Abstract
Gene duplication and divergence via both the loss and gain of gene activities are powerful evolutionary forces underlying the origin of new biological functions. Here a comparative genetics approach was applied to examine the roles of protein kinase A (PKA) catalytic subunits in three closely related varieties or sibling species of the pathogenic fungus genus Cryptococcus. Previous studies revealed that two PKA catalytic subunits, Pka1 and Pka2, control virulence factor production and mating. However, only one of the two plays the predominant physiological role, and this function has been exchanged between Pka1 and Pka2 in strains of the Cryptococcus neoformans var. grubii serotype A lineage compared to divergent C. neoformans var. neoformans serotype D isolates. To understand the basis for this functional plasticity, here the activities of Pka1 and Pka2 were defined in the two varieties and the related sibling species Cryptococcus gattii by gene disruption and characterization, heterologous complementation, and analysis of serotype AD hybrid mutant strains. The findings provide evidence for a shared ancestral role of PKA in governing mating and virulence factor production and indicate that the exchange of catalytic subunit roles is attributable to loss of function. Our studies illustrate the plasticity of signaling networks enabling rapid rewiring during speciation of a clade of common human fungal pathogens.
Gene duplication and divergence are significant forces in the evolution of new genes (21). Gene duplication events can occur both on a macrogenomic scale, such as that involving the ancestral whole genome duplication that gave rise to Saccharomyces cerevisiae and related sensu stricto strains (31), and on the microgenomic scale of individual genes. Several models regarding the mechanisms by which new functions arise following gene duplication have been posited. The model originally postulated by Ohno (21), now termed “neofunctionalization,” suggests that following duplication, one gene of the pair retains the original function while functional constraints on the other gene are relaxed, allowing it to undergo accelerated evolution and eventually develop novel functions.
A well-documented example of neofunctionalization is that of the ORC1/SIR3 gene pair that arose in S. cerevisiae following the whole-genome duplication that occurred approximately 100 to 300 million years ago (mya) (15, 28). In this instance, S. cerevisiae Orc1 shares 48% identity with a protein found in the related species, Saccharomyces kluyveri, while Sir3 shares only 24% identity with this protein, suggesting that Orc1 has retained the ancestral function while Sir3 has been subject to accelerated evolution. Consistent with this is the observation that the S. kluyveri ORC1/SIR3 gene can complement the S. cerevisiae orc1Δ mutant but not the S. cerevisiae sir3Δ mutant.
Another example of gene duplication in S. cerevisiae involves the genes encoding the three protein kinase A (PKA) catalytic subunits, Tpk1, Tpk2, and Tpk3. All three catalytic subunits share a redundant function yet also have novel functions as well. While all three catalytic subunits share redundant roles in viability, as demonstrated by the fact that a tpk1Δ tpk2Δ tpk3Δ triple mutant is inviable, they play opposing roles in pseudohyphal growth, with Tpk2 activating and Tpk1 and Tpk3 repressing the filamentous dimorphic transition (22, 24). The abundance of well-studied gene duplications in S. cerevisiae and the presence of a well-characterized lineage (including the closely related sensu stricto strains that can be used as outgroups) make S. cerevisiae an ideal model system for examining gene duplication events in ascomycete fungi.
Similarly, Cryptococcus neoformans has several attributes that render it a facile system in which to study gene duplication events in basidiomycete fungi. First, there are multiple examples of gene duplications in this fungus, including the laccase genes LAC1 and LAC2, the Ras genes RAS1 and RAS2, the carbonic anhydrase genes CAN1 and CAN2, and the cyclophilin A genes CPA1 and CPA2 (2, 4, 23, 29, 30). Another example of gene duplication in Cryptococcus involves the protein kinase A catalytic subunits, Pka1 and Pka2, which share 35% identity at the protein level. Our previous work showed that both the serotype A and D lineages have both subunits but that the functions of these subunits differ between the two lineages. Deletion of the PKA1 gene in serotype A results in a loss of mating and melanin and capsule production, whereas it is the deletion of the PKA2 gene in serotype D that results in the loss of these functions (3, 11). The second attribute that renders Cryptococcus neoformans an amenable model system for studying gene duplication and evolution in basidiomycete fungi is the presence of full genome sequences for the closely related varieties C. neoformans var. neoformans (serotype D) and C. neoformans var. grubii (serotype A) and sister species Cryptococcus gattii (18). Much like the sensu stricto strains of Saccharomyces, the Cryptococcus species complex enables detailed analyses of gene evolution and function (13).
