Abstract
The maize pathogen Cochliobolus heterostrophus requires two mitogen-activated protein kinases (MAPKs), Chk1 and Mps1, to produce normal pigmentation. Young colonies of mps1 and chk1 deletion mutants have a white and autolytic appearance, which was partially rescued by a hyperosmotic environment. We isolated the transcription factor Cmr1, an ortholog of Colletotrichum lagenarium Cmr1 and Magnaporthe grisea Pig1, which regulates melanin biosynthesis in C. heterostrophus. Deletion of CMR1 in C. heterostrophus resulted in mutants that lacked dark pigmentation and acquired an orange-pink color. In cmr1 deletion strains the expression of putative scytalone dehydratase (SCD1) and hydroxynaphthalene reductase (BRN1 and BRN2) genes involved in melanin biosynthesis was undetectable, whereas expression of PKS18, encoding a polyketide synthase, was only moderately reduced. In chk1 and mps1 mutants expression of PKS18, SCD1, BRN1, BRN2, and the transcription factor CMR1 itself was very low in young colonies, slightly up-regulated in aging colonies, and significantly induced in hyperosmotic conditions, compared to invariably high expression in the wild type. These findings indicate that two MAPKs, Chk1 and Mps1, affect Cmr1 at the transcriptional level and this influence is partially overridden in stress conditions including aging culture and hyperosmotic environment. Surprisingly, we found that the CMR1 gene was transcribed in both sense and antisense directions, apparently producing mRNA as well as a long noncoding RNA transcript. Expression of the antisense CMR1 was also Chk1 and Mps1 dependent. Analysis of chromosomal location of the melanin biosynthesis genes in C. heterostrophus resulted in identification of a small gene cluster comprising BRN1, CMR1, and PKS18. Since expression of all three genes depends on Chk1 and Mps1 MAPKs, we suggest their possible epigenetic regulation.
Fungal pathogens perceive and respond to a variety of environmental signals as well as molecules from the host plant, triggering morphogenetic changes. Mitogen-activated protein kinases (MAPKs) are ubiquitous and evolutionarily conserved enzymes connecting cell surface to intracellular regulatory targets activating various morphogenetic changes. The budding yeast Saccharomyces cerevisiae has five MAPKs, encoded by FUS3, KSS1, HOG1, SLT2, and SMK1. These MAPK cascades regulate fungal growth and differentiation processes such as pheromone response, pseudohyphal growth, osmoadaptation, cell wall integrity, and ascospore formation. Fus3/Kss1 orthologs in filamentous fungi are involved in appressorium formation and pathogenicity in general but have diverse functions in hyphal growth, sexual and asexual reproduction, and conidial germination (4, 18, 22, 24, 25, 27, 29, 39, 41). For example, in the rice blast fungus Magnaporthe grisea Pmk1 is required for appressorium formation and invasive growth on the host, whereas in the anthracnose fungus Colletotrichum lagenarium, Cmk1 is also required for conidiation and conidial germination (29, 39).
Slt2 homologs in filamentous fungi function to maintain cell wall integrity and are essential for the formation of functional appressoria and for virulence (6, 13, 19, 21, 40). mps1 deletion mutants of M. grisea are sensitive to cell wall-digesting enzymes, have reduced conidiation and fertility, form nonfunctional appressoria, and are unable to infect plants (40). C. lagenarium maf1 mutants completely lack appressoria and have reduced conidiation and pathogenicity (13). Mgslt2 mutants of the dothideomycete Mycosphaerella graminicola are sensitive to glucanase and several fungicides and are unable to grow invasively on the plant. Unlike other fungi, Mgslt2 mutants form unmelanized colonies with short aerial hyphae. Mutant mycelium is defective in polarized growth of the tip cells and undergoes progressive autolysis (19).
It can be concluded that in filamentous fungi MAPKs of two classes, Fus3/Kss1 and Slt2, often control the same cellular functions but are not redundant. For example, C. lagenarium Maf1 controls the early differentiation stage of appressorium formation, whereas Cmk1 is involved in the maturation of appressoria (19).
Cochliobolus heterostrophus, the cause of Southern corn leaf blight, is a filamentous necrotrophic ascomycete. Numerous eyespot-like lesions on the leaves are typical of this disease. In the presence of moisture, spores of the pathogen adhere to a leaf or other surface, germinate, and develop small appressoria, unlike the large melanized appressoria formed by M. grisea and by Colletotrichum species (3). C. heterostrophus appressoria are not considered essential for penetration, which occurs directly or through stomata (20).
We have previously characterized the C. heterostrophus MAPK gene CHK1, homologous to the yeast FUS3/KSS1 (18). chk1 deletion mutants are female sterile, lack conidia, and do not form appressoria on an inductive surface, and their virulence is severely reduced. Colonies of chk1 mutants, white and sometimes autolytic, become darkly pigmented during prolonged growth. Recently, we have isolated a C. heterostrophus homolog of yeast SLT2 designated MPS1 (A. Igbaria et al., unpublished data). Interestingly, white colony appearance and formation of autolytic regions were even more pronounced in Δmps1 mutants than in Δchk1 mutants. The aim of this study was to establish how two MAPKs, Chk1 and Mps1, control mycelial melanization.
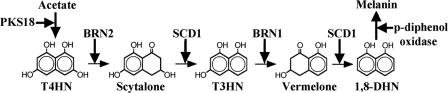
Wild-type colonies of C. heterostrophus undergo melanization and produce abundant conidia when grown under continuous illumination. The melanization is even more pronounced in colonies exposed to hyperosmotic stress. Melanin is a dark pigment tissue considered to protect the fungus from environmental stresses such as UV irradiation (8, 26). Albino strains of C. heterostrophus cannot survive in the field, but under laboratory conditions they cause lesions similar to those of the wild type on the host plant (5). C. heterostrophus produces 1,8-dihydroxynaphthalene (1,8-DHN)-melanin, as do Bipolaris oryzae, Colletotrichum lagenarium, Magnaporthe grisea, Alternaria alternata, and many other fungi (31, 32). DHN-melanin biosynthesis starts with a polyketide synthase (PKS) using acetyl coenzyme A or malonyl coenzyme A as a precursor (Fig. 1). The PKS produces 1,3,6,8-tetrahydroxynaphthalene (T4HN), which is reduced by hydroxynaphthalene reductase to form scytalone. Dehydration of scytalone by scytalone dehydratase forms 1,3,8-trihydroxynaphthalene (T3HN). T3HN reductase converts T3HN to vermelone, which is further dehydrated to 1,8-DHN. Subsequent steps are thought to involve a dimerization of 1,8-DHN molecules, followed by polymerization catalyzed by p-diphenol oxidase (16, 32). The BRN1 gene encoding T3HN reductase has been isolated in C. heterostrophus (28). Genes involved in melanin biosynthesis, including the polyketide synthase (PKS1), T3HN reductase (THR1), and scytalone dehydratase (SCD1) genes, have been isolated in B. oryzae, a rice pathogen closely related to C. heterostrophus, and in C. lagenarium (10, 11, 23, 30).
