Abstract
Cyclin protein behavior has not been systematically investigated in multinucleated cells with asynchronous mitoses. Cyclins are canonical oscillating cell cycle proteins, but it is unclear how fluctuating protein gradients can be established in multinucleated cells where nuclei in different stages of the division cycle share the cytoplasm. Previous work in A. gossypii, a filamentous fungus in which nuclei divide asynchronously in a common cytoplasm, demonstrated that one G1 and one B-type cyclin do not fluctuate in abundance across the division cycle. We have undertaken a comprehensive analysis of all G1 and B-type cyclins in A. gossypii to determine whether any of the cyclins show periodic abundance across the cell cycle and to examine whether cyclins exhibit functional redundancy in such a cellular environment. We localized all G1 and B-type cyclins and notably found that only AgClb5/6p varies in subcellular localization during the division cycle. AgClb5/6p is lost from nuclei at the meta-anaphase transition in a D-box-dependent manner. These data demonstrate that efficient nuclear autonomous protein degradation can occur within multinucleated cells residing in a common cytoplasm. We have shown that three of the five cyclins in A. gossypii are essential genes, indicating that there is minimal functional redundancy in this multinucleated system. In addition, we have identified a cyclin, AgClb3/4p, that is essential only for sporulation. We propose that the cohabitation of different cyclins in nuclei has led to enhanced substrate specificity and limited functional redundancy within classes of cyclins in multinucleated cells.
The timing and fidelity of the eukaryotic cell cycle requires the regulated activity of a central kinase, the cyclin-dependent kinase (CDK), which functions in complex with cyclin proteins. Cyclins work to activate CDKs and to direct the CDK to specific substrates. Most cyclin proteins are controlled through regulated transcription, subcellular localization, and degradation so that they are present for a limited window of time and in a restricted cell compartment (38). The choreographed arrival and disappearance of different cyclins in different cell cycle phases is an integral feature of most eukaryotic cell cycles.
Previous work in the filamentous fungus Ashbya gossypii has demonstrated that this paradigm may not universally describe cyclin behavior in certain multinucleated cells (22). In A. gossypii hyphal cells, multiple nuclei reside in one cytoplasm and divide asynchronously. Nuclei transit the division cycle with different rates and appear to divide without regard for the progression of neighboring nuclei that are on average only 4 to 5 μm apart. Unexpectedly, one B-type cyclin homologue, AgClb1/2p, and one G1 cyclin, AgCln1/2p, do not appear to vary in abundance across the nuclear division cycle. Thus, the B-type cyclin protein is found in anaphase and G1 nuclei, and the G1 cyclin is found in mitotic nuclei. This unique behavior of these two cyclins may stem from cytoplasmic translation in the syncytium combined with nuclear asynchrony. In such situations, newly made cyclin proteins that are expressed from nuclei that are out of phase may be continually supplied to all nuclei residing in the common cytoplasm.
Cyclin protein behavior has been studied in relatively few syncytial cells. In early Drosophila embryos, in which nuclei synchronously divide, the total pool of free cyclin B does not vary, but cyclin destruction is essential for mitosis. It has been shown that this degradation is spatially restricted to along the spindle and thus impacts only a fraction of the total cyclin protein pool (27, 42). In early Arabidopsis seed development, the syncytial endoderm contains domains of synchronously dividing nuclei and the cyclin B protein appears in mitotic but not interphase nuclei, suggesting that cyclin abundance is tightly controlled in these multinucleated compartments (8). Thus, some syncytial cells have been reported to degrade B-type cyclins for mitotic exit; however, in all cases these nuclei are undergoing synchronous division.
Cyclin function has been investigated in several hypha-forming fungi in addition to A. gossypii, including Candida albicans and Ustilago maydis. In C. albicans, the G1 cyclins CaCln3p and CaCln1p are implicated in the initiation and maintenance of normal polarized bud and hyphal growth in response to different environmental stimuli (5, 11, 31). In contrast, the B-type cyclins CaClb2p and CaClb4p were required to limit polarized growth under nonhyphal inducing conditions in C. albicans (7). Thus, in C. albicans both classes of cyclins are needed to varying degrees for normal rates of cell cycle progression and display cell cycle-regulated abundance, although their subcellular localization has not yet been determined. In U. maydis, a maize smut fungus, G1 cyclins contribute to mating and morphogenesis (10). Two U. maydis B-type cyclins are nonredundant and essential for cell cycle progression, and their regulated degradation is required for normal division (10, 19). None of these studies, however, examined the behavior of cyclins in a large, true syncytial form of the fungal cells or in the context of asynchronous mitoses.
The relative paucity of information about the subcellular distribution of cyclins in multinucleated cells and the provocative observation that two different cyclins in A. gossypii did not detectably oscillate led us to comprehensively evaluate all of the cyclins that are predicted to complex with the central CDK in A. gossypii, AgCdc28p. A. gossypii and Saccharomyces cerevisiae are believed to have diverged from a common ancestor over 100 million years ago, and evidence of this relationship is derived from the A. gossypii genome sequence, where more than 90% of the genes are homologous and syntenic to budding yeast genes (14). The whole genome duplication in budding yeast resulted in many pairs of genes where only a single homologue is present in A. gossypii. In the case of cyclins, A. gossypii has two G1 cyclins (AgCLN3 and AgCLN1/2) and three B-type cyclins (AgCLB5/6, AgCLB3/4, and AgCLB1/2), in contrast to S. cerevisiae with three G1 and six B-type cyclins. The A. gossypii cyclins are named according to their syntenic homologue in budding yeast. With these studies we had two main aims. First, we wanted to examine whether the A. gossypii cyclins display functional redundancy when directing asynchronous mitoses. Both S. cerevisiae and Schizosaccharomyces pombe cyclins share many overlapping functions despite carefully controlled and timed abundance, and we wanted to test whether such redundancy may be absent in cells where multiple types of cyclins may cohabitate. Second, we wanted to examine the localization of the uncharacterized A. gossypii cyclins across the nuclear division cycle to determine whether any cyclin proteins can be controlled in abundance during asynchronous division.
MATERIALS AND METHODS
A. gossypii strains and growth conditions.
A. gossypii media, culturing, and transformation protocols are described elsewhere (4, 45). The genotype Δleu2 Δthr4 of the A. gossypii strain ATCC 10895 (reference strain) was used to generate all A. gossypii strains. Either pGEN3 (45) or pGUG (29) was used as a template to generate gene-targeting cassettes encoding Geneticin resistance or green fluorescent protein (GFP) plus Geneticin resistance, respectively. The strains were verified by PCR and are listed in Table 1.
TABLE 1.
A. gossypii and S. cerevisiae strains used in this studya
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| Wild type | Agleu2Δ thr4Δ | 1 |
| AKHAg1 | Agclb1/2Δ::GEN3 leu2Δ thr4Δ/AgCLB1/2 leu2Δ thr4Δ | This study |
| AKHAg3 | Agclb3/4Δ::GEN3 leu2Δ thr4Δ | This study |
| ASG7 | Agcln1/2Δ::GEN3 leu2Δ thr4Δ/AgCLN1/2 leu2Δ thr4Δ | This study |
| ASG29 | Agcln3Δ::GEN3 leu2Δ thr4Δ | This study |
| AKHAg7 | AgCLB1/2-GFP-GEN3 leu2Δ thr4Δ | This study |
| AKHAg8 | Agclb5/6Δ::GEN3 leu2Δ thr4Δ/AgCLB5/6 leu2Δ thr4Δ | This study |
| AgHPH04 | Agleu2Δ thr4 ade2::ADE2-H4-GFP | H. Helfer |
| AKHAg9 | Agclb1/2Δ::GEN3 ade2Δ(310-566)::ADE2-H4-GFP leu2Δ thr4Δ/AgCLB1/2 ade2Δ(310-566)::ADE2-H4-GFP leu2Δ thr4Δ | This study |
| AKHAg31 | AgCLB5/6-GFP-GEN3 leu2Δ thr4Δ | This study |
| AKHAg42 | pAKH52 [AgCLB5/6-13myc-GEN3] leu2Δ thr4Δ | This study |
| AKHAg46 | AgCLB5/6-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg47 | AgCLB3/4-GFP-GEN3 leu2Δ thr4Δ | This study |
| AKHAg48 | AgCLB3/4-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg54 | AgCLN3-GFP-GEN3 leu2Δ thr4Δ | This study |
| AKHAg55 | AgCLN3-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg64 | AgCLB5/6-NESa-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg65 | AgCLB5/6-NESi-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg66 | pAKH72 [AgCLB5/6Δdb1-13myc-GEN3] leu2Δ thr4Δ | This study |
| AKHAg67 | pAKH73 [AgCLB5/6Δdb2-13myc-GEN3] leu2Δ thr4Δ | This study |
| AKHAg68 | pAKH74 [AgCLB5/6Δ(db1-db2)-13myc-GEN3] leu2Δ thr4Δ | This study |
| AKHAg69 | pAKH75 [AgCLB5/6Δdb1Δdb2-13myc-GEN3] leu2Δ thr4Δ | This study |
| AKHAg72 | AgCLB5/6-NLSa-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg73 | AgCLB5/6-NLSi-13myc-GEN3 leu2Δ thr4Δ | This study |
| AKHAg76 | Agclb5/6Δ::GEN3 ade2Δ (310-566)::ADE2-H4-GFP leu2Δ thr4Δ/AgCLB5/6 ade2Δ(310-566)::ADE2-H4-GFP leu2Δ thr4Δ | This study |
| AKHAg78 | Agcln3Δ::GEN3 ade2Δ (310-566)::ADE2-H4-GFP leu2Δ thr4Δ | This study |
| AKHSc1 | pAKH67 [PGAL1AgCLB5/6-13myc] (S. cerevisiae) | This study |
| AKHSc2 | pAKH68 [PGAL1AgCLB5/6Δdb1-13myc] (S. cerevisiae) | This study |
| AKHSc3 | pAKH69 [PGAL1AgCLB5/6Δdb1Δdb2-13myc] (S. cerevisiae) | This study |
| AKHSc4 | pAKH70 [PGAL1AgCLB5/6Δdb2-13myc] (S. cerevisiae) | This study |
| AKHSc5 | pAKH71 [PGAL1AgCLB5/6Δ(db1-db2)-13myc] (S. cerevisiae) | This study |
| AKHSc6 | AY52 + pCLB2-13myc (S. cerevisiae) | This study |
All AKHAg and ASG strains are A. gossypii derived from the reference strain (leu2Δ thr4Δ). Individual nuclei are haploid, and in homokaryotic strains all nuclei have the same genotype. In heterokaryons, where different nuclei have different genotypes, the two different genotypes are written, separated by a slash. A “p” denotes A. gossypii or S. cerevisiae strains transformed with the respective plasmid.