In this study, we conducted a comparative genetics study of the Pka1 and Pka2 catalytic subunits in C. gattii. Based on our results, we propose a model in which Pka1 and Pka2 played a shared ancestral role in mating and melanin and capsule production. In C. gattii, Pka1 has lost its role in melanin production while retaining roles in capsule production and mating, and Pka2 functions in all three pathways. In C. neoformans var. grubii (serotype A), Pka1 has retained its roles in melanin and capsule production and mating, whereas Pka2 has lost all three functional roles. Finally, in C. neoformans var. neoformans (serotype D), Pka1 has lost all of these functions, whereas Pka2 retained all three functions. These findings reveal rapid and plastic rewiring of signaling cascades controlling virulence and development during fungal speciation.
MATERIALS AND METHODS
C. neoformans and C. gattii strains and media.
All strains used in this study are listed in Table 1. C. neoformans (serotypes A and D) and C. gattii (serotypes B and C) strains were grown on standard S. cerevisiae media (26). The selective medium for biolistic transformation, proline medium for serotype AD hybrid selection, Niger seed medium, V8 medium, and Dulbecco's modified Eagle's (DME) medium were prepared as described previously (1, 8, 12, 14, 27).
TABLE 1.
Strains used in this study
| Strain | Relevant genotype/descriptiona | Reference or source |
|---|---|---|
| Serotype A strains | ||
| KN99α | MATα | 20 |
| JKH4 | MATα pka2AΔ::URA5 ura5 | 11 |
| JKH7 | MATα pka1AΔ::URA5 ura5 | This study |
| JKH96 | MATα pka1AΔ::URA5 pka2AΔ::URA5 ura5 | This study |
| JKH165 | MATα pka1AΔ::URA ura5 (FOAr) | This study |
| JKH179 | MATα pka1AΔ::URA5 ura5 (FOAr) + PKA2D | This study |
| JKH242 | MATα pka1AΔ::URA5 ura5 (FOAr) + PKA1D | This study |
| JKH297 | MATα pka1AΔ::URA5 + PKA2B | This study |
| JKH308 | MATα pka1AΔ::URA5 + PKA1B | This study |
| Serotype B and C strains | ||
| R265 | MATα VGII serotype B | 7 |
| NIH312 | MATα VGIII serotype C | 7 |
| B4546 | MATa VGIII serotype C | 7 |
| JKH290 | MATα VGII pka1BΔ::NAT | This study |
| JKH293 | MATα VGII pka2BΔ::NAT | This study |
| JKH311 | MATα VGII pka2BΔ::NAT + PKA2B | This study |
| JKH317 | MATα VGII pka1BΔ::NAT pka2BΔ::NEO | This study |
| Serotype D strains | ||
| JEC21 | MATα | 19 |
| JEC34 | MATaura5 | J. C. Edman |
| CDC85 | MATα pka2DΔ::URA5 ura5 | 11 |
| JKH19 | MATapka2DΔ::URA5 ura5 (FOAr) | 11 |
| JKH247 | MATapka2DΔ::URA5 ura5 (FOAr) + PKA2A | This study |
| JKH287 | MATapka2DΔ::URA5 ura5 (FOAr) + PKA1A | This study |
| JKH299 | MATα pka2DΔ::URA5 + PKA2B | This study |
| JKH313 | MATapka1DΔ::NEO ura5 | This study |
| JKH314 | MATapka1DΔ::NEO pka2DΔ::NAT ura5 | This study |
| JKH315 | MATα pka2DΔ::URA5 + PKA1B | This study |
| Serotype AD diploid strains | ||
| JKH321 | MATα pka1AΔ::URA5 pka2AΔ::URA5 ura5/ | This study |
| MATapka1DΔ::NEO ura5 | ||
| JKH322 | MATα pka1AΔ::URA5 pka2AΔ::URA5 ura5/ | This study |
| MATapka1DΔ::NEO pka2DΔ::NAT ura5 | ||
| JKH323 | MATα pka2AΔ::URA5 ura5/ | This study |
| MATapka1DΔ::NEO ura5 | ||
| JKH324 | MATα pka2AΔ::URA5 ura5/ | This study |
| MATapka1DΔ::NEO pka2DΔ::NAT ura5 |
FOAr, 5-fluoroorotic acid resistant.
pka1BΔ::NAT and pka1DΔ::NAT transformants were selected on yeast extract-peptone-dextrose (YPD) medium containing 100 μg/ml nourseothricin and pka2BΔ::NAT, pka2BΔ::NEO, and pka2DΔ::NEO were selected on YPD medium containing 200 μg/ml G418. Genotypes were confirmed both by Southern hybridization and expression analysis.
