). Membranes were washed in TBST and incubated with a 1:10,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit or rabbit anti-mouse immunoglobulin G (Zymed Labs Inc., South San Francisco, CA) in blocking solution for 1 h at room temperature. Blots were processed using a Western blot chemiluminescence reagent (Perkin-Elmer) and exposed to Kodak X-ray films.
Nucleotide sequence accession number.
The cDNA sequence of PGP1 can be found in GenBank under accession number DQ418790.
RESULTS
Characterization of PGP1.
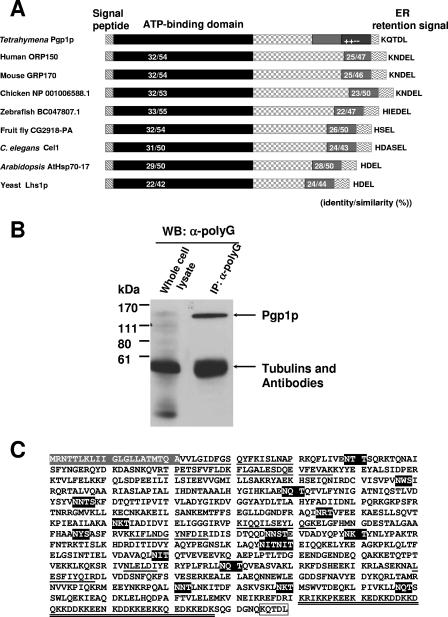
In the course of studies designed to demonstrate the posttranslational glycylation of tubulins in Tetrahymena, we labeled Tetrahymena cells with [3H]glycine in the presence of the protein synthesis inhibitor cycloheximide and analyzed the total protein by fluorography (see Fig. S1 in the supplemental material). In addition to the heavily labeled bands corresponding to the tubulins, a heavily labeled band with an apparent molecular mass of ∼135 kDa was observed, suggesting that another protein was also posttranslationally glycylated. Subsequent Western blot analysis of whole-cell extracts of Tetrahymena thermophila with anti-polyG revealed several weak bands of 65 to 140 kDa, a very strong band of ∼55 kDa (the molecular mass of α- and β-tubulins), and weaker bands that were smaller than 55 kDa (Fig. 1B; also see Fig. 5A). To identify the putatively glycylated nontubulin components, the whole-cell extract was immunoprecipitated with anti-polyG antibody, and the immunoprecipitate was analyzed by Western blotting against anti-polyG. Only the enriched ∼135-kDa band and tubulin bands were clearly observed, indicating that the ∼135-kDa protein was efficiently recognized by anti-polyG antibody (Fig. 1B). The ∼135-kDa protein, named possible glycylated protein 1 (Pgp1p), was recovered from the gel and analyzed by N-terminal protein and peptide sequencing (see Materials and Methods).
FIG. 1.
Characterization of PGP1. (A) Comparison of various members of the GRP170 subfamily of Hsp70-like proteins. The ATP-binding domain is conserved, and a region of similarity is found in the C terminus. All members contain N-terminal signal peptides and C-terminal KDEL-like sequences. Other regions show no significant similarity. (B) Western blot of immunoprecipitates of Pgp1p with the anti-polyG antibody, analyzed by 10% SDS-PAGE and probed with anti-polyG antibody. The ∼135-kDa band was excised from the membrane for protein and peptide sequencing. (C) Predicted Pgp1p amino acid sequence. The gray amino acids at the top correspond to the N-terminal signal peptide. The underlined amino acid sequences are identical to the N-terminal and peptide amino acid sequences of purified Pgp1p. The potential N-linked glycosylation sites are shown in dark gray. The amino acids in the mixed-charge region, where 42 of 47 residues are charged, are shown with double underlines. The ER retention signal-like sequence at the C terminus is boxed. The sequence data are available from GenBank under accession no. DQ418790.
FIG. 5.
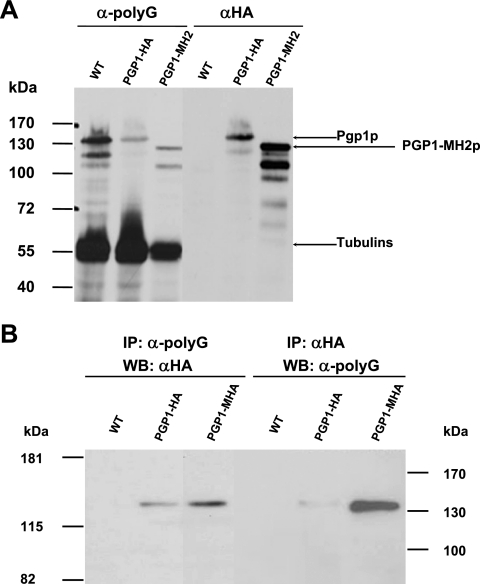
Pgp1p is specifically recognized by anti-polyG antibody. (A) Western blot of whole-cell extracts of wild-type (CU428), PGP1-HA, and PGP1-MH2 cells separated by 7.5% SDS-PAGE, blotted, and probed with anti-polyG antibody. The blot was stripped and reprobed with anti-HA antibody. (B) Whole-cell extracts of wild-type (CU428), PGP1-HA, and PGP1-MHA strains were immunoprecipitated (IP) with anti-polyG antibody or anti-HA antibody. Proteins eluted after immunoprecipitation (from 1 × 106 cells per lane) were subjected to 7.5% SDS-PAGE, followed by Western blot (WB) analysis with anti-HA antibody or anti-polyG antibody (see Materials and Methods for details).