FIG. 1.
Schematic representation of the fungal DHN-melanin biosynthesis pathway. PKS18 (polyketide synthase), BRN2 (T4HN reductase), BRN1 (T3HN reductase), SCD1 (scytalone dehydratase), and p-diphenol oxidase are enzymes presumably involved in the indicated biosynthetic steps.
C. lagenarium and M. grisea produce melanin during appressorium formation, on hyperosmotic media, and in the late stationary stage of mycelial growth. Orthologous transcription factors Cmr1 and Pig1 control melanin biosynthesis genes in C. lagenarium and M. grisea, respectively (34). The expression of transcription factor CMR1 itself was shown to be up-regulated during melanin production. Cmr1 regulates the expression of THR1 and SCD1 genes in vegetative hyphae upon induction of melanization, but during formation of appressoria the expression of THR1 and SCD1 is CMR1 independent (34).
In this study, we determined the influence of two MAPKs of C. heterostrophus, Chk1 and Mps1, on the expression of melanin biosynthesis genes through the C. heterostrophus Cmr1 ortholog.
MATERIALS AND METHODS
Fungal strains and growth media.
Wild-type (C4 and C5) and chk1 and mps1 deletion strains of Cochliobolus heterostrophus and standard growth conditions have been described elsewhere (17, 18, 38; Igbaria et al., unpublished). CMX medium was prepared in the same way as standard complete medium (CM), except that glucose was replaced with the same amount of xylose. For hygromycin B (50 μg/ml; Calbiochem) and nourseothricin (120 μg/ml; Werner BioAgents, Jena, Germany) selection, CM and CMX were prepared without salts.
CMR1 replacement constructs and transformant verification.
The vector for deletion of CMR1 was assembled from the following fragments: 827-bp 5′ flanking sequence (amplified with primers d-SacI-s and d-XhoI-a), 2.1-kb hygromycin resistance cassette, 623-bp 3′ flanking sequence amplified with primers d-NheI-s and BglII-a, and pBluescript cloning vector (Stratagene) (Table 1; see Fig. S1A in the supplemental material). For transformation, the resulting vector was amplified by PCR with primers d-SacI-s and d-BglII-a using BIO-X-ACT Long High Fidelity DNA polymerase (Bioline; BIO-21049). Polyethylene glycol-mediated transformation into fungal protoplasts was performed as described previously (35, 36).
TABLE 1.
Primers used in this study
| Gene or vector | First primer
|
Second primer
|
||
|---|---|---|---|---|
| Name | Sequencea | Name | Sequencea | |
| Actin, RT-PCR | Act1-s | TTCTCCACCACTGCCGAGCG | Act1-a | GCGGTGAACGATGGAGGGAC |
| Actin, RT-PCR | Act2-s | AATGGTTCGGGTATGTGCAAGG | Act2-a | GGGACCGCTCTCGTCGTACTC |
| SCD1, RT-PCR | Scd1-s | TGGGAGGCGATGCCAGCGGAT | Scd1-a | GCTCGCGGCCCTCTGCAAACA |
| BRN1, RT-PCR | Brn1-s | TGGGTGGCCGCATTATCCTCA | Brn1-a | GAAGCAGACGACGCGGGCAAT |
| BRN2, RT-PCR | Brn2-s | CATTGCCGCTGGTCTTCTCGG | Brn2-a | AAGCCACAACCCTCGCAACAT |
| CMR1, RT-PCR, transformant verification | ctrl-s | GCAATCATCGCCGCCCAAGA | V-a | aacaggtaccGACGGCGGGCGAACATCCAG |
| CMR1, transformant verification and intron analysis | s | GCCAATCCAATCTGCCCCAT | a | CAATGCGAGAGGACAGCGAAA |
| CMR1 deletion vector, 5′ flank | d-SacI-s | tacgagcTCCCTGCCATCGCTGAGTCTT | d-XhoI-a | aggctcgAGGGGTTGTTGGTGATGGCTG |
| CMR1 deletion vector, 3′ flank | d-NheI-s | tatgctAGCGGGCGTCTTCGGCGTTG | d-BglII-a | catagatCTGCCAAAGACAATCAACACTGG |
| CMR1 genomic sequence outside 5′ and 3′ flanks | 5′o | CCGCACGCACCTCCACCTCG | 3′o | GCAAGAAGAGGAGGATGGATGG |
| Hygr cassette, promoter and terminator | ptrp | GGTCGTTCACTTACCTTGCTTG | ttrp | GGTGTTCAGGATCTCGATAAG |
| ttrpe | GTGAATGCTCCGTAACACCCAATAC | |||
| CMR1p-GFP vector, GFP RT-PCR | GFP-BamHI-s | gaagGATCCCATGGTGAGCAAGGGC | GFP-SpeI-a | tgaactagtCTTGTACAGCTCGTCCATGCCGT |
| CMR1p-GFP vector, CMR1 promoter | p-XhoI-s | caactcgAGCACCGGGCAGGACAGGACT | p-BamHI-a | caaggATCCGGCTCGAATGTCTACTGCTC |
| CMR1p-GFP vector, 3′ flank | 3′-BglII-s | taccatagatCTCCTCGTTTGTTCATTCGCCC | 3′-SacI-a | taggagcTCAAGTGGGCGGGTGGTTGTA |
| Mating type-specific gene MAT1-1 | MAT1-1s | GTCGTCGATGGTGATGAAAGAAA | MAT1-1a | CCGCACTGGAGCTCAAATGGT |
| Mating type-specific gene MAT1-2 | MAT1-2s | GTTGCATCTCCGTCTGCGCCA | MAT1-2a | GGCTGCAAGGATGACTGGCAT |
The lowercase letters represent sequence with no homology to template DNA, whereas homologous regions are shown in uppercase.