Identification of the C. gattii PKA1 and PKA2 genes.
The C. gattii serotype B PKA1 and PKA2 genes were identified by performing a BLASTn search of the Broad Institute serotype B database (strain R265) by using the C. neoformans serotype A PKA1 and PKA2 open reading frame nucleotide sequences.
Disruption of the C. gattii and C. neoformans serotype D PKA1 and PKA2 genes.
A prototrophic serotype B wild-type strain (R265) was biolistically transformed with the gel-extracted PKA1B and PKA2B disruption alleles as described previously (5) to create strains JKH290 and JKH293, respectively. Details on the construction of the overlap constructs can be found in the supplemental material. An auxotrophic serotype D strain containing a ura5 mutation (JEC34) was transformed with the PKA1D disruption allele to create strain JKH313. To obtain pka1Δ pka2Δ double mutants of serotypes B and D, the pka1BΔ::NAT strain (JKH290) and the pka1DΔ::NEO strain (JKH313) were transformed with pka2BΔ::NEO and pka2DΔ::NAT disruption alleles to create strains JKH317 and JKH314, respectively. The pka1Δ and pka2Δ strains were screened by diagnostic PCR for the 5′ junction and confirmed by Southern blot analysis using specific probes generated by PCR (data not shown).
Complementation experiments.
Complementation of the pka2DΔ mutant with the serotype A PKA genes was tested by transforming strain JKH19 (serotype D pka2DΔ) (11) with plasmid pJH80 (containing the wild-type PKA1A gene) and pJH167 (containing the wild-type PKA2A gene) by using a biolistic apparatus (27) to create strains JKH287 and JKH247, respectively. Construction details of pJH80 and pJH167, as well as the other plasmids in the complementation analysis, are provided in the supplemental material.
Complementation of the pka1AΔ mutant with the serotype D PKA genes was tested by transforming strain JKH165 with pJH26 (PKA2D) and pJH166 (PKA1D) to create strains JKH176 and JKH242, respectively.
Complementation of the pka1AΔ and pka2DΔ mutants with the serotype B PKA1B and PKA2B genes was tested by transforming strain JKH7 (serotype A pka1AΔ) with pJH235 (PKA1B) and pJH240 (PKA2B) to create strains JKH308 and JKH297, respectively, and strain CDC85 (serotype D pka2DΔ) was transformed with pJH235 and pJH240 to create JKH315 and JKH299, respectively. The serotype B pka2BΔ mutant (JKH293) was also transformed with pJH240 to create strain JKH311. RNA analysis was performed on all transformants to confirm the expression of the heterologous gene.
Serotype A and D hybrid assays.
Hybrid assays were accomplished by mixing 1 × 107 cells/ml of each of the mating partners and then plating 20 μl of the mixed cell suspension onto V8 medium (pH 7.0). After 2 days of growth in the dark, the cells were scraped off the V8 medium (pH 7.0) and replated on selective proline medium containing either G418 (JKH96 × JKH313 and JKH4 × JKH313) or G418 and nourseothricin (JKH96 × JKH314 and JKH4 × JKH314). The plates were then incubated at 37°C for 4 days. Fusion products were then examined for melanin and capsule production. The test crosses were as follows: JKH324, MATα PKA1A pka2AΔ::URA5 (JKH4) × MATa pka1DΔ::NEO pka2DΔ::NAT ura5 (JKH314); JKH321, MATα pka1AΔ::URA5 pka2AΔ::URA5 (JKH96) × MATa pka1DΔ::NEO PKA2D ura5 (JKH313); JKH323, MATα PKA1A pka2AΔ::URA5 (JKH4) × MATa pka1DΔ::NEO PKA2D ura5 (JKH313); JKH322, MATα pka1AΔ::URA5 pka2AΔ::URA5 (JKH96) × MATa pka1DΔ::NEO pka2DΔ::NAT ura5 (JKH314). PCRs with serotype A MATa- and MATα-specific primers (7270/7271 and 7264/7266, respectively) and serotype D MATa- and MATα-specific primers (7273/7274 and 7267/7269, respectively) were utilized to confirm the hybrid diploid status of the fusion products.