A unique 19-amino-acid N-terminal sequence, VVLGIDFGSQYFKISLNAP, was obtained that exactly matched residues 22 to 40 of a predicted protein encoded by a cDNA from the Tetrahymena expressed sequence tag cDNA library (12). The full-length cDNA encoded by PGP1, the gene encoding Pgp1p, was obtained by 5′ and 3′ rapid amplification of cDNA ends. Other sequenced peptides also exactly matched the predicted protein sequences (Table 2 and Fig. 1C). The sequence of the PGP1 cDNA predicts a protein of 879 amino acids (Fig. 1C) whose sequence is highly similar (Fig. 1A) to those of the GRP170 subfamily proteins of the Hsp70s in the ER (5).
TABLE 2.
N-terminal sequencing results and peptide sequencing results for Pgp1p protein
| Sample | Sequence |
|---|---|
| Pgp1p N terminus | VVLGIDFGSQYFKISLNAP |
| Peptides | (R/K)TPETS(F/OX-M)V(F/OX-M)(I/L)DK |
| (R/K)(I/L)(F/OX-M)(I/L)NDGYN(F/OX-M)D(I/L)R | |
| (R/K)(I/L)QQ(I/L)SEY(I/L)QGK | |
| (R/K)(F/OX-M)(I/L)GA(I/L)ESDQEV(F/OX-M)EVAK | |
| (I/L)ES(F/OX-M)(I/L)YQ(I/L)R | |
| N(I/L)E(I/L)D(I/L) |
Analyzing the N-terminal sequence of Pgp1p with SignalP 3.0 (2) showed that the sequence contains a putative cleavable signal peptide with a predicted cleavage site between amino acids A21 and V22. Since the N-terminal amino acid sequence obtained from purified Pgp1p corresponded to amino acids 22 to 40 deduced from the cDNA (Fig. 1C), it is likely that the first 21 residues represent the signal peptide. This sequence also includes a sequence composed of 12 hydrophobic amino acids, which is characteristic of signal peptides required for initial transfer to the ER. The C-terminal sequence of Pgp1p is KQTDL (Fig. 1C), which resembles the ER retention signal KDEL (27). These two recognizable motifs in the protein argue that Pgp1p resides in the ER.
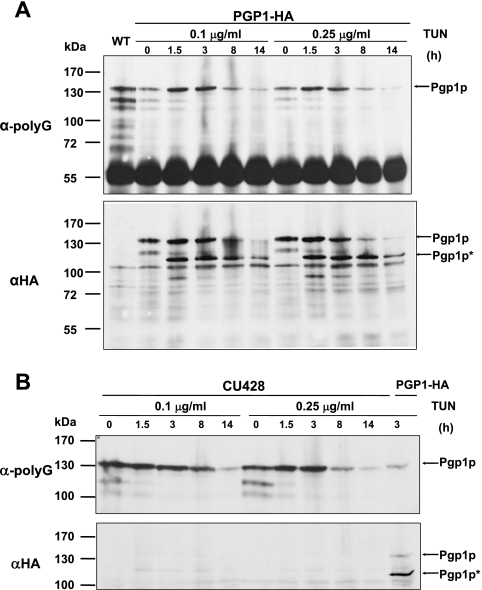
Site-specific enzymatic glycosylation of the amide nitrogens of asparagine side chains (N-linked glycosylation) occurs in eukaryotes and archaea and, rarely, in eubacteria and is a coposttranslational modification thought to be important for the proper folding, sorting, oligomerization, or stability of many ER, membrane, and secreted proteins (17, 25) The primary sequence of Pgp1p contains 16 potential sites for N-linked glycosylation (Fig. 1C; Asn-X-Ser/Thr consensus sequence, where X can be any amino acid but proline) (17), suggesting that the protein might be an N-linked glycoprotein. Consistent with this, the Pgp1p band detected by an anti-HA monoclonal antibody in a Western blot of whole-cell extract from cells expressing HA-tagged Pgp1p had a relative molecular mass of ∼135 kDa (see Fig. 5A and Fig. 7A), while the calculated molecular mass of Pgp1p (including the N-terminal signal peptide) is about 100 kDa. When cells expressing the PGP1-HA strain were treated with TUN, an inhibitor of N-linked glycosylation, the HA-staining band was reduced to ∼110 kDa (Pgp1p*), which likely represents the nonglycosylated HA-tagged Pgp1p protein (see Fig. 6A). These results argue that PGP1 encodes an N-linked glycoprotein.