DNA manipulations.
All DNA manipulations were carried out as described previously (40) using DH5αF′ as host (25). The plasmids generated and used here are listed in Fig. S2 in the supplemental material. PCR was performed by using standard methods with Taq polymerase, the Expand High-Fidelity PCR system, and the GC-Rich PCR system from Roche (Basel, Switzerland). Oligonucleotides were synthesized at MWG (Ebersberg, Germany) or Microsynth (Balgach, Switzerland) for polyacrylamide gel electrophoresis-purified oligonucleotides. Oligonucleotide primers are listed in Fig. S3 in the supplemental material. All restriction enzymes came from New England Biolabs or Roche.
Plasmid isolation from yeast was performed as described previously (41) using DHD5 as a yeast reference strain (3). All sequencing to verify plasmids was done at MWG.
Generation of A. gossypii deletion strains.
A. gossypii deletion mutants were generated by using a PCR-based one-step gene targeting approach (6, 43, 45). A. gossypii deletion mutants were made in Δleu2 Δthr4 using pairs of “gene name”-S1 or -S2 oligonucleotides, which contained a 20-amino-acid (aa) binding region to pGEN3 and at least 45-bp homology upstream and downstream of the open reading frames. Δleu2 Δthr4 young mycelia were transformed with PCR products amplified off the pGEN3 template (containing the open reading frame of the kanamycin resistance gene under the control of promoter and terminator sequences of the ScTEF2 gene). The primary transformation produced heterokaryon mycelia, which contained a mixture of wild-type and transformed nuclei; thus, even mutations in essential genes produce viable transformants. Heterokaryon and homokaryon transformants were verified by analytical PCR with oligonucleotides “gene name”-G1 or -G2 (Geneticin marker) and “gene name”-G4 with G3 (Geneticin marker). For each deletion, three independent transformants were characterized. To evaluate phenotypes of lethal mutants, the heterokaryon was sporulated to produce uninucleated spores, which were then germinated under selective conditions.
To generate AKHAg9, AKHAg76, and AKHAg78, the deletions of AgCLB1/2, AgCLB5/6, and AgCLN3 were constructed in the background of AgHPH04 (H. Helfer, unpublished data), which contains HH1-GFP (histone H4-GFP), stably integrated at the ADE2 locus.
Generation of the tagged A. gossypii strains.
The tagging of the cyclins was performed via cotransformation in yeast by using a PCR fragment containing 45-bp homology to the cyclin gene cloned in pRS416. This plasmid was either taken from the A. gossypii pAG clone collection (pAG9687 for AgCLB5/6 or pAG7578 for AgCLB1/2) (14), or the plasmid was constructed by amplifying the gene from genomic DNA. To construct pAKH53 (containing AgCLN3), genomic DNA was amplified with the oligonucleotides CLN3-P1 and CLN3-P2, containing at the 5′ ends restriction sites for BamHI and XbaI, respectively. The PCR fragment obtained (2,296 bp, containing AgCLN3) was cut with these enzymes resulting in a 2,273-bp fragment and ligated into cut and column-purified pRS416 vector. The resulting pAKH53 was sequenced to avoid mutations due to random PCR errors. pAKH54 (containing AgCLB3/4) was constructed by amplifying the gene from genomic DNA with the oligonucleotides CLB3/4-P1 and CLB3/4-P2, containing at the 5′ ends restriction sites for HindIII and XbaI, respectively. The PCR product (2,148 bp, containing AgCLB3/4) was cut with HindIII and XbaI, resulting in a fragment of 2,125 bp, which was ligated into a HindIII- and XbaI-cleaved and column-purified pRS416 plasmid and transformed into Escherichia coli. This yielded pAKH54, which was verified by sequencing.
AgCLB5/6 was subcloned from pAG9687 by digestion with NotI and XhoI, resulting in a fragment of 2,120 bp. This fragment was gel purified and ligated into the NotI- and XhoI-cut pRS415, leading to pAKH1, which was verified by restriction digests.
To C-terminally GFP tag the cyclin genes, the pGUG cassette (29) was amplified with pairs of “gene name”-GS1/GS2 oligonucleotides containing 45-bp homology to the C terminus of the gene. The resulting PCR product (∼2,850 bp, containing the GFP with the ScURA3 terminator and GEN3) was cotransformed in yeast with the plasmid pAG9687 (for AgCLB5/6), pAg7578 (for AgCLB1/2), pAKH53 (for AgCLN3), or pAKH54 (for AgCLB3/4) to produce pAKH40, pAKH7, pAKH58, and pAKH55, respectively. pAKH40 was digested with SalI and StuI, pAKH7 was digested with FspI and SpeI, pAKH58 was digested with PstI, and pAKH55 was digested with NheI and AatII before transformation into A. gossypii mycelia. This produced AKHAg31, AKHAg7, AKHAg54, and AKHAg47, which were verified by PCR using oligonucleotides “gene name”-I1 and GG2 (in GFP) and “gene name”-G4 and -G3 (in the Geneticin marker).
To C-terminally myc tag the cyclin genes, the pAg13-myc cassette (22, 32) was amplified with oligonucleotides CLB5mycF and CLB5mycR for AgCLB5/6, nucleotides CLN3-M1 and CLN3-M2 for AgCLN3, and nucleotides CLB3/4-myc-P1 and CLB3/4-myc-P2 for AgCLB3/4. The resulting PCR products were cotransformed in yeast with the plasmids pAG9687 (for AgCLB5/6), pAKH53 (for AgCLN3), and pAKH54 (for AgCLB3/4) to produce pAgCLB5/6-13myc (pAKH52), pAgCLN3-13myc (pAKH59), and pAgCLB3/4-13myc (pAKH56). pAKH52 was digested with NheI, KpnI, and NdeI, pAKH59 was digested with AatII and PpuMI, and pAKH56 was digested with NheI and AatII before transformation of A. gossypii mycelia to tag the endogenous cyclin genes. This created the homokaryotic AKHAg46 (AgCLB5/6-13myc), AKHAg55 (AgCLN3-13myc), and AKHAg48 (AgCLB3/4-13myc) strains, which were verified with the oligonucleotides CLB5MycI, CLN3-I1, CLB3/4-I1, and MycI (in 13myc) and CLB5-G4, CLN3-G4, CLB3/4-G4, and -G3 (in the Geneticin marker).