Genomic DNA was isolated from the fungal mycelium using the Extract-N-Amp Plant PCR Kit (Sigma; XNAP2). The deletion of CMR1 in hygromycin-resistant colonies was confirmed by PCR amplification crossing the junction points of the vector with genomic DNA formed following a homologous recombination event. Primer pairs used for PCR were 5′o and ptrp, 3′o and ttrp (or ttrpe), and s and a (or ctrl-s and V-a) (see Fig. S1A, B, and D in the supplemental material; Table 1).
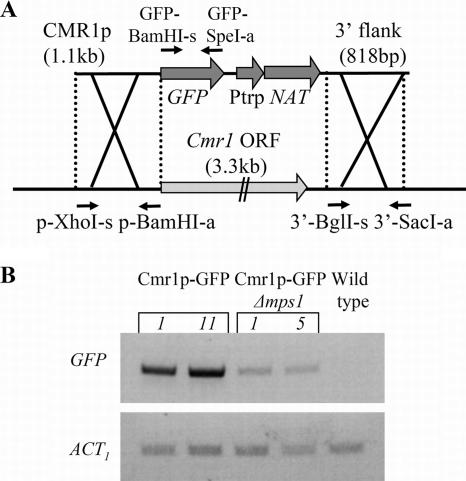
CMR1p-green fluorescent protein (GFP) vector was assembled by ligation of the following DNA fragments: 1.1-kb CMR1 promoter (amplified with primers p-XhoI-s and p-BamHI-a), GFP amplified with GFP-BamHI-s and GFP-SpeI-a, CMR1 3′ untranslated region/terminator, nourseothricin resistance cassette including the Streptomyces noursei nourseothricin acetyltransferase-encoding gene (GenBank accession number X73149), 818-bp 3′ flanking sequence amplified using primers 3′-BglII-s and 3′-SacI-a, and pBluescript vector (see Fig. 7A; Table 1). For transformation, CMR1p-GFP vector was linearized.
FIG. 7.
Construction of CMR1p-GFP reporter strain and analysis of CMR1 promoter activity in the absence of Cmr1 protein. (A) CMR1p-GFP transformation vector. Abbreviations: Ptrp, constitutive promoter; NAT, nourseothricin acetyltransferase-encoding gene; p-XhoI-s, p-BamHI-a, 3′-BglI-s, 3′-SacI-a, GFP-BamHI-s, and GFP-SpeI-a, primers used for vector construction. (B) RT-PCR analysis of GFP reporter expression in parental CMR1p-GFP strain and in mps1-deleted progeny. Expression of the actin-encoding gene indicates relative RNA quantities in each sample. Numbers 1, 11, and 5 represent different transformants.
Mutant characterization.
To compare formation of appressoria by the mutants and that by the wild type, water-suspended conidia were placed in drops on the surface of a 9-cm polystyrene petri dish (Miniplast, Ein Shemer, Israel). The dishes were incubated for 3 to 5 h at 30°C and then studied under an inverted microscope (Olympus). For virulence tests, sweet maize plants (Zea mays hybrid ‘Grand Jubilee’, purchased locally) were grown for about 2 weeks at 22 to 24°C. Intact leaves were inoculated with conidial suspension in 0.05% Tween 80, and the plants were sealed in moist plastic bags and incubated for 2 days at 30°C under continuous light. For inoculation of detached leaves, they were placed on 1% agar plates just before solidification. Mycelial mats (1 × 1 mm) were placed on the agar near the leaves. The dishes were sealed and incubated under continuous illumination at 30°C for 2 days.
Crosses between fungal strains C4 and C5 were set up according to the method in reference 17. The plates were incubated in darkness at 25°C for 23 to 24 days, and then the ascospores were isolated. The mating type of the progeny was determined by PCR using primer pairs MAT1-1s-MAT1-1a and MAT1-2s-MAT1-2a (Table 1) corresponding to the genes MAT1-1 and MAT1-2, respectively.
For the UV sensitivity test, water-suspended conidia and mycelial fragments were incubated on glass slides until conidia became firmly attached to the glass surface and started to germinate (45 min). Then the slides were washed with water to eliminate unattached mycelium and unviable conidia. The slides were irradiated at 254 nm at energy levels of 20 to 70 mJ/cm2 in a UV cross-linker and incubated in aluminum-foil-wrapped petri dishes overnight until examination.
RNA isolation and reverse transcription-PCR (RT-PCR).
RNA was isolated from mycelia ground in liquid nitrogen followed by extraction with TRI reagent (Molecular Research Center) using the manufacturer's protocol. Routinely, 2 μg RNA was treated with RQ DNase (Promega; M610A) and then used for cDNA synthesis with oligo(dT)17N primer (protocol supplied with Moloney murine leukemia virus reverse transcriptase; Promega; M170A). To synthesize cDNA with a sequence-specific primer, the primer was added to a final concentration of 2 μM, along with 0.2 μM of act2-a primer. Semiquantitative PCR was performed with Ready-To-Use mix (Larova; PCR-0540). We performed a series of PCRs for each primer pair to establish the optimal cycle number, which was then used in the actual experiment (Table 1). Each PCR was done at least twice, and data shown are representative of two independent experiments.
Identification of melanin biosynthesis gene clusters.
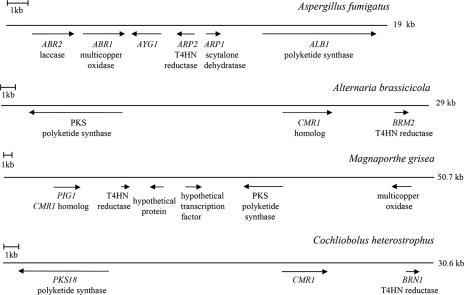
The Magnaporthe grisea genome sequence was obtained from the PEDANT database (http://pedant.gsf.de/cgi-bin/wwwfly.pl?Set=Magnaporthe_grisea_BI&Page=index). The Alternaria brassicicola complete genome was downloaded from http://genome.wustl.edu/pub/organism/Fungi/Alternaria_brassicicola/assembly/draft/Alternaria_brassicicola-1.0/. For the analysis in Fig. 8, sequences from four contigs (1.244 to 1.247) were combined. A. brassicicola melanin biosynthesis genes were identified by similarity to other fungal species. GenBank accession numbers of the annotated genes are listed in Table 2.
FIG. 8.
Organization of the melanin biosynthesis gene clusters in Aspergillus fumigatus, Alternaria brassicicola, Magnaporthe grisea, and Cochliobolus heterostrophus. The accession numbers of the indicated genes are listed in Table 2.
TABLE 2.