Mating assays.
α and a strains were cocultured on V8 medium (pH 7) and incubated at room temperature for 4 days in the dark prior to photography (×100 magnification). These strains included wild-type VGII α × wild-type VGIII a (R265 × B4546); pka1BΔ VGII α × VGIII a (JKH290 × B4546); pka2BΔ VGII α × VGIII a (JKH293 × B4546); and pka1BΔ pka2BΔ VGII α × VGIII a (JKH317 × B4546).
Expression analysis.
Fungal strains were inoculated into 5 ml YPD medium and grown overnight at 30°C. Fifty-milliliter amounts of YPD medium in 125-ml flasks were inoculated with 500 μl of the overnight cultures and grown at 250 rpm and 30°C for 5 h prior to harvesting. RNA was isolated from the harvested cultures with TRIzol (Gibco-BRL) following the manufacturer's instructions. Fifteen micrograms (based on spectrophotometric measurement) of RNA was separated on a denaturing gel and transferred to nylon membrane. The resulting blots were probed with PKA1B, PKA1D, PKA2B, or PKA2D gene-specific probes and with actin as a loading control.
Microscopy.
All images of mating and confrontation assays were captured with a Nikon Eclipse E400 microscope equipped with a Nikon DXM1200F camera. Images of melanized colonies were captured with a Nikon CoolPix digital camera. Differential interference microscopy images were taken with the ×1,000 objective of a Zeiss Axioskop 2 Plus fluorescence microscope equipped with an AxioCam MRM digital camera.
RESULTS
Identification of the C. gattii serotype B Pka1 and Pka2.
We have previously shown that, despite sharing only 35% identity at the amino acid level, the serotype A Pka1 and serotype D Pka2 catalytic subunits share similar roles in regulating melanin and capsule production and mating in their respective serotypes (11). To better understand the functional divergence of Pka1 and Pka2, we identified the Pka1 and Pka2 catalytic subunits from the genomic sequence of the R265 strain (molecular group VGII) (6) of C. gattii, a species closely related to C. neoformans (32) and the cause of an outbreak on Vancouver Island (6, 16). At the amino acid level, the Pka1 protein from C. gattii (Pka1B) shares 86% identity with the serotype A Pka1 subunit (Pka1A) and 84% identity with the serotype D Pka1 subunit (Pka1D); Pka1A and Pka1D are 93% identical. The C. gattii Pka2 subunit (Pka2B) shares 87% identity with the serotype A Pka2 subunit (Pka2A) and 88% identity with the serotype D Pka2 subunit (Pka2D), similar to the 93% identity shared between Pka2A and Pka2D. In contrast, the Pka1B subunit shares only 34% identity with the Pka2B subunit. This is similar to the identity comparison of Pka1A and Pka2A (35%) and that of Pka1D and Pka2D (35%).
Complementation of pka1AΔ and pka2DΔ mutations with the PKA1 and PKA2 genes.
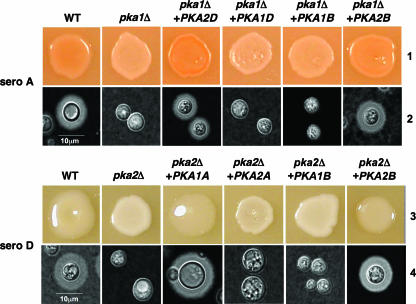
We were interested in determining whether the wild-type PKA1A, PKA2B, or PKA2D gene could complement the pka2DΔ and pka1AΔ mutations in serotypes D and A, respectively. To test this, wild-type PKA1A, PKA2A, PKA1B, PKA2B, PKA1D, and PKA2D genes were ligated into transformation vectors (plasmids pJH80, pJH167, pJH235, pJH240, pJH166, and pJH26, respectively) and biolistically transformed into pka2DΔ and pka1AΔ mutant strains. The resulting transformants were examined for melanin and capsule production. The wild-type PKA1A gene, but not PKA2A, complemented the melanin and capsule defects of a pka2DΔ mutant strain (Fig. 1, lanes 3 and 4), while the wild-type PKA2D gene, but not the PKA1D gene, complemented the melanin and capsule defects of a pka1AΔ mutant strain (Fig. 1, lanes 1 and 2). The PKA2B gene, but not the PKA1B gene, complemented the capsule and melanin defect in both pka1AΔ and pka2DΔ mutant strains (Fig. 1). In control experiments, when the pka1AΔ, pka2DΔ, and pka2BΔ mutant strains were transformed with the plasmids containing the wild-type PKA1A (pJH80), PKA2D (pJH26), and PKA2B (pJH240) genes, respectively, the melanin and capsule defects of the resulting transformants were complemented (data not shown). Expression analysis by Northern blot analysis confirmed that all heterologous genes were expressed (data not shown).