To mislocalize AgClb5/6p, two exogenous nuclear localization signal (NLS) or nuclear export signal (NES) sequences were integrated at the 3′end of AgCLB5/6. Forced localization cassettes pAKH18 (containing NLSactive-13myc) and pAKH19 (containing NLSinactive-13myc) cassettes (22) were amplified with the oligonucleotides CLB5/6-NLS-A-P1 (for NLSa) or CLB5/6-NLS-I-P1 (for NLSi) and CLB5/6-NES-Myc-P2, and the pAKH20 (containing NESa-13myc) and pAKH21 (containing NESi-13myc) cassettes were amplified with the oligonucleotides CLB5/6-NES-P1 and CLB5/6-NES-Myc-P2, each giving rise to a 2,800-bp fragment. These PCR fragments were cotransformed into yeast cells together with pAG9687 to generate pAKH76 (AgCLB5/6-NLSa-13myc) and pAKH77 (AgCLB5/6-NLSi-13myc), both of which were verified by sequencing. About 30 μg of pAKH76 and pAKH77 was digested with ApaI and BsrGI, and the cut DNA was transformed into small A. gossypii mycelia, giving rise to the strains AKHAg72, AKHAg73, AKHAg64, and AKHAg65, which were controlled by verification PCR with the primers CLB5-I1 and MycI (in 13myc) and CLB5-G1-G2 (in the Geneticin marker).
The Agclb5/6 D-box mutants were made by using an overlap PCR approach, which deleted aa 22 to 31 (Δdb1), aa 38 to 47 (Δdb2), aa 22 to 31 and aa 38 to 47 (Δdb1 Δdb2), or aa 22 to 47 (Δ[db1-db2]) in AgClb5/6p, and all were confirmed by sequencing. AgCLB5/6Δdb2, AgCLB5/6Δdb1Δdb2, and AgCLB5/6Δ(db1db2) were lethal in S. cerevisiae; therefore, in order to construct these plasmids, the inducible promoter GAL1 was placed in front of AgCLB5/6-13myc. The pFA6a-his3mx6-Pgal1 cassette was amplified with the oligonucleotides Gal_CLB5/6-F and Gal_CLB5/6-R, resulting in a PCR fragment of 1,996 bp. This PCR fragment was cotransformed into yeast together with pAKH63 (pAgCLB5/6-13myc) to generate pAKH67, which was verified by sequencing. To generate pAKH68 (PGAL1AgCLB5/6Δdb1-13myc), pAKH67 was used as a template for a PCR with GAL1-CLB5-dbA and CLB5db1B to produce a 470-bp product and a second reaction with CLB5db1C and CLB5dbD to create a 428-bp product. Oligonucleotide GAL-CLB5-dbA was homologous to a region 400 bp upstream, and clb5dbD was homologous to a region 400 bp downstream of the D-box1. Oligonucleotide CLB5dbB contained homology to AgCLB5/6 immediately upstream of the D-box1 but lacked the D-box1 sequences between nucleotides 64 and 93. In addition, at the 3′ end this oligonucleotide has homology to the region immediately downstream of the D-box. This sequence is also present in the 5′ half of oligonucleotide CLB5db1C. This overlapping homology was then used in a second PCR involving the two products of A+B and C+D and additional GAL-CLB5-dbA and CLB5dbD primers to create a 845-bp fragment containing an in-frame deletion of the D-box1. To generate a full-length AgCLB5/6Δdb1 mutant, this fragment was cotransformed into yeast with previously Nhe1 cut, dephosphorylated and gel purified pAKH67. The gap-repaired plasmid containing the deleted D-box1 was confirmed by sequencing and was called pCLB5/6Δdb1-13myc (pAKH68). Similarly, to generate pAKH70 (PGAL1AgCLB5/6Δdb2-myc), oligonucleotides GAL-CLB5-dbA and CLB5db2B were used with pAKH67 as a template in the first reaction and CLB5db2C and CLB5dbD in the second reaction, resulting in two PCR fragments of 485 and 380 bp, which were then handled just as for AgCLB5/6Δdb1.
For the construction of pAKH69 (PGAL1AgCLB5/6Δdb1Δdb2-13myc), the same procedure was performed as for the Δdb1 and Δdb2 but with the template pAKH68 and CLB5db2B_in_db1. To delete both D-boxes (Δ[db1-db2]), a similar approach used pAKH67 as a template and the oligonucleotides GAL-CLB5dbA and CLB5dbdB in the first PCR and CLB5db2C and CLBdbD in the second PCR. Since these D-box mutants are all expressed from the inducible GAL1 promoter, which is not functional in A. gossypii, the GAL1 was removed by digestion with AatII. A 1,900-bp fragment was excised from pAKH68 (Δdb1), pAKH70 (Δdb2), pAKH71 (Δ[db1-db2]), and pAKH69 (Δdb1 Δdb2), and each plasmid was self-religated, resulting in pAKH72, pAKH73, pAKH74, and pAKH75, respectively, which are expressed from their endogenous A. gossypii promoters.
Immunofluorescence and Hoechst staining.
A. gossypii cells were processed for immunofluorescence as described earlier (22). Anti-myc and anti-tubulin immunofluorescence were performed sequentially so as to limit cross-reactivity, beginning with mouse anti-myc antibody (Santa Cruz Biotechnology, Inc.) at 1:100, Alexa Fluor 488 anti-mouse antibody (Invitrogen) at 1:200, rat anti-alpha-tubulin antibody (YOL34; Serotec) at 1:50, and Alexa Fluor 568 anti-rat antibody (Invitrogen) at 1:200 with Hoechst (Invitrogen) dye to visualize nuclei at 5 μg/ml. Antibody dilutions and washes were performed with phosphate-buffered saline plus 1.0 mg of bovine serum albumin/ml.
Microscopy.
Microscopy pictures were taken with an Axioplan 2 imaging microscope (Carl Zeiss) with a Plan Neofluar 100× Ph3 N.A. 1.3 objective lens. It was equipped with a 75 WXBO and a 100 WHBO illumination source controlled by a MAC2000 shutter and filter wheel system (Ludl Electronics). The camera was a TE/CCD-1000PB back-illuminated cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ). The following filter sets for different fluorophores were used: #02 for Hoechst, #15 for Rhodamine 568, and #09 for GFP (Carl Zeiss). The excitation intensity was controlled with different neutral density filters (Chroma Technology). MetaMorph 4.6r9 software (Universal Imaging Corp.) controlled the microscope, camera, and Ludl shutter controller (Ludl Electronics) and was used for processing images.
Stacks of tubulin images were processed by using “no-neighbors” deconvolution within MetaMorph and, like all other fixed specimens, the Z-stacks were compressed using “stack arithmetic,” the brightness and contrast were adjusted, and the images were overlaid using color alignment.
RESULTS
Functional analysis of cyclins in A. gossypii.
Single-deletion mutants were generated for each of the five cyclin genes in A. gossypii (Table 2) and were evaluated for defects in nuclear division and morphology. Altered nuclear division cycle phenotypes in these mutants confirmed predictions based on sequence and synteny that these proteins are involved in nuclear cycle progression. However, some of these mutants displayed additional, surprising phenotypes indicating roles in sporulation and morphogenesis (Fig. 1).
TABLE 2.
Overview of the A. gossypii cyclins and their homologues in S. cerevisiaea
| A. gossypii cyclin | Homologue in S. cerevisiae
|
% Identity to S. cerevisiae homologue
|
A. gossypii protein size (aa) | Size (aa) of homologue in S. cerevisiae
|
A. gossypii deletion | Deletion in S. cerevisiae homologueb
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1° | 2° | 1° | 2° | 1° | 2° | 1° | 2° | |||
| AgCLN1/2 (AFL174C) | ScCLN1 (YMR199W) | ScCLN2 (YPL256C) | 63 | 60 | 513 | 545 | 546 | Lethal | Viable* | Viable* |
| AgCLN3 (ADR384W) | ScCLN3 (YAL040C) | 34 | 459 | 580 | Viable | Viable | ||||
| AgCLB1/2 (AAR099W) | ScCLB2 (YPR119W) | ScCLB1 (YGR108W) | 65 | 56 | 556 | 491 | 471 | Lethal | Viable† | Viable† |
| AgCLB3/4 (ADR068W) | ScCLB4 (YLR210W) | ScCLB3 (YDL155W) | 52 | 51 | 376 | 460 | 427 | Viable | Viable* | Viable* |
| AgCLB5/6 (AAR100C) | ScCLB5 (YPR120C) | ScCLB6 (YGR109C) | 44 | 39 | 405 | 435 | 380 | Lethal | Viable* | Viable* |
In the cases where a single A. gossypii gene is homologous to two S. cerevisiae genes due to the whole genome duplication in budding yeast, 1° and 2° refer to the homologue with the highest and second highest degrees of homology, respectively, to the A. gossypii sequence.
*, Double deletion viable; †, double deletion lethal.