FIG. 7.
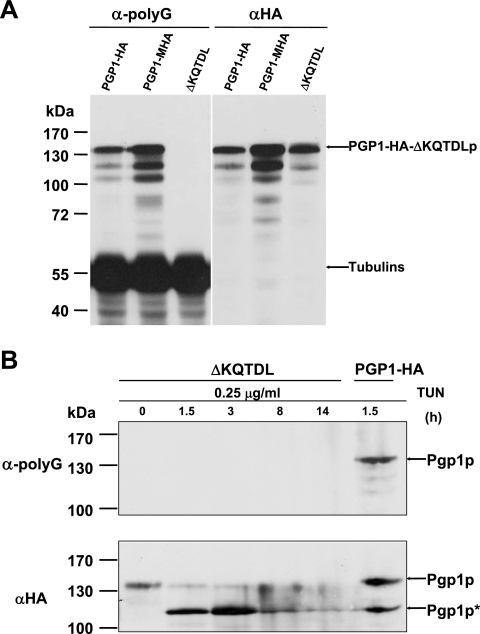
Pgp1p missing the putative ER retention site is not recognized by anti-polyG antibody. (A) Western blot analysis of whole-cell extracts from PGP1-HA, PGP1-MHA, and PGP1-HA-ΔKQTDL (ΔKQTDL) strains, using anti-polyG or anti-HA antibody. Equal amounts of total protein were loaded into each lane. The electrophoretic mobility of Pgp1p without KQTDL is not detectably different from that of full-length Pgp1p. (B) Western blot staining of whole-cell extracts from PGP1-HA-ΔKQTDL cells treated for increasing times with 0.25 μg/ml tunicamycin. After being stained with anti-polyG antibody, the blot was stripped and reprobed with anti-HA antibody. The whole-cell proteins from PGP1-HA cells treated with tunicamycin were the control.
FIG. 6.
Nonglycosylated Pgp1p (Pgp1p*) is not recognized by anti-polyG antibody. (A) Western blot of Pgp1p in the PGP1-HA strain at the indicated times after tunicamycin treatment (0.1 μg/ml and 0.25 μg/ml), using anti-polyG or (after stripping of the membrane) anti-HA antibody. The control extract is from wild-type (WT) CU428 cells. Approximately the same amounts of total protein were loaded in each lane. The glycosylated (Pgp1p) and nonglycosylated (Pgp1p*) forms are indicated. (B) Expression of Pgp1p in CU428 cells treated with tunicamycin, as analyzed by Western blotting with anti-polyG or anti-HA. The control protein extract was prepared from the PGP1-HA strain treated with tunicamycin.
As in GRP170 subfamily proteins in other organisms, the N-terminal region of Pgp1p contains a conserved ATP-binding domain (Fig. 1A), and a region in the C-terminal domain of Pgp1p shows high similarity to various members of the GRP170 subfamily (Fig. 1A). Near the C terminus (residues K821 to K867), we found a mixed-charge region with 42 positively and negatively charged residues among 47 amino acids (Fig. 1C) that is not found in other GRP170 proteins. The function of this region is not known. Cells containing a PGP1 gene in which this region was deleted grew well and appeared normal (data not shown).
Expression of PGP1.
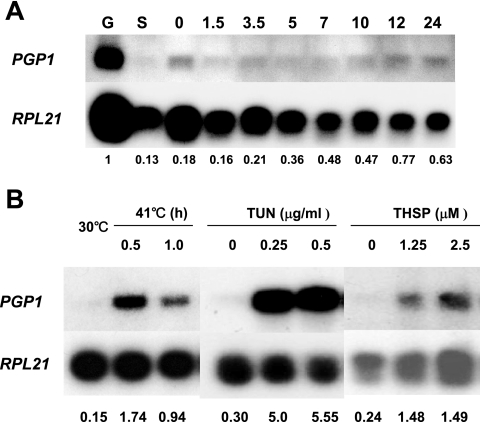
Using Northern blot analyses (Fig. 2A), we found that PGP1 was transcribed in all life cycle stages examined (log-phase growing cells, starved cells, and different stages of conjugating cells).
FIG. 2.