GenBank accession numbers
| Organisma | Gene | Description | GenBank accession no. |
|---|---|---|---|
| Cochliobolus heterostrophus | CMR1 | Transcription factor containing two Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster domains | DQ902714 |
| BRN1 | T3HN reductase | AB001564 | |
| BRN2 | T4HN reductase | EF060260 | |
| SCD1 | Scytalone dehydratase | EF060261 | |
| PKS18 | Polyketide synthase | AY495659 | |
| ACT1 | Actin | AY748990 | |
| MAT1-1 | Mating-type gene | X68399 | |
| MAT1-2 | Mating-type gene | X68398 | |
| Aspergillus fumigatus | ABR1 | Multicopper oxidase | AF116901 |
| ABR2 | Laccase | AF104823 | |
| AYG1 | Encodes DUF1100 family protein of unknown function | AF116902 | |
| ARP1 | Scytalone dehydratase | U95042 | |
| ARP2 | T4HN reductase | AF099736 | |
| ALB1 | Polyketide synthase | AF025541 | |
| Magnaporthe grisea | PIG1 | CMR1 homolog, Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster-containing transcription factor | AF230811 |
| T4HN reductase | AF290182 | ||
| Hypothetical protein-encoding gene | XM_367292 | ||
| Hypothetical GAL4-like transcription factor containing Zn2Cys6 binuclear cluster domain | XM_367293 | ||
| Polyketide synthase | XM_367294 | ||
| Multicopper oxidase | XM_367295 |
Alternaria brassicicola melanin biosynthesis genes have not been annotated.
RESULTS
Identification and structural analysis of the C. heterostrophus CMR1 gene.
We searched the genome of C. heterostrophus with C. lagenarium Cmr1 and M. grisea Pig1 amino acid sequences to identify their homolog in C. heterostrophus. The gene with the best score showed significant homology to both CMR1 and PIG1 over a large part of the coding sequence. The C. heterostrophus CMR1 coding region is interrupted by three introns. The deduced Cmr1 protein is 1,014 amino acids long and shares 69% and 70% similarity with Cmr1 of C. lagenarium and Pig1 of M. grisea, respectively. The predicted C. heterostrophus Cmr1 protein has two Cys2His2 zinc finger domains and one Zn(II)2Cys6 binuclear cluster domain close to its N terminus, similar to Pig1 and C. lagenarium Cmr1 (Fig. 2). Interestingly, all three proteins are highly similar at their N termini and their C-terminal halves, but a region of low similarity separates these conserved regions.
FIG. 2.
CMR1 gene features and domain architecture of the protein product.
Deletion of the CMR1 gene.
We designed a vector to delete the CMR1 gene by double-crossover integration of the hygromycin resistance cassette (see Fig. S1A in the supplemental material). All emerging transformants lacked the typical gray-green color of the wild-type Cochliobolus when grown on plates. We verified the CMR1 deletion by PCR using primers homologous to the hygromycin resistance cassette and genomic sequence outside the flank regions (see Fig. S1B in the supplemental material). Thus, only transformants in which the CMR1 coding region was replaced with the hygromycin resistance cassette would provide the correct template. The deletion of the CMR1 coding region in the transformants was confirmed using two internal primers. Notably, even in the first days of hygromycin selection on the transformation plates, when new transformants emerge as heterokaryons, complete loss of the black pigment was observed. We can speculate that production of melanin requires some critical quantity of CMR1 transcripts. Reduction in this amount leads to complete loss (at least visibly) of melanin production.
Characterization of the mutants.
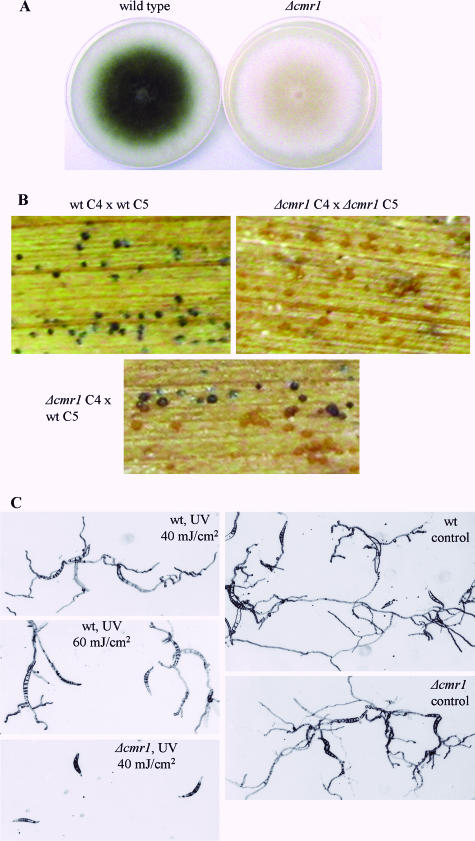
Wild-type Cochliobolus acquires a gray-green color when grown on plates, along with the onset of conidiation. Colonies of the cmr1 deletion mutants developed a light orange-pink color during maturation (Fig. 3A). This color was different from the near-white of mps1 and chk1 mutants, and characteristic autolytic-appearing areas of MAPK mutants were absent in the Δcmr1 strain. The conidiation of mutant colonies was significantly decreased compared to the wild type. To ensure that conidiation of mutants was not due to residual wild-type nuclei, we isolated single spores from the mutant fungus and plated them separately. These single spores developed into orange-pink colonies which also produced a small number of conidia. These conidia germinated and formed appressoria on a glass surface undistinguishable from those of the wild type. Inoculation of intact maize plants and detached leaves with Δcmr1 strain conidial suspension and mycelial mats, respectively, resulted in disease symptoms similar to those caused by the wild-type strain.
FIG. 3.
Characterization of the cmr1 mutant phenotype. (A) Colonies of wild type and Δcmr1 mutant; (B) Δcmr1 mutants form light brown pseudothecia when crossed with one another and with the wild-type strain; (C) growth inhibition of mutant and wild-type strains as a result of UV irradiation. Wild-type and mutant conidia were allowed to attach to the slides (45 min); mycelial debris and conidia that remained unattached were washed off. Following UV irradiation germ tubes of the mutant stopped further growth. The energy of irradiation is indicated. wt, wild type.