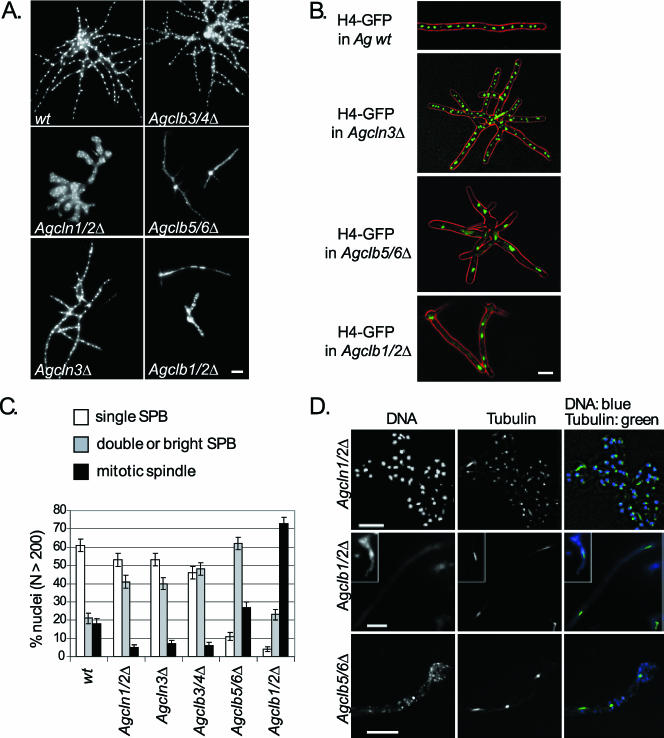
FIG. 1.
Phenotypes of cyclin gene deletions in A. gossypii. (A) Spores from wild type (wt), Agcln1/2Δ, Agcln3Δ, Agclb3/4Δ, Agclb5/6Δ, and Agclb1/2Δ strains were inoculated in liquid media, grown under selection (except for the wild type) for 14 to 16 h, and evaluated for morphology and nuclear density using Hoechst dye. Bar, 10 μm. (B) In vivo nuclear staining with H4-GFP in A. gossypii wild type (wt) and selected cyclin gene deletions. Strains were grown for 14 to 16 h in liquid media at 30°C. In the overlays, the differential interference contrast is red and the H4-GFP is green. Bar, 10 μm. (C) The strains described in panel A were scored for nuclear cycle stage based on tubulin immunofluorescence and categorized as having either a single SPB, a duplicated or brighter SPB, or a mitotic spindle. More than 200 nuclei were scored for each mutant. (D) Tubulin in cyclin mutants. In the top panel, Agcln1/2Δ cells show predominantly nuclei with either single and duplicated SPBs in aberrantly shaped hyphae. In the middle panel, low nuclear density in Agclb1/2Δ cells shows arrest with mitotic spindles and one to four nuclei. In the bottom panel, Agclb5/6Δ cells show nuclei arrested with predominantly duplicated SPBs and mitotic spindles. In overlays, DNA is blue, and tubulin is green. Bars, 10 μm.
Deletion of the G1 cyclin Agcln1/2Δ led to gross morphology defects in mutant cells. Agcln1/2Δ cells cycled through many rounds of nuclear division after germination but ultimately arrested growth as microcolonies. These hyphae were swollen and had aberrant branching patterns, which may be the consequence of a diffuse or unstable polarity site at hyphal tips (Fig. 1A). The localization of AgCln1/2p at the tip of hyphae shown by Gladfelter et al. (22) supports a potential role for this cyclin in polarity control. In contrast, the loss of AgCLN3, the only other gene encoding a G1 cyclin predicted to complex with AgCdc28p in A. gossypii, led to no obvious growth defects, but the distribution of nuclei across stages of the division cycle differed from that of wild-type cells. Nuclei were observed both in fixed Agcln3Δ cells using Hoechst dye and in living cells with a histone-GFP label. By both methods, nuclear morphology and distribution were comparable to wild-type cells (Fig. 1A and B). The nuclear division cycle stage was assayed in Agcln3Δ mutant cells by using anti-alpha-tubulin immunofluorescence to visualize spindles and spindle pole bodies (SPBs). Nuclei were categorized by SPB and/or spindle appearance and evaluated as having (i) a single SPB; (ii) adjacent, duplicated SPBs or SPBs that are larger in diameter and ≥2-fold brighter than single SPBs; (iii) a spindle aligned across the middle of the DNA (meta); or (iv) a spindle >2.8 μm long connecting two separated DNA signals (ana). Deletion of either G1 cyclin led to a decrease in the proportion of mitotic spindles and an increase in the proportion of nuclei with bright SPBs (category ii), suggesting a delay early in the division cycle in the G1/S transition (Fig. 1C and D).
Three B-type cyclin genes that are homologous and syntenic to budding yeast genes are found in the A. gossypii genome. Deletion of the predicted “S-phase cyclin” gene AgCLB5/6 led to an arrest of growth with stunted hyphal formation and fragmented nuclei (Fig. 1A). Up to 10 nuclei could be observed in mutant cells expressing histone-GFP that were imaged before lethal arrest, suggesting that only a few rounds of mitoses are possible in the absence of AgClb5/6p (Fig. 1B). Where it was possible to still observe SPBs and tubulin, the majority of nuclei appeared to have duplicated SPBs or short mitotic spindles. The inviability of Agclb5/6Δ cells suggests that there is little functional redundancy between the B-type cyclins in A. gossypii. This may be due to significantly different substrate specificities between the B-type cyclins or to limitations due to the spatial organization or compartmentalization of nuclei within multinucleated cells.
Unlike Agclb5/6Δ mutants, Agclb3/4Δ cells formed mature mycelia with normal nuclear density; however, the nuclear cycle appears to be delayed prior to spindle assembly and spore formation was completely abolished, suggesting a specific function for this cyclin in sporulation (Fig. 1). Finally, the Agclb1/2Δ mutants generally arrested growth with a primary germ tube and fragmented nuclei (Fig. 1). Many of these inviable hyphae contained one to two short mitotic spindles. Two to four nuclei were observed in newly germinated mutants expressing histone-GFP, suggesting that one or at most two mitoses occur before this arrest in early mitosis (Fig. 1B). These limited mitoses may result from mitotic cyclin protein which was packed into spores in the heterokaryon, where only a subset of nuclei contain the deleted cyclin gene. Thus, three of the five cyclin deletions were lethal, indicating that there is little functional redundancy within the G1 or B-type classes of cyclins in A. gossypii. These data are especially striking because both S. cerevisiae and S. pombe cells are able to proliferate with only a single B-type cyclin, suggesting that these multinucleated filamentous hyphal cells may have different regulatory requirements for cell cycle progression (18, 24).
The S-phase cyclin AgClb5/6p levels oscillate during the cell cycle.
The AgClb1/2p and AgCln1/2p do not undergo major fluctuations in abundance across the nuclear division cycle (22), and here we evaluated whether the other cyclins AgCln3p, AgClb3/4p, and AgClb5/6p oscillate across the nuclear division cycle. The predicted G1 cyclin AgCLN3 and the B-type cyclins AgCLB3/4 and AgCLB5/6 were epitope tagged at their endogenous loci with 13 copies of the c-myc epitope, and the tagged strains were evaluated for growth and nuclear division. Unfortunately, AgCLB3/4-13myc did not display normal growth and failed to sporulate. Thus, localization of this cyclin cannot be determined. However, AgCLN3-13myc and AgCLB5/6-13myc strains grew normally, and the localization of these tagged cyclin proteins was investigated by indirect immunofluorescence. AgCln3p-13myc was concentrated in nuclei and present during all stages of the nuclear cycle (Fig. 2A, top, and B, left). The AgCln3p signal was very weak compared to other proteins visualized thus far in A. gossypii, and changes in abundance of the protein in the nucleus through time could not be detected, but we do not know whether this is due to problems resolving low levels of protein by immunofluorescence.
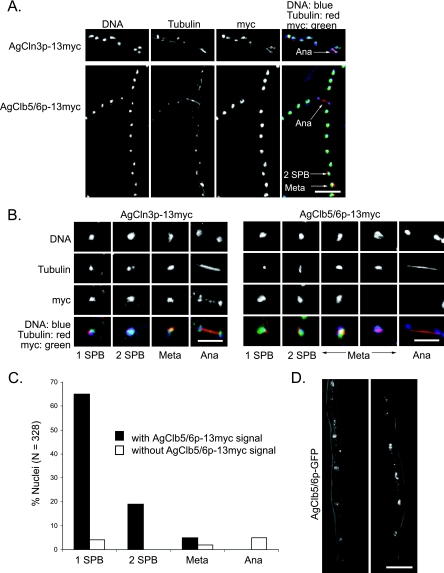
FIG. 2.