FIG. 1.
PKA1A, PKA2D, and PKA2B genes complement defects conferred by pka1AΔ and pka2DΔ mutations. The ability of wild-type (WT) PKA catalytic subunits from serotypes (sero) A, D, and B to complement melanin and capsule production defects in strains bearing mutations in their counterparts from other serotypes was tested. (Lane 1) A wild-type serotype A strain (KN99α), a pka1AΔ mutant strain (JKH7), and pka1AΔ mutant strains carrying wild-type alleles of PKA1 and PKA2 from serotypes D and B (strains JKH242, JKH308, JKH179, and JKH297, respectively) were grown on Niger seed agar and incubated for 2 days at 37°C prior to being photographed. (Lane 2) The strains described for lane 1 were inoculated onto agar-based DME medium and incubated at 37°C for 2 days prior to being examined for capsule production by exclusion of India ink (×1,000 magnification). (Lane 3) A wild-type serotype D strain (JEC21), a pka2DΔ mutant strain (CDC85), and pka2DΔ mutant strains carrying wild-type alleles of PKA1 and PKA2 from serotypes A and B (strains JKH287, JKH315, JKH247, and JKH299, respectively) were grown on Niger seed agar and incubated for 2 days at 30°C prior to being photographed. (Lane 4) The strains described for lane 3 were inoculated onto agar-based DME medium and incubated at 37°C for 2 days prior to being examined by exclusion of India ink (×1,000 magnification).
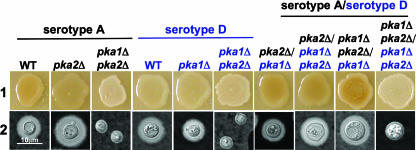
To independently confirm these findings by a complementary approach, we isolated a series of serotype AD hybrid diploid strains that contained combinations of the pka1AΔ, pka2AΔ, pka1DΔ, and pka2DΔ mutations and determined which PKA catalytic subunit was functional in the hybrid by examining melanin and capsule production. AD hybrids were isolated following cell-cell fusions, and PCR analysis using both serotype A- and serotype D-specific primers confirmed the presence of both the serotype A MATα mating type and the serotype D MATa mating type in each hybrid. Melanin and capsule were produced by pka2AΔ/pka1DΔ pka2DΔ (JKH324), pka1AΔ pka2AΔ/pka1DΔ (JKH321), and pka2AΔ/pka1DΔ (JKH323) hybrids, all of which express PKA1A, PKA2D, or both. Only the hybrid with no functional PKA catalytic subunits (pka1AΔ pka2AΔ/pka1DΔ pka2DΔ; JKH322) was unable to produce melanin and capsule (Fig. 2). These results confirm that Pka1A and Pka2D are able to cross-complement the pka2DΔ and pka1AΔ mutations in serotypes D and A, respectively.
FIG. 2.
Pka1A and Pka2D are functional in serotype AD hybrids. To test if PKA1A and PKA2D can complement mutations in serotypes D and A, respectively, hybrid diploid serotype AD strains bearing various combinations of pka1AΔ and pka2DΔ mutations were isolated. (Lane 1) Wild-type (WT) serotype A and D strains (KN99α and JEC20), as well as pka2AΔ (JKH4), pka1AΔ pka2AΔ (JKH96), pka1DΔ (JKH313), and pka1DΔ pka2DΔ (JKH314) mutant strains and serotype AD diploids with the various combinations of pka1Δ and pka2Δ mutations (JKH321 to JKH324), were grown on Niger seed medium and incubated for 2 days at 30°C prior to photographing. (Lane 2) The strains described for lane 1 were inoculated onto agar-based DME medium and incubated at 37°C for 2 days prior to being examined for capsule production by exclusion of India ink (×1,000 magnification). Three independent diploid strains with each genotype were examined in two independent experiments, and representative data are shown.