We made a cross between Δcmr1 mutants (mating type MAT1-2) and wild-type C5 (MAT1-1). Black pseudothecia were formed from the wild-type side of maize leaf, and light brown pseudothecia were formed on the side of the mutant (Fig. 3B). Both types of fruiting bodies contained viable ascospores. The progeny of the cross had a wild-type or mutant appearance identical to the parents, and hygromycin resistance segregated together with the pigment deficiency trait. Out of the six pigment-deficient progeny strains, four belonged to the mating type MAT1-2 (as their Δcmr1 parent) and two were of mating type MAT1-1 (see Fig. S1C in the supplemental material). No traces of CMR1 DNA were detected in the mutants, as verified by PCR (see Fig. S1D in the supplemental material). Mutant progeny of the Δcmr1 cross with the wild type were used in subsequent gene expression studies. When crossed with one another, Δcmr1 strains were able to form only light brown pseudothecia (Fig. 3B).
Since melanin is known to protect fungal cells from UV irradiation, we compared the sensitivities of the cmr1 mutant and the wild type to UV light at different intensities. Conidia germinating on a glass surface were UV irradiated, and hyphal growth was observed after overnight incubation in darkness. Irradiation with 40 mJ/cm2 caused complete inhibition of growth in cmr1 mutants, whereas wild-type germ tubes continued to elongate even after 60 mJ/cm2 (Fig. 3C).
Identification of the melanin biosynthesis genes and their expression.
The genome of C. heterostrophus is unusually rich in polyketide synthase-encoding genes (15). C. heterostrophus PKS18 has 96% similarity to the PKS1 gene of B. oryzae, which was shown to be involved in melanin biosynthesis (23).
We searched the C. heterostrophus genome for hydroxynaphthalene reductases with the BRN1 (T3HN reductase) gene sequence. As a result of this search we identified another hydroxynaphthalene reductase, designated BRN2, which showed a significant degree of homology to the fungal T4HN reductases. Another genome search was performed with the SCD1 gene of B. oryzae. As result of this search we identified a C. heterostrophus gene, also designated SCD1, which had 99% similarity with SCD1 of B. oryzae (GenBank accession numbers are given in Table 2).
Wild-type C. heterostrophus and cmr1, mps1, and chk1 deletion mutants were grown for 3 to 5 days in stationary liquid culture (complete medium, CMX). Pigment deficiency and autolytic appearance of the Δchk1 and Δmps1 mutants were very pronounced under these conditions (Fig. 4). Partial restoration of pigmentation and decrease of the autolytic phenotype (especially in the Δchk1 mutant) were observed in cultures supplemented with 1.5 M sorbitol (hyperosmotic stress). We tested the expression of PKS18, BRN1, BRN2, and SCD1 in all cultures. In the wild-type strain the expression of PKS18, BRN1, BRN2, and SCD1 was high in 3-day-old stationary culture, as well as in 5-day-old cultures with or without sorbitol. In cmr1 deletion strains the expression of BRN1, BRN2, and SCD1 was undetectable under all growth conditions tested, indicating that these genes absolutely require Cmr1 for their expression (Fig. 5A). Expression of PKS18 was only moderately reduced in 3-day-old Δcmr1 culture compared to the wild type. Aging and hyperosmotic stress caused further decrease in PKS18 expression in both Δcmr1 and wild-type strains (Fig. 5B).
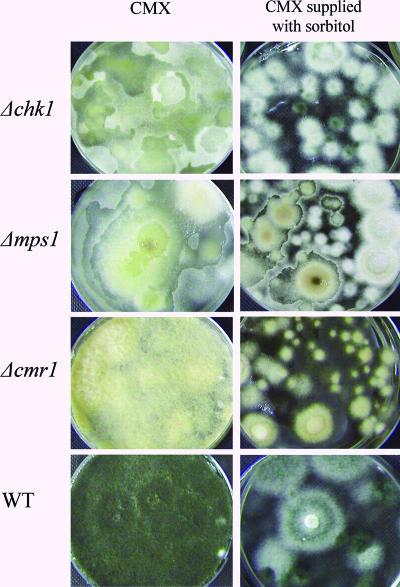
FIG. 4.
Pigmentation and autolytic phenotypes. Five-day-old stationary liquid cultures of Δcmr1, Δchk1, and Δmps1 mutants and the wild-type strain (WT) were grown in complete medium with xylose (CMX) with or without addition of 1.5 M sorbitol.
FIG. 5.
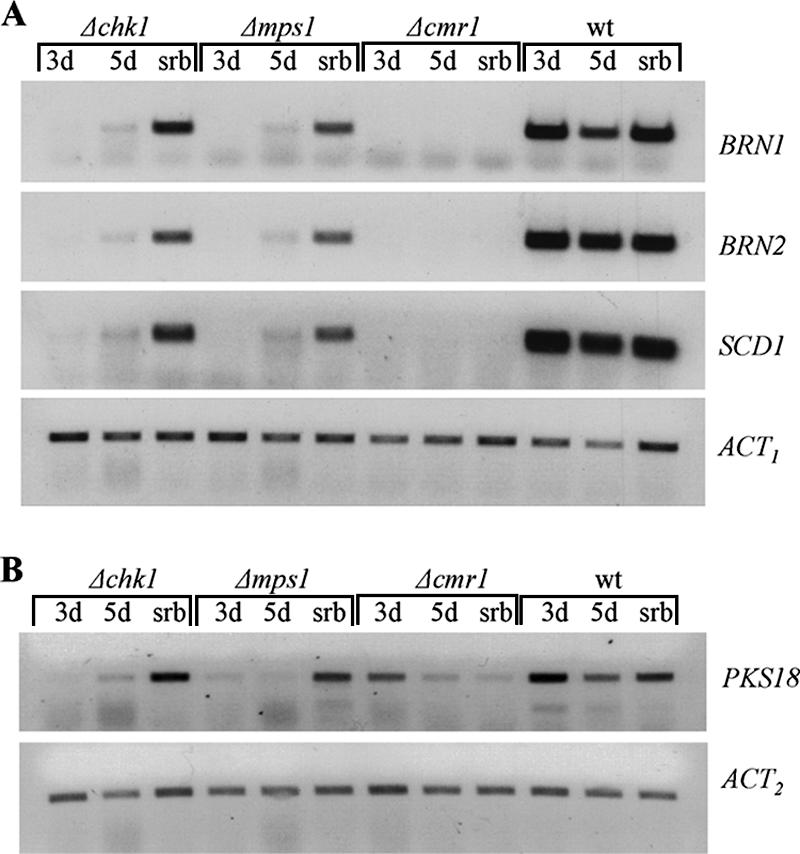
RT-PCR analysis of expression of the melanin biosynthesis genes BRN1, BRN2, and SCD1 (A) and PKS18 (B) in Δcmr1, Δchk1, and Δmps1 mutants and the wild-type strain (wt). The stationary liquid cultures were grown for 3 and 5 days in CMX (3d and 5d, respectively) or 5 days in CMX supplemented with 1.5 M sorbitol (srb). Expression of the actin-encoding gene (ACT) indicates relative RNA quantities in each sample. ACT1 and ACT2 indicate different primer pairs used for PCR (Table 1).