Localization of the G1 cyclin AgCln3p and the S-phase cyclin AgClb5/6p during the nuclear cycle. (A) Spores from the strains AgCLN3-13myc and AgCLB5/6-13myc were grown in liquid medium for 14 to 16 h at 30°C under selection, followed by fixation and immunostaining. Arrows highlight the nuclei of different nuclear stages. (B) Individual nuclei representing different spindle stages from the strains described in panel A. In overlays, tubulin is red, cyclins are green, and nuclei are blue; therefore, nuclei containing cyclins appear turquoise. (C) Percent nuclei in each spindle stage with or without AgClb5/6p-13myc signal, based on anti-tubulin and anti-myc immunostaining. More than 300 nuclei were scored. (D) Two examples of AgClb5/6p-GFP in vivo fluorescence. Localization of AgClb5/6p-GFP expressed from the genome under the control of the endogenous promoter in the hyphae of AgClb5/6p-GFP that were grown under selection in liquid medium for 14 to 16 h prior to microscopy. Bars: A and D, 10 μm; B, 5 μm.
In contrast, AgClb5/6p-13myc displayed a uniform and intense nuclear signal, which was visible in all nuclear cycle stages except late metaphase and anaphase (Fig. 2A, bottom, and B, right). AgClb5/6p was not visible in 38% of all metaphase nuclei and in 100% of anaphase nuclei (defined by spindles longer than 2.8 μm). However, 94% of all nuclei with a single SPB had visible AgClb5/6p (Fig. 2C), suggesting a very efficient accumulation of AgClb5/6p in G1 nuclei (n = 328 nuclei scored).
To confirm that these localizations were not due to artifacts from fixation and immunofluorescence, we also tried to evaluate GFP-tagged cyclin distribution in living cells. Unfortunately, the signal of AgCln3p-GFP was not detectable, presumably due to the low abundance of AgCln3p. In addition, AgClb3/4p-GFP was not functional, as observed for AgClb3/4p-13myc (data not shown). AgClb5/6p-GFP could be observed in nuclei (Fig. 2D); however, the signal was very weak and sensitive to photobleaching, so we were not able to capture the localization of the protein across the progression of an entire division cycle in living cells. In summary, AgClb5/6p was excluded from nuclei beginning in metaphase and throughout anaphase before it reappeared in early G1 phase. These localization data show that at least one cyclin can oscillate in this unique nuclear division cycle, demonstrating that a multinucleated organism such as A. gossypii can control subcellular localization and/or protein abundance in individual nuclei.
Role of nuclear localization in the function of AgClb5/6p.
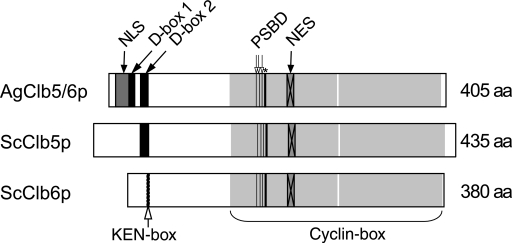
Based on domains identified in the protein sequence of AgClb5/6p, the disappearance of AgClb5/6p during metaphase could be explained in two ways. AgClb5/6p may be specifically degraded for mitotic progression. In support of this, AgClb5/6p, which is 44% identical to ScClb5p and 39% identical to ScClb6p, contains two N-terminal D-boxes (Fig. 3 and Fig. S1 in the supplemental material), a motif that targets proteins to the anaphase-promoting complex for ubiquitination and ultimately destruction by the proteosome (23). Alternatively, AgClb5/6p might be exported from the nucleus to the cytoplasm during the metaphase-to-anaphase transition. AgClb5/6p has a bipartite NLS and a leucine-rich NES, supporting a possible role for dynamic translocation across the nuclear membrane for the timely loss of AgClb5/6p from nuclei. The second possibility is appealing because it may account for the observation that nearly all nuclei with a single SPB have abundant AgClb5/6 protein. In this model, there would be no delay due to translation of new protein, which would support the presumably swift reaccumulation of the cyclin in G1 nuclei. Based on the conserved protein domains and the localization, degradation and/or nuclear export may be possible explanations for the behavior of the AgClb5/6p-13myc signal.
FIG. 3.
Domain comparison between AgClb5/6p, ScClb5p, and ScClb6p. The D-box (db2 [indicated by 2, aa 38 to 47] (23), the C-and N-terminal cyclin box (Pfam scan, N [aa 145 to 274], C [aa 276 to 399]), and the putative NES [aa 214 to 222] (26), as well as the potential substrate-binding domains (PSBD) (12) as the hydrophobic patch (open arrows [177, 181, and 184] and the salt bridge (asterisk [187]), are conserved in A. gossypii. AgClb5/6p appears to have an additional putative D-box sequence (db1 [indicated by 1], aa 22 to 31) and an additional putative bipartite NLS (aa 8 to 24 [Prosite scan]). Numbers in brackets always refer to the A. gossypii domains.
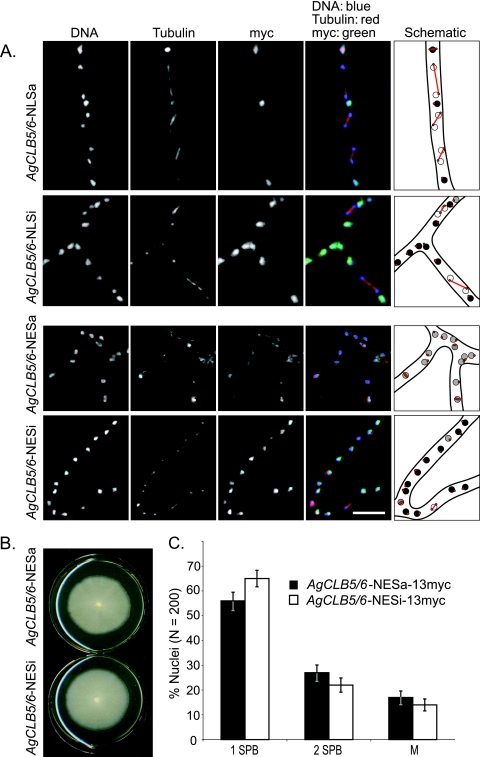
To distinguish between these two possible explanations for the localization patterns of AgClb5/6p, we first attempted to shift the balance of the pools of AgClb5/6p between the nucleus and cytoplasm by the addition of exogenous NESs or NLSs to the C terminus expressed from the endogenous genomic locus. We attempted to increase the nuclear levels of AgClb5/6p by the addition of two NLSs (17). We reasoned that enhancing the nuclear import of this protein and potentially extending the nuclear residence time and/or concentration may cause a mitotic delay if complete nuclear export is required for efficient progression through anaphase. AgClb5/6p-NLSa-13myc cells grew comparably to the wild-type strain and did not display obvious defects in morphology or nuclear density. The additional NLSs did not detectably change the localization of AgClb5/6p, and no significant change in nuclear cycle stage or asynchrony could be observed (Fig. 4A). With these results we cannot distinguish between the possibilities that nuclear export is highly efficient or that nuclear autonomous degradation is occurring.
FIG. 4.
Displacement of AgClb5/6p from the nucleus disturbs nuclear cycle progression. (A) AgCLB5/6-NLSa-13myc (active NLS), AgCLB5/6-NLSi-13myc (inactive NLS control), AgCLB5/6-NESa-13myc (active NES), and AgCLB5/6-NESi-13myc (inactive NES control) were grown for 14 to 16 h in liquid selective medium and then processed for anti-tubulin and anti-myc immunofluorescence. False coloring: DNA is blue, tubulin is red, and cyclins are green; therefore, nuclei containing cyclins appear turquoise. Bars, 10 μm. (B) Radial growth assay. A small patch of mycelium from AgCLB5/6-NESa-13myc or AgCLB5/6-NESi-13myc was transferred to the center of plates containing complete medium (+G418). The radial diameters of colonies were measured after 7 days of growth at 30°C. (C) Percentage of nuclei in different spindle stages based on tubulin staining. A total of 200 nuclei were scored for each strain.