The serotype B Pka2B subunit is responsible for melanin production, while both Pka1B and Pka2B play roles in capsule production.
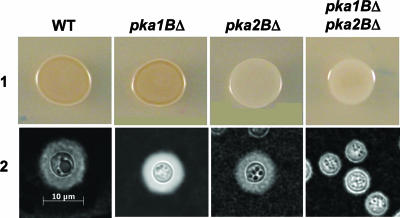
To determine which of the C. gattii PKA catalytic subunits, Pka1B or Pka2B, was responsible for melanin production in C. gattii, we deleted the entire open reading frames of both the PKA1B and the PKA2B genes. The deletions were confirmed by Southern blot and Northern analyses. The deletion strains were then grown on l-DOPA (l-, 4-dihydroxyphenylalanine) and Niger seed medium and incubated at 37°C for 12 to 14 h. The pka2BΔ strain exhibited a severe defect in melanin production, whereas melanin production by the pka1BΔ strain was indistinguishable from that of the serotype B wild-type strain (Fig. 3, lane 1). These findings indicate that the Pka2 catalytic subunit is responsible for positively regulating melanin production in serotype B.
FIG. 3.
Pka2B functions in regulating melanin, and both Pka1B and Pka2B regulate capsule production. The pka1BΔ, pka2BΔ, and pka1BΔ pka2BΔ mutants were examined for the ability to produce melanin and capsule. (Lane 1) A wild-type (WT) strain (R265) and strains containing pka1BΔ (JKH290), pka2BΔ (JKH293), and pka1BΔ pka2BΔ (JKH317) mutations were grown on l-DOPA agar medium and incubated for 12 to 14 h prior to being photographed. (Lane 2) The strains described for lane 1 were inoculated onto agar-based DME medium and incubated at 37°C for 2 days prior to being examined for capsule production by exclusion of India ink (×1,000 magnification). Three independent transformants with each genotype were examined in two independent experiments, and representative data are shown.
In contrast, neither the pka1BΔ mutant strain nor the pka2BΔ mutant strain exhibited a demonstrable capsule defect (Fig. 3, lane 2). We hypothesized that Pka1B and Pka2B might play a shared role in regulating capsule production. To test this, a pka1BΔ pka2BΔ double-mutant strain was created via transformation of the pka1BΔ mutant with a pka2BΔ disruption allele. As with the pka2BΔ single mutant, the double mutant had a severe defect in melanin production (Fig. 3, lane 1). In addition, several independent double-mutant strains also exhibited a profound capsule defect that was not observed in either single-mutant strain, indicating that Pka1B and Pka2B play a redundant role in capsule production (Fig. 3, lane 2).
Pka1B and Pka2B have degenerate roles in mating.
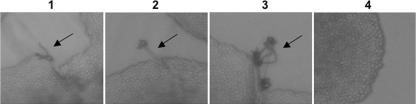
To examine the roles of Pka1B and Pka2B in mating, the pka1BΔ and pka2BΔ single-mutant strains and the pka1BΔ pka2BΔ double-mutant strain were tested as mating partners with the C. gattii MATa strain B4546 (molecular group VGIII) (6, 7). After 4 days of growth on V8 medium (pH 7.0), profuse filamentation and basidial formation was observed in the mating between the pka2BΔ mutant and wild-type strain B4546. This was in comparison to the mating between the wild-type strains (R265 × B4546) and the mating between the pka1BΔ mutant and wild-type strain B4546, in which only minimal filamentation and basidial formation were observed. With the pka1BΔ pka2BΔ double mutant crossed to the wild-type strain, no filamentation or basidial formation was observed (Fig. 4). These data provide evidence that the PKA catalytic subunits in C. gattii have overlapping roles and at least one functional PKA catalytic subunit is required for mating. In addition, the Pka2 catalytic subunit appears to play an additional role in repressing mating in C. gattii.
FIG. 4.