Expression of PGP1, as analyzed by Northern blotting. (A) PGP1 is expressed under normal physiological conditions. Total RNAs were obtained from log-phase growing (G), starved (S), and conjugating cells (0, 1.5, 3, 5, 7, 10, 12, and 24 h after being mixed). (B) PGP1 mRNA is induced by heat shock and ER stress agents. RNAs were isolated from CU428 cells grown at 30°C or after 0.5- and 1.0-h shifts to 41°C. Total RNA was isolated from CU428 cells that had been grown to log phase in 1× SPP medium at 30°C and then incubated in the presence of the indicated concentrations of TUN or THSP for 6 h prior to RNA extraction. Hybridization was carried out with radiolabeled probes for PGP1 and for RPL21, which encodes a ribosomal protein and was used as a loading control.
Since PGP1 encodes a member of a subfamily of Hsp70s found in the ER, we next analyzed PGP1 mRNA levels under stress conditions. TUN inhibits N-linked glycosylation and causes defects in glycoprotein trafficking between the ER and the Golgi apparatus, producing ER stress (4). PGP1 mRNA was dramatically induced when wild-type CU428 cells were treated with TUN (Fig. 2B). Treatment of CU428 cells with THPS, a selective inhibitor of the Ca2+-ATPase of the ER (4), also increased the PGP1 mRNA level compared to that in untreated cells (Fig. 2B). Thus, PGP1 expression is strongly induced by agents that cause ER stress.
Northern blot analyses also showed that PGP1 mRNA increased when wild-type cells were shifted to 41°C from 30°C (Fig. 2B). Consistent with this result, a putative heat shock element, consisting of two or more contiguous inverted repeats of the five-base-pair sequence 5′-NGAAN-3′ (24), is present at nucleotides −1081 to −1067 (GGAATATTCTTTTCA) in the PGP1 sequence.
PGP1 is an essential gene in Tetrahymena.
To examine the role of the PGP1 gene in vivo, we created PGP1 knockout cells by targeted gene disruption. The PGP1 knockout construct (Fig. 3A) was introduced into conjugating Tetrahymena cells to produce germ line heterozygous knockout heterokaryon strains. We then created germ line homozygous knockout heterokaryon strains (PGP1KO1 and PGP1KO4) with both copies of the PGP1 gene disrupted in micronuclei but with wild-type PGP1 genes still present in macronuclei.
When two such homozygous heterokaryon strains are mated, their progeny are homozygous homokaryons (ΔPGP1 cells) whose macronuclei and micronuclei both lack PGP1 genes. All of the exconjugants (∼5 × 106 cells) derived from mating two homozygous heterokaryon strains died, while control cells derived from mating wild-type cells with either of the two knockout heterokaryons produced viable progeny. Individual pairs from a mating between the two PGP1 knockout homozygous heterokaryon strains were isolated into drops of 1× SPP medium (no drug), and the cells in each drop were counted before they died. Most of the exconjugants divided two or three times (Fig. 3B), and cells became smaller and smaller until they died. When the mating homozygous PGP1 knockout heterokaryon cells were transformed with a wild-type PGP1 gene, progeny cells grew normally. These results demonstrate that PGP1 is essential for vegetative growth.
Localization of PGP1.
To determine the cellular localization of Pgp1p, the progeny cells of a mating of the homozygous knockout heterokaryons PGP1KO1 and PGP1KO4 were rescued with a C-terminally HA-tagged PGP1 gene (PGP1-HA construct) (Fig. 4A). The growth of the rescued progeny was indistinguishable from that of wild-type cells. In addition, when the HA-tagged PGP1 gene (PGP1-MHA construct) (Fig. 4A) was overexpressed, no obvious effect on cell growth under normal conditions was observed (data not shown). These observations indicate that the HA-tagged gene functions normally. The PGP1-HA construct was detected by indirect immunofluorescence microscopy with anti-HA antibody. In vegetative cells, staining was observed in most of the cytoplasm. Particularly strong staining was detected around the outside of macronuclei and in vesicles that appeared to be organized beneath the cell surface in diffuse rows similar to the basal body rows (Fig. 4B). These patterns are similar to the localization of the ER in Tetrahymena (13), consistent with the homology of the Pgp1p sequence to those of a known class of ER resident proteins.
Pgp1p is glycylated.
Although Pgp1p was identified by Western blot analysis of whole-cell extracts and of proteins immunoprecipitated with anti-polyG antibody (Fig. 1B), it was still possible that the protein we used to clone the PGP1 gene was a contaminant that comigrated with the anti-polyG-staining protein. To confirm that Pgp1p was the protein recognized by the anti-polyG antibody, we carried out Western blot analyses of whole-cell proteins from HA-tagged PGP1-HA and wild-type strains probed with anti-polyG or anti-HA antibody. With anti-polyG, besides the α- and β-tubulin bands (∼55 kDa), an ∼135-kDa band was clearly observed in both wild-type and PGP1-HA cells. When the same membrane was stained with anti-HA, an ∼135-kDa band was detected in PGP1-HA but not in control extracts (Fig. 5A), indicating that the ∼135-kDa band detected by the anti-polyG antibody comigrates with the Pgp1p protein. Except for the tubulin bands, bands running faster than the 135-kDa band are likely to be degradation products of Pgp1p (Fig. 5A).