As an alternative way to examine whether AgClb5/6p nucleocytoplasmic shuttling is important for mitosis, we next displaced a fraction of the AgClb5/6 protein from nuclei to the cytoplasm by the fusion of two exogenous NESs to AgCLB5/6. If nuclear export is tightly regulated in the cell cycle, then enhancing the proportion of cyclin in the cytoplasm and/or potentially altering the timing of export of the cyclin protein may impact the rate of nuclear progression or the levels of mitotic asynchrony. The additional NESs (NESa) led to an increase in the cytoplasmic levels of AgClb5/6p-13myc that was not observed in a control strain with inactive mutant NESs (NESi, Fig. 4A, bottom, measured by immunofluorescence and calculation of the ratio of the nucleus to cytoplasmic signal intensity; the NESa ratio was 2.6 compared to 9.6 for NESi cells). Sufficient protein may have been left in the nucleus to allow normal vegetative growth; however, spore formation was inhibited by this partial shift of AgClb5/6p out of the nucleus (Fig. 4B). Hyphae with elevated cytoplasmic pools of AgClb5/6p had a slightly lower but significantly different (P < 0.05, based on two proportion Z-tests) proportion of nuclei with single SPBs compared to the inactive NES fusion. This suggests altered nuclear cycle progression kinetics when this protein is displaced; however, it is not clear whether these different proportions are due to a delay in passage through mitosis or to an increased frequency of mitosis (Fig. 4C). Interestingly, nuclei were also less asynchronous with enhanced levels of cytoplasmic AgClb5/6p (AgCLB5/6-NESa-13myc [36% ± 3.3% asynchronous] compared to AgCLB5/6-NESi-13myc [46% ± 3.4% asynchronous] as determined by comparing the spindle stage of neighboring nuclei; n.b. synchronous nuclei in wild-type A. gossypii are nearly all G1 nuclei beside other G1 nuclei), indicating that nuclear sequestration of AgClb5/6p may be important for asynchrony. This higher degree of synchrony may have resulted from an increased diffusion of protein between nuclei due to the elevated levels of AgClb5/6p in the cytoplasm. Thus, perturbing the nucleocytoplasmic balance of AgClb5/6p only led to minor defects in nuclear division, suggesting that the disappearance of AgClb5/6p at the metaphase-to-anaphase transition may be controlled by mechanisms other than regulated localization.
Deletion of AgClb5/6 D-boxes inhibits cell growth and alters protein localization.
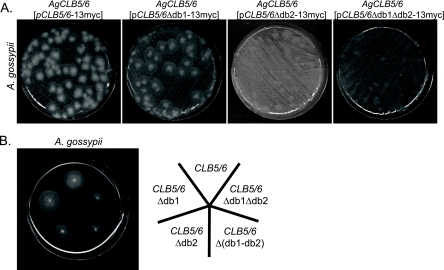
The presence of two destruction box (D-box) elements in A. gossypii, one at a site homologous to the D-box in S. cerevisiae (db2) and a second that is located more N terminal from db2 (db1, Fig. 3), suggests a possible role for degradation in the regulation of this protein. To test whether degradation is the basis for the loss of AgClb5/6p from nuclei and whether this contributes to normal cell cycle progression, low-copy plasmids were constructed that contained mutant versions of AgCLB5/6 lacking D-boxes and under control of the native AgCLB5/6 promoter [AgCLB5/6-13myc with Δdb1, Δdb2, Δdb1Δdb2, or Δ(db1-db2) in which the entire region between Δdb1 and Δdb2 was deleted]. If AgClb5/6p degradation is crucial for normal division, then the expression of a stabilized version of AgClb5/6p would be expected to have a dominant-negative effect on the cells.
Young A. gossypii cells transformed with pAgCLB5/6-13myc or pAgCLB5/6Δdb1-13myc formed small colonies (5 to 10 mm in size) within 2 days after transformation, whereas cells harboring pAgCLB5/6Δdb2-13myc or pAgCLB5/6Δdb1Δdb2-13myc produced microcolonies that were barely visible (Fig. 5A). Similar amounts of mycelia from these strains were repicked on selective plates to compare growth rates. Cells carrying plasmids with the deletion of D-box2 [Δdb2, Δdb1Δdb2, and Δ(db1-db2)] grew ca. 70% slower than cells carrying the wild-type allele or Δdb1 (Fig. 5B). These slow-growing cells also displayed nuclear and morphology defects. Specifically, lateral branches and apical tip splitting branches (Y-shaped tips) were formed in the same hyphae after 24 h of growth (Fig. 6). This is in contrast to wild-type cells, which exclusively grow by tip splitting at this time during development. These aberrant hyphae contained regions with normally shaped and distributed nuclei alternating with regions containing clumps of fragmented nuclei (Fig. 6B). This heterogeneity in nuclear phenotype is likely because plasmids are not present in all nuclei, and thus the dosage of the mutant protein will vary from region to region depending on the local plasmid copy number.
FIG. 5.
AgCLB5/6 D-box mutant alleles are dominant and impede normal growth. (A) Young A. gossypii mycelia were transformed with 6 μg of plasmid containing either AgCLB5/6-13myc, AgCLB5/6Δdb1-13myc, AgCLB5/6Δdb2-13myc, or AgCLB5/6Δdb1Δdb2-13myc and plated on selective, solid medium. The growth of primary transformants under selection was evaluated after 2 days. (B) A small patch of the mycelium of AgCLB5/6 (pAgCLB5/6-13myc), AgCLB5/6 (pAgCLB5/6Δdb1-13myc), AgCLB5/6 (pAgCLB5/6Δdb2-13myc), AgCLB5/6 [pAgCLB5/6Δ(db1-db2)-13myc], or AgCLB5/6 (pAgCLB5/6Δdb1Δdb2-13myc) was inoculated onto plates containing G418, and the diameters of the mycelial colonies were determined after 3 days of growth at 30°C.
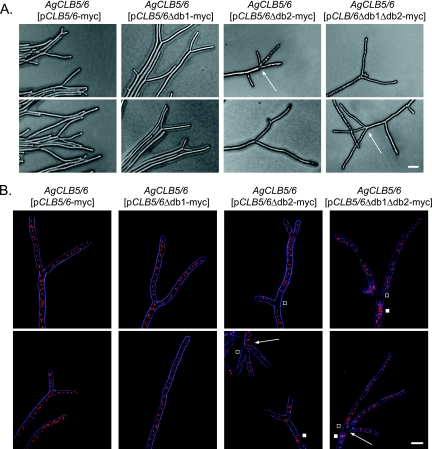
FIG. 6.
Phenotypic analysis of AgCLB5/6 D-box mutants. Mycelia of strains carrying plasmids with D-box mutant alleles (AgCLB5/6 [pAgCLB5/6-13myc], AgCLB5/6 [pAgCLB5/6Δdb1-13myc], AgCLB5/6 [pAgCLB5/6Δdb2-13myc], and AgCLB5/6 [pAgCLB5/6Δdb1Δdb2-13myc]) were grown on solid, complete medium under selection for 2 days. (A) Phase-contrast images of mycelia carrying D-box mutant alleles. Tip splitting of D-box2 and D-box1/D-box2 deletion strains was often accompanied by lateral branching and occasionally by triple tip splitting. (B) Nucleus staining of mycelia carrying D-box mutant alleles with Hoechst dye. Arrows indicate triple-tip splitting, white squares show sites with clustered nuclei, and open squares show regions without nuclei. Artificial coloring: bright field in blue, and nuclei in red. Bars: A, 20 μm; B, 10 μm.
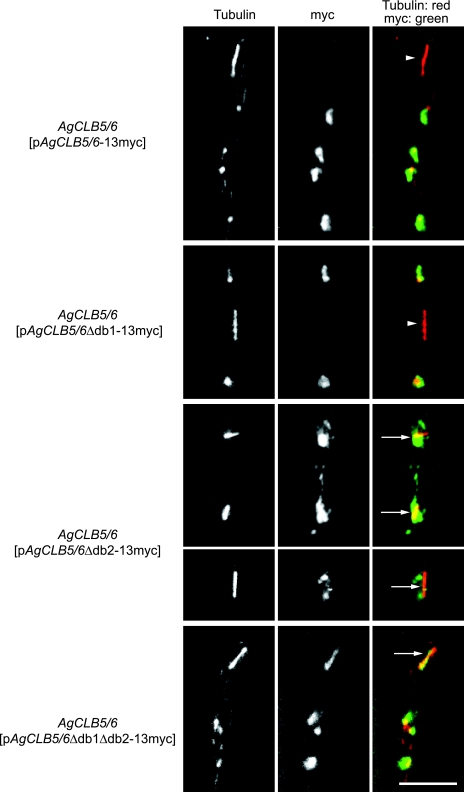
These dominant phenotypes suggest that the D-box2 of AgClb5/6p is functionally important. To evaluate whether AgClb5/6p is stabilized during anaphase due to the D-box mutations, the panel of D-box mutant proteins were localized. In AgCLB5/6Δdb1-13myc, the myc signal was detected in all stages of the nuclear cycle except in some metaphase and all anaphase nuclei (Fig. 7). This suggests that the D-box1 only plays a minor role, if any role at all, in the degradation of AgClb5/6p. In contrast, the protein was clearly visible in anaphase nuclei in strains lacking D-box2 and/or both D-boxes (Fig. 7), suggesting that stabilization of AgClb5/6p in anaphase contributes to the growth defect of strains expressing these mutant proteins. When combined, these data suggest that cell cycle-dependent AgClb5/6p disappearance in anaphase nuclei is due to degradation mediated by the D-box2 sequence.