In MAPK mutants the expression of BRN1, BRN2, and SCD1 was undetectable (very low for PKS18) in 3-day-old cultures, barely detectable in 5-day-old CMX cultures, and significantly induced in the presence of sorbitol (Fig. 5A and B). These data indicate that both MAPKs, Chk1 and Mps1, are required for the expression of the melanin biosynthesis genes, but this dependence is overridden by the hyperosmotic stress. Nevertheless, even under conditions of hyperosmotic stress the expression of SCD1, BRN1, and BRN2 was decreased in MAPK mutants compared to the wild type. It appears that hyperosmotic stress signaling may bypass the MAPKs but not Cmr1 for the activation of melanin biosynthesis genes.
Coordinated expression of the sense and antisense CMR1 transcripts.
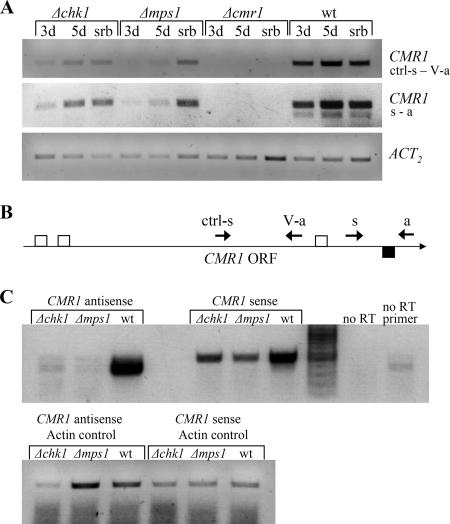
We tested the expression of the CMR1 gene in wild-type and chk1 and mps1 deletion strains using two different primer pairs. Surprisingly, the expression of CMR1 was in accordance with the expression of its target genes: it was reduced in 3-day-old cultures of MAPK mutants compared to the wild type and up-regulated in the aging cultures and hyperosmotic medium (Fig. 6A).
FIG. 6.
Expression of sense and antisense CMR1. (A) Semiquantitative RT-PCR analysis of CMR1 gene expression in wild type and Δchk1 and Δmpk1 mutants grown as described in the Fig. 5 legend. Polyadenylated mRNA was primed with standard oligo(dT) primer. Two primer pairs were used for the subsequent PCR: ctrl-s and V-a, and s and a (Table 1). Expression of the actin-encoding gene (ACT2) indicates relative RNA quantities in each sample. (B) Schematic representation of the primer and intron positions as related to the CMR1 open reading frame (ORF). Small arrows indicate primers used for RT and subsequent PCR. Open squares denote introns in the sense CMR1 transcript, whereas the filled square indicates an intron in the antisense CMR1. (C) Analysis of sense and antisense CMR1 transcript levels by directional cDNA synthesis. RNA used for RT-PCR was prepared from wild type and Δchk1 and Δmps1 mutants grown for 3 days in CMX, as described in the Fig. 5 legend. To examine antisense CMR1 expression, cDNA was synthesized with CMR1 primer s and actin primer act2-a to serve as an internal control. For sense CMR1 expression, RT was performed with CMR1 primer a and actin primer act2-a. Primers s and a (and act2-s and act2-a for actin) were used for the subsequent PCRs. Note that for detection of the antisense CMR1 34 PCR cycles were required, compared to 28 cycles for the sense transcript. wt, wild type.
We noticed that RT-PCR with primer pair s and a repeatedly produced two products in the wild-type strain, even at the highest annealing temperature. The main product was considerably more abundant than the other, shorter one (Fig. 6A). Sequencing of the shorter PCR product revealed the presence of an intron with reverse and complement splice sequences. This finding implies that CMR1 is transcribed in both the sense and antisense directions. To test this possibility, we performed directional CMR1 cDNA synthesis using different primers for RT: ctrl-s and s (antisense transcript), and a (sense transcript) (Fig. 6B). Antisense actin primer was added to each cDNA synthesis reaction to serve as an internal standard to indicate RNA quantity. Resulting cDNA was amplified by PCR using primers s and a for CMR1 and act2-s and act2-a for actin. Antisense CMR1 transcript was detected in the wild-type strain but was almost undetectable in the MAPK deletion strains (Fig. 6C). Note the different sizes of the product obtained from the sense and antisense transcripts: in the antisense, but not the sense, transcript an intron between primers s and a is spliced (Fig. 6B and C). The overall antisense CMR1 expression level was significantly lower than that of the sense CMR1. A BlastP search with putative protein products of the antisense CMR1 transcript produced no significant hits, suggesting that the antisense CMR1 is a noncoding transcript.
Activity of CMR1 promoter in absence of Cmr1 protein.
Since Cmr1 is a transcription factor, it is possible that it participates in the regulation of its own expression. We therefore replaced the CMR1 coding sequence with GFP to report CMR1 promoter activity (Fig. 7A). A CMR1p-GFP reporter strain was crossed with the mps1 deletion mutant to obtain Δmps1/CMR1p-GFP progeny. If Cmr1 is involved in its own regulation, we would expect the same low expression of GFP in both strains, CMR1p-GFP and Δmps1/CMR1p-GFP. On the other hand, in the case of regulation of CMR1 via another factor (MAPK dependent), we would expect decreased expression of GFP in a Δmps1/CMR1p-GFP strain compared to CMR1p-GFP. Our results show that the GFP transcript accumulated in the absence of the CMR1 coding sequence and to lower levels in the absence of MPS1 (Fig. 7B). This result implies that the decreased expression of CMR1 in Δmps1 seen in Fig. 6A cannot be explained by decreased levels of the Cmr1 protein. Thus, if CMR1 transcription is activated by the Mps1 pathway, a factor other than Cmr1 must be involved.
Melanin biosynthesis genes are clustered in several fungal species.