When the whole-cell extracts from PGP1-HA or PGP1-MHA cells were immunoprecipitated with anti-polyG or anti-HA antibody and the immunoprecipitates were analyzed by Western blotting using anti-HA or anti-polyG antibody, reciprocal enrichment of the ∼135-kDa band was detected in HA-tagged cells but not in wild-type cells (Fig. 5B). In addition, when we deleted the mixed-charge region (47 amino acids) from the C-terminal end of Pgp1p (Fig. 1C and 4A), the ∼135-kDa band shifted to a band of ∼128 kDa in the Western blots stained with anti-HA or anti-polyG (Fig. 5A). These data demonstrate that Pgp1p is specifically recognized by anti-polyG antibody and that the mixed-charge region is not required for glycylation.
We next sought to determine whether the recognition of Pgp1p by the anti-polyG antibody was dependent on any of the other known properties of Pgp1p. When we treated the PGP1-HA cells with TUN, the ∼135-kDa band, which should represent glycosylated Pgp1p, was detected in Western blot analysis with either anti-HA or anti-polyG, and staining with both antibodies showed the same decreases in signal intensity with increased times of TUN treatment (Fig. 6A). Interestingly, TUN treatment strongly increased the amount of a nonglycosylated, ∼110-kDa form of Pgp1p (Pgp1p*) in the Western blot stained with anti-HA but not with anti-polyG (Fig. 6A). In wild-type cells treated with TUN, the ∼135-kDa band was detected on the membrane stained with anti-polyG but, as expected, not with anti-HA, and the ∼110-kDa band was not observed with either anti-HA or anti-polyG antibody (Fig. 6B). These results argue that the nonglycosylated Pgp1p* protein is not recognized by the anti-polyG antibody and, therefore, that it cannot be the amino acid sequence of Pgp1p that is recognized by anti-polyG. Rather, Pgp1p binding of the anti-polyG antibody must be due to a posttranslational modification of Pgp1p that is somehow linked to N-linked glycosylation, either directly or because glycosylation is required for folding of the protein into a glycylatable conformation.
The ER retention signal is required for glycylation of PGP1.
ORP150/Grp170, the Pgp1p homologous proteins in mammalian cells (20, 23), and Lhs1p, a Grp170 family member in Saccharomyces cerevisiae (6), have been demonstrated to be ER resident proteins. Soluble ER resident proteins are retained in the ER by a retrograde transport system that recognizes the specific retrieval signal KDEL or closely related sequences (27), and all of the Pgp1p orthologues contain a similar sequence (Fig. 1A). The C-terminal sequence of Pgp1p is KQTDL (Fig. 1C), which is similar to KDEL.
When the C-terminal KQTDL sequence in Pgp1p was deleted (PGP1-HA-ΔKQTDL) (Fig. 4A), the truncated protein stained weakly and more diffusely than the HA-tagged wild-type protein, and no deleterious effect on cell growth was observed under normal conditions (data not shown), indicating that the ER retention signal is not essential for growth. Similarly, removal of the HDEL signal of Bip, another ER protein, had no deleterious effect on the growth of yeast, even through Bip is essential for viability (16). Presumably, the essential function of Pgp1p (and of Bip in yeast), if it occurs in the ER, can be carried out by the initial targeting of the protein to the ER and does not require its reentry and accumulation in the ER. Surprisingly, for the KQTDL deletion mutants, the ∼135-kDa glycosylated Pgp1p band was not observed on Western blots stained with anti-polyG antibody but was clearly visible when similar amounts of protein were probed with anti-HA antibody (Fig. 7A). As noted above, the bands below the 135-kDa band, except for the tubulin bands, are likely to be the degradation products of Pgp1p (Fig. 7A).
These results indicate that Pgp1p lacking the ER retention signal cannot be recognized by the anti-polyG antibody, even though the size of this protein is indistinguishable from that of the protein from the PGP1-HA strain (Fig. 7A and B) and should therefore be glycosylated Pgp1p. When cells of the PGP1-HA-ΔKQTDL strain were treated with TUN, Western blot analyses showed that neither the glycosylated form (∼135 kDa; Pgp1p) nor the nonglycosylated form (110 kDa; Pgp1p*) of Pgp1p was detected with anti-polyG antibody, but staining the same membrane with anti-HA demonstrated that the glycosylated form was detected and decreased in amount with increased times of TUN treatment (Fig. 7B). The nonglycosylated form (Pgp1p*) strongly increased when the PGP1-HA-ΔKQTDL strain was treated with TUN (Fig. 7B), consistent with the above results (Fig. 6A). These results suggest that recognition of Pgp1p by anti-polyG antibody requires the ER retention signal KQTDL. When we deleted the ER retention signal KQTDL from the carboxyl terminus of untagged Pgp1p, the glycosylated Pgp1p protein also was not recognized by the anti-polyG antibody (data not shown). Taken together, these data argue that the N-linked glycans associated with Pgp1p do not react with anti-polyG and that Pgp1p is glycylated postsynthetically within the ER.