FIG. 7.
D-box2 is required for the degradation of AgClb5/6p. A. gossypii wild-type strains carrying autonomously replicating plasmids (pAgCLB5/6-13myc, pAgCLB5/6Δdb1-13myc, pAgCLB5/6Δdb2-13myc, or pAgCLB5/6Δdb1Δdb2-13myc) were incubated on solid medium for 4 days under selection, scraped off the plate, and grown in liquid AFM plus G418 for 14 to 16 h prior to processing for immunofluorescence. Representative examples of the four strains visualizing microtubules and AgClb5/6p-13myc are shown. A. gossypii strains lacking D-box2 or both D-boxes in AgCLB5/6 show a clear signal of AgClb5/6p-13myc during anaphase. Anaphase nuclei lacking the 13myc signal are marked with an arrowhead; those showing a 13myc signal are marked with an arrow. False coloring: tubulin is shown in red, and Myc is shown in green. Bars, 10 μm.
DISCUSSION
We present here a comprehensive study of the function and localization of previously uncharacterized G1 and B-type cyclins in A. gossypii. We conducted these experiments with two goals in mind. Namely, we wanted to explore whether functional redundancy, as is found to a large degree in cyclins from uninucleate cells, would be a trait of cyclins directing division in multinucleated cells. Second, we had previously observed that AgClb1/2p and AgCln1/2p failed to detectably oscillate, and we wanted to determine whether this apparent protein stability was a general feature of all cyclins during the vegetative growth of A. gossypii. Here we have shown that there is minimal functional redundancy among cyclins such that AgCln1/2p, AgClb5/6p, and AgClb1/2p are all essential proteins. Furthermore, we found that the cyclin AgClb5/6p is specifically degraded late in mitosis, demonstrating that nuclear autonomous protein degradation can occur in the midst of asynchronous division in a syncytium.
Cyclin functional redundancy.
Why do eukaryotic cells utilize such a variety of cyclins to progress through the cell cycle? The unicellular budding yeast has six different B-type cyclins that complex with the CDK, ScCdc28p to drive cell cycle progression, and yet these cells can actually proliferate with only a single B-type cyclin and the G1 cyclins (24). Displaying even more frugality, the fission yeast can proliferate with only a single B-type cyclin driving the cell cycle (18). This observation in fission yeast led to a quantitative model for CDK activity in which the absolute level of activity of the CDK directs progression. In this model, low levels of activity would trigger S phase, while higher levels of activity would initiate mitosis rather than different cyclins being required for each stage. This model is appealing considering the likely evolution of cyclins from a single common ancestral gene and the prevailing idea that in primitive division cycles the CDK could be directed through the distinct cell cycle phases without variety in cyclin subunits (38, 39). Certainly, at least the CLB1/2/5/6 genes in S. cerevisiae share a single ancestral cyclin gene based on comparisons with recently sequenced fungal genomes (2). Nevertheless, the numbers of cyclins clearly multiply along with organismal complexity and, while yeast cells can proliferate with abridged collections of cyclins, this division is in many respects abnormal (see below and references 12, 13, 15, 16, and 28).
Current discussion about why cells express a multitude of related cyclin proteins across a single division cycle focuses on whether differences between cyclin proteins are due to variable timing of expression, localization, CDK complex activity, and/or intrinsic substrate specificity (34, 38, 39). There is experimental support for all of these explanations in the literature. For example, in higher eukaryotes, a mitotic cyclin B can take on functions in DNA replication as long as it is targeted to the nucleus (37). Similarly, the S. cerevisiae G1 cyclins ScCln2p (cytoplasmic) and ScCln3p (nuclear) exchange functional roles when their subcellular localization is switched (35, 36). Furthermore, mice lacking cyclin D have very minimal phenotypes, and these can be rescued completely by knocking in cyclin E at the cyclin D locus (20). These data support the idea that cyclin subcellular compartmentalization or tissue-specific expression yields different functions for otherwise biochemically similar proteins.
In contrast, the mitotic ScClb2p in yeast can direct DNA replication in the absence of ScClb5p and ScClb6p but with significant S-phase delays even when expressed prematurely from the ScCLB5 promoter, suggesting that the B-type cyclins in budding yeast are not truly created equal in terms of substrate specificity (12, 13, 15, 16, 28). There is growing evidence that some of this specificity is mediated by the conserved hydrophobic patch (HP) region of ScClb2p and ScClb5p proteins that lies at the interface between cyclins and substrates. Notably, the HP sequence diverges between ScClb2p and ScClb5p in budding yeast, and these differences are preserved in orthologues in related Saccharomyces yeast species, suggesting the patch differences between B-cyclins are functionally relevant (2). Furthermore, screens for direct substrates of the CDK/Clb5p and Cdk/Clb2p complexes in budding yeast yielded a set of targets specific to ScClb5p that interact via this HP motif and that appear to be poor or unlikely substrates for CDK/Clb2p (33).
Potential differences in cyclin specificity and substrate targeting may be tightly linked to cell architecture; however, the functional redundancy of different cyclins had not been investigated in multinucleated, asynchronous cells prior to the present study. We predicted that the continuous cytoplasm and cohabitation of nuclei from different stages of the cell cycle may create a situation wherein many different cyclins coexist in time and space in A. gossypii. In fact, for much of the cell cycle, AgCln1/2p, AgCln3p, AgClb1/2p, and AgClb5/6p are present in nearly all nuclei (with the exception of AgClb5/6p disappearing at the end of mitosis [see below]). Thus, AgClb1/2p and AgClb5/6p appear to share the essentially same expression and localization and yet, remarkably, neither B-type cyclin can functionally complement for the loss of the other, suggesting that these cyclins have evolved very strict substrate specificities in A. gossypii. Similarly, the G1 cyclin AgCln3p cannot complement for the loss of AgCln1/2p.
While in uninucleate budding yeast both the G1 and the B-type cyclins can complement for each other at least to support viability, albeit imperfectly, in these multinucleated cells all redundancy in function appears to be lost. This suggests that the cell organization, in this case multinucleated compartments with asynchronous nuclei, may influence the specificity of cyclin/CDK complexes. Presumably, the coexistence of the different cyclins within the nucleus has selected for enhanced substrate specificity among the cyclins to ensure order in the execution of S and M phases in the absence of spatial and temporal control of cyclins.
Interestingly, in previous work AgCln1/2p was shown to be localized to hyphal tips in addition to nuclei, and here the characterized phenotype of null mutants is a pronounced polarity defect in addition to altered nuclear progression, leading to inviable cells. The contribution of ScCln1/2p in directing actin polarity in bud emergence has been known for over a decade, but the molecular mechanism for integrating CDK/Cln signaling to the polarity establishment machinery remains elusive in yeast (9, 30). These experiments in A. gossypii suggest that the role of the G1 cyclins in directing polarity is ancient and important in hyphal growth.
Protein degradation in a syncytium.
Timed destruction of cyclins is a central component of cell cycle control. Degradation and/or inhibition of B-type cyclins is a prerequisite for mitotic exit in most systems, and periods of low CDK/Clb activity are required for the assembly of prereplication complexes. Our previous experiments in A. gossypii suggested that, rather than by degradation, AgClb1/2p was kept inactive by an inhibitor AgSic1p (22). We hypothesized that this mode of control was more secure in a syncytium where newly made proteins would be continuously supplied from the cytosol, thus making protein degradation a potentially inefficient mode of negative regulation. This prompted us to investigate the behavior of the other cyclins in these cells to see whether noncycling cyclins were an intrinsic feature of asynchronous mitosis.
In fact, we determined that AgClb5/6p is lost from nuclei at the metaphase-to-anaphase transition and is not detected in any anaphase nuclei. However, nearly all G1 nuclei have abundant AgClb5/6p, suggesting that this absence from the nuclei is quite transient. We have shown that the AgClb5/6p disappearance is not likely due to regulated nuclear export but rather appears to be through destruction box-mediated anaphase-promoting complex (APC) targeting and proteasomal degradation. We recently showed that the APC is essential for mitotic progression in A. gossypii and acts to degrade the Securin homologue (21). With these experiments, we now add AgClb5/6 as an essential target for the APC in A. gossypii. Expression of a mutant Agclb5/6 allele lacking one of the destruction boxes from the endogenous promoter leads to severely crippled mycelia with AgClb5/6p present in anaphase nuclei. Notably, ScClb5Δdb mutants (with protein expressed at endogenous levels) are viable with minimal mitotic exit defects and minor delays early in the cell cycle, potentially due to problems with replication origin establishment (44). Thus, the consequences of misregulation of this cyclin orthologue are more profound in multinucleated cells. One enigma from our studies was the severity of the phenotype when wild-type AgClb5/6p was overexpressed in otherwise wild-type yeast cells, which normally have no difficulty buffering overexpressed wild-type ScClb5p (44) (see Fig. S4 in the supplemental material). This suggests that the AgClb5/6p is not properly degraded in yeast and/or that it has neomorphic, deleterious functions when heterologously expressed in yeast.