In fungi, genes encoding proteins related to the same developmental function or metabolic process are sometimes clustered. It was found that in Aspergillus fumigatus and Alternaria alternata some of the melanin biosynthesis genes are located in close groups (12, 33). We therefore investigated the chromosomal location of the C. heterostrophus melanin biosynthesis genes, compared to other fungal species. Genes BRN1, PKS18, and CMR1 of C. heterostrophus are located together on a 30-kb chromosomal fragment, and their mutual orientation is the same as in Alternaria brassicicola, a close relative of C. heterostrophus (Fig. 8). In Aspergillus fumigatus and Magnaporthe grisea melanin biosynthesis gene clusters also include multicopper oxidases which are probably required for the final 1,4-DHN polymerization step. In general, the most complete cluster among the species tested was found in A. fumigatus. This cluster might be the closest to some original ancestral melanin biosynthesis gene cluster (it is also the shortest cluster with the maximal number of genes), which apparently underwent significant shuffling during evolution, losing some of the genes in the process. Interestingly, the A. fumigatus cluster does not include a transcriptional regulator, unlike M. grisea, A. brassicicola, and C. heterostrophus.
DISCUSSION
One of the most prominent features in chk1 and mps1 MAPK mutants is loss of pigmentation. In this study we established that, at least in part, this loss of pigmentation is the result of underexpression of transcription factor CMR1 and its target genes involved in melanin biosynthesis.
C. heterostrophus produces melanin of the DHN type. It is likely that the first enzyme in the biosynthesis pathway is Pks18, based on its close homology to the Pks1 of B. oryzae (23). The C. heterostrophus genome includes tetra- and trihydroxynaphthalene reductases, BRN2 and BRN1, respectively, and scytalone dehydratase, SCD1. The last step in melanin biosynthesis is the polymerization of 1,8-DHN. In C. heterostrophus this step is catalyzed by p-diphenol oxidase, which has not been identified yet (32).
Cmr1 regulates transcription of BRN1, BRN2, and SCD1 genes, whereas the expression of PKS18 is only partially Cmr1 dependent. It is possible that upon accumulation of melanin biosynthesis intermediates in Δcmr1 mutants, feedback inhibition of PKS18 occurs. Another signaling pathway may participate in PKS18 regulation: in Aspergillus fumigatus the cyclic AMP/protein kinase A pathway regulates the expression of polyketide synthase, PKSP (2).
Mutants with mutations in cmr1 have a light orange-pink color, different from the white of the MAPK mutants. The different color may result from the accumulation of different melanin biosynthesis intermediates by cmr1 and MAPK mutants. We can hypothesize that in the absence of Cmr1 Pks18 activity is reduced, but not completely lost, and the melanin biosynthesis pathway is blocked after the Pks18 step, resulting in accumulation of T4HN and its derivative flaviolin. In MAPK mutants all genes of the melanin biosynthesis pathway, including PKS18, are coordinately down-regulated, leading to an overall decrease in melanin biosynthesis. Another possibility is that MAPKs control production of an additional reddish-colored secondary metabolite. In melanin-deficient cmr1 mutants the production of reddish pigment is normal, whereas MAPK mutants are deficient in both melanin and the reddish pigment.
The color of the chk1 and mps1 mutants darkened slightly during prolonged growth and was significantly darker in hyperosmotic medium. Correspondingly, the expression of the melanin biosynthesis genes was up-regulated under these conditions. Interestingly, the color of the cmr1 mutant changed to deep orange in hyperosmotic medium, but the expression of melanin biosynthesis genes was undetectable in this mutant under all conditions tested. We suggest that one of the stress-activated signaling pathways bypasses Chk1 and Mps1, but not Cmr1, to activate expression of melanin biosynthesis genes. This stress-activated pathway does not include the C. heterostrophus HOG1 homolog because in the absence of HOG1 the pigmentation significantly increases, indicating that HOG1 has rather an inhibitory effect on the expression of melanin biosynthesis genes (1; Igbaria et al., unpublished).
We have noticed that sometimes chk1 mutants formed dark-centered colonies with wide white margins on complete medium with xylose even in the absence of any stress condition. The expression of CMR1 and its downstream target genes was completely restored in these dark regions. Melanin biosynthesis genes are clustered in several fungal species, including A. fumigatus, M. grisea, and A. brassicicola and, as we established, in C. heterostrophus (Fig. 8). It is possible that MAPKs control the epigenetic state of chromatin through regulators like LaeA of Aspergillus spp. (9), and restoration of expression of melanin biosynthesis genes in chk1 mutants is caused by gradual heterochromatin-euchromatin transition in response to a stress-related signal, rather than by direct transcriptional activation. Further evidence to support this hypothesis comes from the identification of the probably noncoding CMR1 antisense transcript, which was almost undetectable in chk1 and mps1 deletion mutants. Notably, the ratio between CMR1 expression in wild type and in chk1 (mps1) deletion mutants is much greater for the antisense transcript than for the sense CMR1. The phenomenon of antisense transcription was extensively studied in mammalian genomes (37). In fungi, the Neurospora crassa FRQ gene is transcribed in sense and antisense directions, but only sense transcript codes for a protein product (14). Interestingly, coordinate regulation of the sense and antisense transcripts is common in the mammalian transcriptome, arguing against a model where the antisense transcripts silence the cognate sense RNA (7).
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the Israel Academy of Sciences (ISF 773 04).
We are grateful to Syngenta for access to sequence from the C. heterostrophus genome project and to B. Gillian Turgeon for the nourseothricin resistance cassette. We also thank Amir Sharon and his group for help and support during the final stages of work on this project.