Pka1B and Pka2B share a role in promoting C. gattii mating, and Pka2B has an additional role in repressing mating. The abilities of the α wild type and pka1BΔ, pka2BΔ, and pka1BΔ pka2BΔ mutant strains to mate with the a strain B4546 were tested. Both the wild-type mating (R265) (panel 1) and the pka1BΔ mating (JKH290) (panel 2) produced only a few basidiospores. The pka2BΔ mating (JKH293) (panel 3) was more robust, producing many more basidiospores. The pka1BΔ pka2BΔ mating (JKH317) (panel 4) was completely devoid of basidiospore formation. Matings were performed as described in Materials and Methods. Three independent transformants from each genotype were tested in three independent experiments. Representative data are shown. Arrows point to the filaments, basidia, and basidiospores.
Protein kinase A catalytic and regulatory subunits interact in the two-hybrid system.
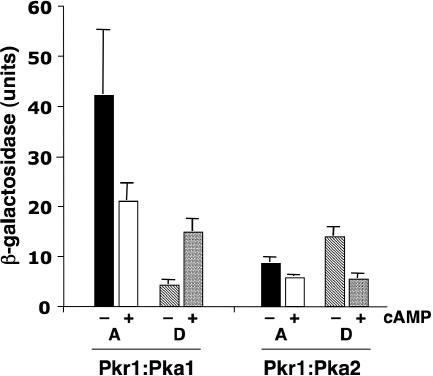
To begin to address the mechanistic basis for the functional differences between the Pka1 and Pka2 catalytic subunits in divergent varieties and species, the yeast two-hybrid system was employed to assess interactions between the subunits. First, we found that the protein kinase A regulatory subunit Pkr1 was capable of interacting with itself when fused to both the Gal4 DNA binding domain and the activation domain, and the magnitude of this interaction was reduced by the provision of 10 mM exogenous cyclic AMP (cAMP) (data not shown). Additionally, we found that the serotype A Pka1 catalytic subunit interacted much more robustly with the Pkr1 regulatory subunit than did the Pka2 catalytic subunit, and the magnitudes of both the Pka1-Pkr1 and the Pka2-Pkr1 interaction were reduced by 10 mM cAMP, as assessed by monitoring Gal4-driven β-galactosidase expression (Fig. 5). Finally, the serotype D Pka1 and Pka2 subunits interacted to a more modest extent with the Pkr1 regulatory subunit, and the provision of cAMP reduced the Pkr1-Pka2 interaction but not the Pkr1-Pka1 interaction (Fig. 5). Taken together, these findings suggest that differences in the interaction of the catalytic and regulatory subunits of protein kinase A or in the cAMP responsiveness of the complex could contribute to functional differences between subunits.
FIG. 5.
Protein kinase A regulatory and catalytic subunit interactions in the yeast two-hybrid assay. Yeast two-hybrid reporter strains expressing Gal4 domains fused to Pkr1 and Pka1 or Pka2 from serotype A or D as indicated were tested for β-galatactosidase expression in the presence (+) and absence (−) of 10 mM exogenous cAMP. Samples were assayed in triplicate, and the standard errors of the means are presented as error bars.
DISCUSSION
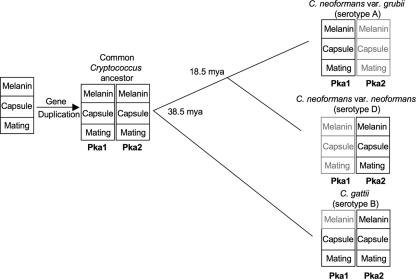
We have taken advantage of the availability of the total genome sequences from three Cryptococcus serotypes (serotype A, C. neoformans var. grubii; serotype D, C. neoformans var. neoformans; and serotype B, C. gattii) to examine the evolution of two PKA catalytic subunits, Pka1 and Pka2. C. gattii diverged from C. neoformans approximately 38.5 mya, while the two C. neoformans varieties (C. neoformans var. grubii and C. neoformans var. neoformans) diverged from each other around 18.5 mya (32).