DISCUSSION
PGP1 encodes an ER glycoprotein.
The N-terminal sequence of Pgp1p has the properties of a cleavable signal peptide, and the N-terminal sequence of purified Pgp1p begins exactly at the predicted cleavage site for the signal peptide (Fig. 1C and Table 2). The Pgp1p C-terminal KQTDL sequence is similar to other KDEL-like ER retention signal motifs and to similarly located sequences in the known Pgp1p homologues (Fig. 1A). In yeast, mice, and humans, the homologous proteins have been localized to the ER lumen (8). PGP1 mRNA was strongly induced by drugs (TUN and THSP) that stress the ER. A fully functional HA-tagged Pgp1p fusion protein had a perinuclear and cytosolic distribution (Fig. 4B), consistent with the location of the ER in Tetrahymena. Pgp1p is also a glycoprotein, as indicated by the high frequency of N-glycosylation sites (Fig. 1C) and the large increase in electrophoretic mobility that occurs when cells are treated with tunicamycin (Fig. 6A). All of these results argue strongly that Pgp1p is an ER glycoprotein, the first to be described for this organism.
Potential functions of Pgp1p.
The Pgp1p protein shares sequence similarity with members of a distinct subfamily of the Hsp70 class of molecular chaperones (5, 8). Members of this family have been found in all eukaryotic cells (Fig. 1A) and include Lhs1p in yeast (6) and Grp170/Orp150 in mammalian cells (20, 23). Lhs1p is required for the correct processing of unfolded polypeptide substrates (6). LSH1 is not essential for the viability of yeast (6). The functions of Lhs1p overlap with those of Bip (Kar2p). These two chaperones interact, and their coordinated regulation is essential for normal function in the yeast ER (31). PGP1 is expressed throughout the Tetrahymena life cycle (Fig. 2A) and is essential for vegetative growth. Thus, unlike the case for yeast, in Tetrahymena the Bip homologue (ORF 14.m00361 in the Tetrahymena genome database [http://db.ciliate.org/cgi-bin/protein/protein?locus=14.m00361]) cannot compensate for the function of Pgp1p in normal growth. ORP150−/− mice have been reported to display an embryonic lethal phenotype (19). Thus, the functions of Pgp1p may be more similar to those of its mammalian homologues than to those of Lhs1p in yeast.
Grp170 has been shown to be involved in peptide transport into the ER via the transporter associated with antigen processing (23, 26). ORP150 is an inducible ER chaperone with cytoprotective properties during cell stress, such as ischemia/reperfusion (19, 20, 32). Unlike Lhs1p and ORP150 (6, 20), PGP1 mRNA was strongly induced by heat shock (Fig. 2B). PGP1 mRNA was also dramatically induced by agents that stress the ER (Fig. 2B). These results argue that Pgp1p has a constitutive function as an ER molecular chaperone and that its functions are enhanced under conditions that stress the ER.
A novel posttranslational modification occurs in the ER.
To date, glycylation has been identified on tubulins in diverse organisms and on g14-3-3, a protein of unknown function in Giardia duodenalis. Here we provide two independent lines of evidence that Pgp1p is a glycylated protein. Pgp1p and tubulins are the major proteins that are labeled by radioactive glycine in the absence of protein synthesis and are the only proteins immunoprecipitated from Tetrahymena whole-cell extracts that are detected by an anti-polyG antibody directed against a Cys-(Gly)9 peptide. Pgp1p immunoprecipitated from cells containing HA-tagged Pgp1p with either anti-HA or anti-polyG was recognized by both antibodies (Fig. 5B). When the mixed-charge domain of Pgp1p was deleted, changing the protein size, the truncated fragment showed the same altered patterns of mobilities in Western blots stained with anti-polyG and anti-HA (Fig. 5A). These results demonstrate that Pgp1p is specifically recognized by the anti-polyG antibody.