These studies show that protein degradation can play a role in regulating syncytial asynchronous mitosis. Given the tremendous conservation of the DNA replication machinery, we predict that the transient loss of AgClb5/6p from nuclei is essential for the establishment of prereplication complexes at origins. It will be of interest to monitor origin loading and map it temporally relative to the disappearance of AgClb5/6p. AgClb5/6p reappears in many nuclei presumably well before S phase begins, therefore limiting the window of time available for resetting origins. This presence of AgClb5/6p very early in the division cycle suggests that additional tight controls of the CDK/AgClb5/6p activity must modulate the firing of origins to prevent premature entry into S phase. In addition, it is mysterious how the APC can be activated in a nuclear autonomous manner, but clearly there are at least two APC substrates that are degraded in only a subset of nuclei in a common cytoplasm. In summary, these studies underscore the importance of examining a variety of types of cells when testing paradigms of cell cycle control and demonstrate that the cell cycle machinery diverged in certain fundamental mechanisms to support multinucleated nuclear division.
Supplementary Material
Acknowledgments
We thank Hanspeter Helfer for constructing the A. gossypii histone H4-GFP strain and Simon Braun for deleting AgCLN3 and AgCLB5/6 in this strain. We also thank Hanspeter Helfer, Mark Borsuk, and Hans-Peter Schmitz for useful technical comments.
This study was supported by an NSF postdoctoral fellowship and a Roche Research Foundation grant to A.S.G. and a Swiss National Fund grant (3100A0-100734) to P.P. and A.S.G.
Footnotes
Published ahead of print on 22 November 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altmann-Johl, R., and P. Philippsen. 1996. AgTHR4, a new selection marker for transformation of the filamentous fungus Ashbya gossypii, maps in a four-gene cluster that is conserved between A. gossypii and Saccharomyces cerevisiae. Mol. Gen. Genet. 250:69-80. [DOI] [PubMed] [Google Scholar]
- 2.Archambault, V., N. E. Buchler, G. M. Wilmes, M. D. Jacobson, and F. R. Cross. 2005. Two-faced cyclins with eyes on the targets. Cell Cycle 4:125-130. [DOI] [PubMed] [Google Scholar]
- 3.Arvanitidis, A., and J. J. Heinisch. 1994. Studies on the function of yeast phosphofructokinase subunits by in vitro mutagenesis. J. Biol. Chem. 269:8911-8918. [PubMed] [Google Scholar]
- 4.Ayad-Durieux, Y., P. Knechtle, S. Goff, F. Dietrich, and P. Philippsen. 2000. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 113(Pt. 24):4563-4575. [DOI] [PubMed] [Google Scholar]
- 5.Bachewich, C., and M. Whiteway. 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensen, E. S., A. Clemente-Blanco, K. R. Finley, J. Correa-Bordes, and J. Berman. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisnard-Lorig, C., A. Colon-Carmona, M. Bauch, S. Hodge, P. Doerner, E. Bancharel, C. Dumas, J. Haseloff, and F. Berger. 2001. Dynamic analyses of the expression of the HISTONE:YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13:495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bose, I., J. Irazoqui, J. J. Moskow, E. S. G. Bardes, T. R. Zyla, and D. J. Lew. 2001. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle regulated phosphorylation of Cdc24p. J. Biol. Chem. 276:7176-7186. [DOI] [PubMed] [Google Scholar]
- 10.Castillo-Lluva, S., and J. Perez-Martin. 2005. The induction of the mating program in the phytopathogen Ustilago maydis is controlled by a G1 cyclin. Plant Cell 17:3544-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapa y Lazo, B., S. Bates, and P. Sudbery. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4:90-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, F. R., and M. D. Jacobson. 2000. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 20:4782-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross, F. R., M. Yuste-Rojas, S. Gray, and M. D. Jacobson. 1999. Specialization and targeting of B-type cyclins. Mol. Cell 4:11-19. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich, F. S., S. Voegeli, S. Brachat, A. Lerch, K. Gates, S. Steiner, C. Mohr, R. Pohlmann, P. Luedi, S. Choi, R. A. Wing, A. Flavier, T. D. Gaffney, and P. Philippsen. 2004. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304:304-307. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson, A. D. 2000. The yeast mitotic cyclin Clb2 cannot substitute for S phase cyclins in replication origin firing. EMBO Rep. 1:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in Saccharomyces cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 17.Edgington, N. P., and B. Futcher. 2001. Relationship between the function and the location of G1 cyclins in Saccharomyces cerevisiae. J. Cell Sci. 114:4599-4611. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, D. L., and P. Nurse. 1996. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 15:850-860. [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Muse, T., G. Steinberg, and J. Perez-Martin. 2004. Characterization of B-type cyclins in the smut fungus Ustilago maydis: roles in morphogenesis and pathogenicity. J. Cell Sci. 117:487-506. [DOI] [PubMed] [Google Scholar]
- 20.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 21.Gladfelter, A. S., N. Sustreanu, A. K. Hungerbuehler, S. Voegeli, V. Galati, and P. Philippsen. 2007. The anaphase-promoting complex/cyclosome is required for anaphase progression in multinucleated Ashbya gossypii cells. Eukaryot. Cell 6:182-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gladfelter, A. S., A. K. Hungerbuehler, and P. Philippsen. 2006. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172:347-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glotzer, M., A. Murray, and M. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349:132-138. [DOI] [PubMed] [Google Scholar]
- 24.Haase, S. B., and S. I. Reed. 1999. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature 401:394-397. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 26.Hood, J. K., W. W. Hwang, and P. A. Silver. 2001. The Saccharomyces cerevisiae cyclin Clb2p is targeted to multiple subcellular locations by cis- and trans-acting determinants. J. Cell Sci. 114:589-597. [DOI] [PubMed] [Google Scholar]
- 27.Huang, J., and J. W. Raff. 1999. The disappearance of cyclin B at the end of mitosis is regulated spatially in Drosophila cells. EMBO J. 18:2184-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobson, M. D., S. Gray, M. Yuste-Rojas, and F. R. Cross. 2000. Testing cyclin specificity in the exit from mitosis. Mol. Cell. Biol. 20:4483-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knechtle, P., F. Dietrich, and P. Philippsen. 2003. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol. Biol. Cell 14:4140-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lew, D. J., and S. I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loeb, J. D., M. Sepulveda-Becerra, I. Hazan, and H. Liu. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19:4019-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longtine, M. S., A. McKenzie III, D. J. DeMarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 33.Loog, M., and D. O. Morgan. 2005. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature 434:104-108. [DOI] [PubMed] [Google Scholar]
- 34.Miller, M. E., and F. R. Cross. 2001. Cyclin specificity: how many wheels do you need on a unicycle? J. Cell Sci. 114:1811-1820. [DOI] [PubMed] [Google Scholar]
- 35.Miller, M. E., and F. R. Cross. 2000. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol. Cell. Biol. 20:542-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, M. E., and F. R. Cross. 2001. Mechanisms controlling subcellular localization of the G(1) cyclins Cln2p and Cln3p in budding yeast. Mol. Cell. Biol. 21:6292-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, J. D., J. A. Kirk, and T. Hunt. 2003. Unmasking the S-phase-promoting potential of cyclin B1. Science 300:987-990. [DOI] [PubMed] [Google Scholar]
- 38.Murray, A. W. 2004. Recycling the cell cycle: cyclins revisited. Cell 116:221-234. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, J. M. 1999. Evolving ideas about cyclins. Cell 98:129-132. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Schmitz, H. P., A. Kaufmann, M. Kohli, P. P. Laissue, and P. Philippsen. 2006. From function to shape: a novel role of a formin in morphogenesis of the fungus Ashbya gossypii. Mol. Biol. Cell 17:130-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su, T. T., F. Sprenger, P. J. DiGregorio, S. D. Campbell, and P. H. O'Farrell. 1998. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 12:1495-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 44.Wasch, R., and F. R. Cross. 2002. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418:556-562. [DOI] [PubMed] [Google Scholar]
- 45.Wendland, J., Y. Ayad-Durieux, P. Knechtle, C. Rebischung, and P. Philippsen. 2000. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene 242:381-391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.