Footnotes
Published ahead of print on 19 January 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bahn, Y. S., K. Kojima, G. M. Cox, and J. Heitman. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brakhage, A. A., and B. Liebmann. 2005. Aspergillus fumigatus conidial pigment and cAMP signal transduction: significance for virulence. Med. Mycol. 43(Suppl. 1):S75-S82. [DOI] [PubMed] [Google Scholar]
- 3.Braun, E., and R. Howard. 1994. Adhesion of Cochliobolus heterostrophus conidia and germlings to leaves and artificial surfaces. Exp. Mycol. 18:211-220. [Google Scholar]
- 4.Di Pietro, A., F. I. Garcia-MacEira, E. Meglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 5.Guillen, A., B. G. Turgeon, P. R. Thorson, C. R. Bronson, and O. C. Yoder. 1994. Linkage among melanin biosynthetic mutations in Cochliobolus heterostrophus. Fungal Genet. Newsl. 41:40. [Google Scholar]
- 6.Hou, Z., C. Xue, Y. Peng, T. Katan, H. C. Kistler, and J. R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 15:1119-1127. [DOI] [PubMed] [Google Scholar]
- 7.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 8.Kawamura, C., T. Tsujimoto, and T. Tsuge. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 9.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 10.Kihara, J., A. Moriwaki, M. Ito, S. Arase, and Y. Honda. 2004. Expression of THR1, a 1,3,8-trihydroxynaphthalene reductase gene involved in melanin biosynthesis in the phytopathogenic fungus Bipolaris oryzae, is enhanced by near-ultraviolet radiation. Pigment Cell Res. 17:15-23. [DOI] [PubMed] [Google Scholar]
- 11.Kihara, J., A. Moriwaki, M. Ueno, T. Tokunaga, S. Arase, and Y. Honda. 2004. Cloning, functional analysis and expression of a scytalone dehydratase gene (SCD1) involved in melanin biosynthesis of the phytopathogenic fungus Bipolaris oryzae. Curr. Genet. 45:197-204. [DOI] [PubMed] [Google Scholar]
- 12.Kimura, N., and T. Tsuge. 1993. Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175:4427-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kojima, K., T. Kikuchi, Y. Takano, E. Oshiro, and T. Okuno. 2002. The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant-Microbe Interact. 15:1268-1276. [DOI] [PubMed] [Google Scholar]
- 14.Kramer, C., J. J. Loros, J. C. Dunlap, and S. K. Crosthwaite. 2003. Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421:948-952. [DOI] [PubMed] [Google Scholar]
- 15.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langfelder, K., M. Streibel, B. Jahn, G. Haase, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 17.Leach, J., B. Lang, and O. Yoder. 1982. Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J. Gen. Microbiol. 128:1719-1729. [Google Scholar]
- 18.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96:13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrabi, R., T. Van der Lee, C. Waalwijk, and H. J. Gert. 2006. MgSlt2, a cellular integrity MAP kinase gene of the fungal wheat pathogen Mycosphaerella graminicola, is dispensable for penetration but essential for invasive growth. Mol. Plant-Microbe Interact. 19:389-398. [DOI] [PubMed] [Google Scholar]
- 20.Mendgen, K., M. Hahn, and H. Deising. 1996. Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu. Rev. Phytopathol. 34:367-386. [DOI] [PubMed] [Google Scholar]
- 21.Mey, G., K. Held, J. Scheffer, K. B. Tenberge, and P. Tudzynski. 2002. CPMK2, an SLT2-homologous mitogen-activated protein (MAP) kinase, is essential for pathogenesis of Claviceps purpurea on rye: evidence for a second conserved pathogenesis-related MAP kinase cascade in phytopathogenic fungi. Mol. Microbiol. 46:305-318. [DOI] [PubMed] [Google Scholar]
- 22.Mey, G., B. Oeser, M. H. Lebrun, and P. Tudzynski. 2002. The biotrophic, non-appressorium-forming grass pathogen Claviceps purpurea needs a Fus3/Pmk1 homologous mitogen-activated protein kinase for colonization of rye ovarian tissue. Mol. Plant-Microbe Interact. 15:303-312. [DOI] [PubMed] [Google Scholar]
- 23.Moriwaki, A., J. Kihara, T. Kobayashi, T. Tokunaga, S. Arase, and Y. Honda. 2004. Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 238:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Moriwaki, A., J. Kihara, C. Mori, and S. Arase. 16 March 2006. A MAP kinase gene, BMK1, is required for conidiation and pathogenicity in the rice leaf spot pathogen Bipolaris oryzae. Microbiol. Res. [Epub ahead of print.] doi: 10.1016/j.micres.2006.01.014. [DOI] [PubMed]
- 25.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nosanchuk, J. D., and A. Casadevall. 2003. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5:203-223. [DOI] [PubMed] [Google Scholar]
- 27.Pandey, A., M. G. Roca, N. D. Read, and N. L. Glass. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell 3:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimizu, K., C. Tanaka, and M. Tsuda. 1997. Cloning of Brn1, a reductase gene involved in melanin biosynthesis in Cochliobolus heterostrophus. J. Gen. Appl. Microbiol. 43:145-150. [DOI] [PubMed] [Google Scholar]
- 29.Takano, Y., T. Kikuchi, Y. Kubo, J. E. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13:374-383. [DOI] [PubMed] [Google Scholar]
- 30.Takano, Y., Y. Kubo, I. Kuroda, and I. Furusawa. 1997. Temporal transcriptional pattern of three melanin biosynthesis genes, PKS1, SCD1, and THR1, in appressorium-differentiating and nondifferentiating conidia of Colletotrichum lagenarium. Appl. Environ. Microbiol. 63:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka, C., Y. Kubo, and M. Tsuda. 1991. Genetic analysis and characterization of Cochliobolus heterostrophus colour mutants. Mycol. Res. 95:49-56. [Google Scholar]
- 32.Tanaka, C., S. Tajima, I. Furusawa, and M. Tsuda. 1992. The Pgr1 mutant of Cochliobolus heterostrophus lacks a p-diphenol oxidase involved in naphthalenediol melanin synthesis. Mycol. Res. 96:959-964. [Google Scholar]
- 33.Tsai, H. F., M. H. Wheeler, Y. C. Chang, and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuji, G., Y. Kenmochi, Y. Takano, J. Sweigard, L. Farrall, I. Furusawa, O. Horino, and Y. Kubo. 2000. Novel fungal transcriptional activators, Cmr1p of Colletotrichum lagenarium and pig1p of Magnaporthe grisea, contain Cys2His2 zinc finger and Zn(II)2Cys6 binuclear cluster DNA-binding motifs and regulate transcription of melanin biosynthesis genes in a developmentally specific manner. Mol. Microbiol. 38:940-954. [DOI] [PubMed] [Google Scholar]
- 35.Turgeon, B. G., H. Bohlmann, L. M. Ciuffetti, S. K. Christiansen, G. Yang, W. Schaefer, and O. C. Yoder. 1993. Cloning and analysis of the mating type genes from Cochliobolus heterostrophus. Mol. Gen. Genet. 238:270-284. [DOI] [PubMed] [Google Scholar]
- 36.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7:3297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Werner, A., and A. Berdal. 2005. Natural antisense transcripts: sound or silence? Physiol. Genomics 23:125-131. [DOI] [PubMed] [Google Scholar]
- 38.Wirsel, S., B. G. Turgeon, and O. C. Yoder. 1996. Deletion of the Cochliobolus heterostrophus mating-type (MAT) locus promotes the function of MAT transgenes. Curr. Genet. 29:241-249. [PubMed] [Google Scholar]
- 39.Xu, J. R., and J. E. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10:2696-2706. [DOI] [PubMed] [Google Scholar]
- 40.Xu, J. R., C. J. Staiger, and J. E. Hamer. 1998. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 95:12713-12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng, L., M. Campbell, J. Murphy, S. Lam, and J. R. Xu. 2000. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13:724-732. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.