Additional evidence supports the conclusion that anti-polyG staining of Pgp1p is due to a posttranslational modification and suggests that Pgp1p must be properly glycosylated to be glycylated. We found that when PGP1-HA strains were treated with TUN, nonglycosylated Pgp1p* was not recognized by anti-polyG (Fig. 6A). Pgp1p* should have the same primary amino acid sequence as Pgp1p, indicating that anti-polyG does not recognize the polypeptide chain of Pgp1p. Several posttranslational modifications take place in the ER but not in the cytosol, such as signal peptide cleavage, N-linked glycosylation, disulfide bond formation, and glycophosphatidylinositol anchor addition (10, 22). The observation that inhibition of N-linked glycosylation eliminated staining with anti-polyG made this modification a candidate for recognition by the antibody. However, when the KQTDL ER retention signal of Pgp1p was deleted, the glycosylated form of Pgp1p was still produced but was not recognized by the anti-polyG antibody (Fig. 7A and B). Thus, while some N-linked glycosylation of Pgp1p may be necessary for glycylation to occur, it is not sufficient, and direct recognition of N-linked glycosyl groups by anti-polyG is highly unlikely. While the antibody studies alone cannot rule out the possibility that some other unknown posttranslational modification of Pgp1p that occurs in the ER is specifically recognized by anti-polyG and is dependent on ER retention, when coupled with the glycine labeling results, the simplest interpretation of our data is that Pgp1p is glycylated in the ER.
We propose that an enzyme(s) that catalyzes glycylation of Pgp1p is present in the ER and that this enzyme requires correctly glycosylated Pgp1p that must be retained in the ER long enough to be modified. Recently, two types of glutamylases were identified in Tetrahymena (18). They are members of a large family of proteins with a TTL homology domain. Other members of the TTLL family may catalyze different types of posttranslational additions of an amino acid, such as glycylation (18). Consistent with our proposal that there is a glycylase(s) in the ER, SignalP 3.0 (2) predicts that two members of the TTLL family in Tetrahymena have ER signal peptides.
The glycylation of tubulin is essential for cell survival and is required for the maintenance of specific stable microtubular organelles in Tetrahymena (33, 35). The function of glycylation of Pgp1p is likely to be distinct from that of glycylation of tubulin, and our results indicate that while Pgp1p is essential, its glycylation, like its ER retention, is not. Future studies should reveal whether specific TTLL proteins with glycylase activity reside in the ER and whether they have any essential functions.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM-26973.
We thank Josephine Bowen for technical assistance and for a critical reading of the manuscript.
Footnotes
Published ahead of print on 22 December 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1988. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 2.Bendtsen, J. D., H. Nielsen, G. Von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Cassidy-Hanley, J., D. Bowen, J. H. Lee, E. Cole, L. A. VerPlank, J. Gaertig, M. A. Gorovsky, and P. J. Bruns. 1997. Germ line and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 146:135-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chae, H. J., H. R. Kim, C. Xu, B. Bailly-Maitre, M. Krajewska, S. Krajewski, S. Banares, J. Cui, M. Digicaylioglu, N. Ke, S. Kitada, E. Monosov, M. Thomas, C. L. Kress, J. R. Babendure, R. Y. Tsien, S. A. Lipton, and J. C. Reed. 2004. BI-1 regulates an apoptosis pathway linked to endoplasmic reticulum stress. Mol. Cell 15:355-366. [DOI] [PubMed] [Google Scholar]
- 5.Craven, R. A., J. R. Tyson, and C. J. Stirling. 1997. A novel subfamily of Hsp70s in the endoplasmic reticulum. Trends Cell Biol. 7:277-282. [DOI] [PubMed] [Google Scholar]
- 6.Craven, R. A., M. Egerton, and C. J. Stirling. 1996. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 15:2640-2650. [PMC free article] [PubMed] [Google Scholar]
- 7.Duan, J., and M. A. Gorovsky. 2002. Both carboxy-terminal tails of alpha- and beta-tubulin are essential, but either one will suffice. Curr. Biol. 12:313-316. [DOI] [PubMed] [Google Scholar]
- 8.Easton, D. P., Y. Kaneko, and J. R. Subjeck. 2000. The Hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5:276-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddé, B., J. Rossier, J.-P. Le Caer, E. Desbruyeres, F. Gros, and P. Denoulet. 1990. Posttranslational glutamylation of α-tubulin. Science 247:83-84. [DOI] [PubMed] [Google Scholar]
- 10.Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181-191. [DOI] [PubMed] [Google Scholar]
- 11.Ersfeld, K., J. Wehland, U. Plessmann, H. Dodemont, V. Gerke, and K. Weber. 1993. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120:725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fillingham, J. S., N. D. Chilcoat, A. P. Turkewitz, E. Orias, M. Reith, and R. E. Pearlman. 2002. Analysis of expressed sequence tags (ETSs) in the ciliated protozoan Tetrahymena thermophila. J. Eukaryot. Microbiol. 49:99-107. [DOI] [PubMed] [Google Scholar]
- 13.Frankel, J. 2000. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62:28-103. [DOI] [PubMed] [Google Scholar]
- 14.Gorovsky, M. A., M. C. Yao, J. B. Keevert, and G. L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 9:311-327. [DOI] [PubMed] [Google Scholar]
- 15.Hai, B., J. Gaertig, and M. A. Gorovsky. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62:513-531. [DOI] [PubMed] [Google Scholar]
- 16.Hardwick, K. G., M. J. Lewis, J. C. Semenza, N. Dean, and H. R. B. Pelham. 1990. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J. 9:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019-1049. [DOI] [PubMed] [Google Scholar]
- 18.Janke, C., K. Rogowski, D. Wloga, C. Renard, A. V. Kajava, J. M. Strub, N. Temurak, J. van Dijk, D. Boucher, A. van Dorsselaer, S. Suryavanshi, J. Garertig, and B. Eddé. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science 308:1758-1762. [DOI] [PubMed] [Google Scholar]
- 19.Kitao, Y., K. Hashimoto, T. Matsuyama, H. Iso, T. Tamatani, O. Hori, D. M. Stern, M. Kano, K. Ozawa, and S. Ogawa. 2004. ORP150/HSP12A regulates Purkinje cell survival: a role for endoplasmic reticulum stress in cerebellar development. J. Neurosci. 24:1486-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwabara, K., M. Matsumoto, J. Ikeda, O. Hori, S. Ogawa, Y. Maeda, K. Kitagawa, N. Imuta, T. Kinoshita, D. M. Stern, H. Yanagi, and T. Kamada. 1996. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J. Biol. Chem. 271:5025-5032. [DOI] [PubMed] [Google Scholar]
- 21.Lalle, M., A. M. Salzano, M. Crescenzi, and E. Pozio. 2006. The Giardia duodenalis 14-3-3 protein is post-translationally modified by phosphorylation and polyglycylation of the C-terminal tail. J. Biol. Chem. 281:5137-5148. [DOI] [PubMed] [Google Scholar]
- 22.Lee, M. C. S., E. A. Miller, J. Goldberg, L. Orci, and R. Schekman. 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20:87-123. [DOI] [PubMed] [Google Scholar]
- 23.Lin, H. Y., P. Masso-Welch, Y. P. Di, J. W. Cai, J. W. Shen, and J. R. Subjeck. 1993. The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol. Biol. Cell 4:1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, X. D., P. C. Liu, N. Santoro, and D. J. Thiele. 1997. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 16:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra, N., S. Sinha, T. N. C. Ramya, and A. Surolia. 2006. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31:156-163. [DOI] [PubMed] [Google Scholar]
- 26.Park, J., J. Facciponte, X. Chen, I. MacDonald, E. A. Repasky, M. H. Manjili, X.-Y. Wang, and J. R. Subjeck. 2006. Chaperoning function of stress protein grp170, a member of the hsp70 superfamily, is responsible for its immunoadjuvant activity. Cancer Res. 66:1161-1168. [DOI] [PubMed] [Google Scholar]
- 27.Pelham, H. R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15:483-486. [DOI] [PubMed] [Google Scholar]
- 28.Redeker, V., N. Levilliers, J.-M. Schmitter, J.-P. Le Caer, J. Rossier, A. Adoutte, and M.-H. Bré. 1994. Polyglycylation of tubulin: a post-translational modification in axonemal microtubules. Science 266:1688-1691. [DOI] [PubMed] [Google Scholar]
- 29.Regnard, C., E. Desbruyeres, J.-C. Huet, C. Beauvallet, J.-C. Pernollet, and B. Eddé. 2000. Polyglutamylation of nucleosome assembly proteins. J. Biol. Chem. 275:15969-15976. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., X. Song, J. Bowen, R. Corstanje, Y. Gao, J. Gaertig, and M. A. Gorovsky. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 99:3734-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steel, G. J., D. M. Fullerton, J. R. Tyson, and C. J. Stirling. 2004. Coordinated activation of Hsp70 chaperones. Science 303:98-101. [DOI] [PubMed] [Google Scholar]
- 32.Tamatani, M., T. Matsuyama, A. Yamaguchi, N. Mitsuda, Y. Tsukamoto, M. Taniguchi, Y. H. Che, K. Ozawa, O. Hori, H. Nishimura, A. Yamashita, M. Okabe, H. Yanagi, D. M. Stern, S. Ogawa, and M. Tohyama. 2001. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat. Med. 7:317-323. [DOI] [PubMed] [Google Scholar]
- 33.Thazhath, R., C. Liu, and J. Gaertig. 2002. Polyglycylation domain of β-tubulin maintains axonemal architecture and affects cytokinesis in Tetrahymena. Nat. Cell Biol. 4:256-259. [DOI] [PubMed] [Google Scholar]
- 34.Westermann, S., and K. Weber. 2003. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 4:938-947. [DOI] [PubMed] [Google Scholar]
- 35.Xia, L., B. Hai, Y. Gao, D. Burnette, R. Thazhath, J. Duan, M. H. Bré, N. Levilliers, M. A. Gorovsky, and J. Gaertig. 2000. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 149:1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30:892-897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.