An examination of the C. gattii serotype B strain R265 genome revealed that C. gattii also has two catalytic subunits that share 84% (Pka1) and 87% (Pka2) identity with their serotype A and D counterparts. The presence of two catalytic subunits in C. gattii suggests that a gene duplication event occurred over 38.5 mya, prior to the divergence of C. gattii and C. neoformans, resulting in two catalytic subunits in both species. We were interested in characterizing the C. gattii Pka1 and Pka2 subunits to determine how the roles in regulating melanin and capsule production and mating were allocated between the two subunits in this more divergent species. The deletion of both the PKA1B and PKA2B genes revealed that solely Pka2B is responsible for regulating melanin production (Fig. 3, lane 1). However, a defect in capsule production was observed only when both the PKA1B and PKA2B genes were deleted (Fig. 3, lane 2), indicating that Pka1B and Pka2B share redundant roles in regulating capsule production.
Similar to the opposing roles played by Tpk1/3 and Tpk2 in regulating pseudohyphal growth in Saccharomyces cerevisiae (22, 24), the diverse roles of the PKA catalytic subunits in mating in Cryptococcus serotypes A, D, and B are consummate examples of the complexity of the PKA signaling pathway. In serotype A, the deletion of the PKA1 gene (but not PKA2) in either parental strain results in loss of mating. The situation is more complex in serotype D, wherein a mating defect is detected only when both parental strains harbor a pka2DΔ mutation. Furthermore, unlike serotype A pka1AΔ mutants, pka2DΔ mutants (but not pka1DΔ mutants) have defects in cell and nuclear fusion, leading to production of aberrant filaments (11). In C. gattii, the situation is different than that in the lineage of either serotype A or D because both the Pka1B and Pka2B subunits are required for mating, as indicated by the observation that mating still occurs when either the PKA1B or PKA2B gene was individually deleted but no mating was observed when both the PKA1B and PKA2B genes were deleted (Fig. 4). In addition to playing a redundant role that it shares with Pka1B in activating mating, Pka2B may play a role in repressing mating, as suggested by the observation that the mating of the pka2BΔ mutant with a tester strain resulted in more robust filamentation than was seen with the wild type (Fig. 4).
Our heterologous complementation studies, as well as our serotype AD hybrid diploid studies, showed that the wild-type PKA2D gene complemented the melanin and capsule defects resulting from a pka1AΔ mutation. Similarly, a wild-type PKA1A gene complemented the melanin and capsule defects of a pka2DΔ mutation. The wild-type PKA2B gene, but not PKA1B, was able to complement the melanin defect of the pka1AΔ and pka2DΔ mutants. Interestingly, even though our data show that both the C. gattii Pka1B and Pka2B subunits have roles in capsule production, only the wild-type PKA2B gene, and not PKA1B, was able to complement the capsule defect of the pka1AΔ and pka2DΔ mutants. The failure of the PKA1B gene to complement was not due to a defect in expression or attributable to a mutation in the complementation allele, based on sequence analysis. One remaining possibility is that the lack of complementation of the capsule defect of the pka1AΔ and the pka2DΔ mutant by the PKA1B gene is the result of the Pka1B subunit having, over time, lost its ability to promote capsule production in the divergent C. neoformans serotype A and D lineages.
Based on the data presented here and our previous data showing that the serotype A Pka1A subunit and the serotype D Pka2D subunit have the same function despite having only 35% identity at the amino acid level (11), we propose a model in which the C. gattii Pka1B and Pka2B subunits are representative of the PKA catalytic subunits shortly after the gene duplication event, when the two subunits both still retained ancestral shared functions (Fig. 5). Over time, the serotype A Pka2A subunit lost many of its original functions, with the exception of a minor role in melanin production, whereas the Pka1A subunit retained all of its original functions. On the contrary, in serotype D, the Pka1D subunit lost most of its original functions, whereas the Pka2D subunit retained its original functions (Fig. 6).
FIG. 6.
Model for functional loss and speciation following duplication of the PKA catalytic subunits in Cryptococcus. Prior to the divergence of C. gattii from C. neoformans around 38.5 mya, a gene duplication event that resulted in two PKA catalytic subunits, Pka1 and Pka2, occurred. The catalytic subunits are involved in the regulation of at least three virulence factors that include melanin production, capsule production, and mating. In C. gattii, Pka1 and Pka2 both retain the majority of their original functions, with the exception of Pka1 having lost its ability to regulate melanin (indicated by gray shading). With the exception of a minor role for Pka2A in melanin production, serotype A Pka2A and serotype D Pka1D have lost their original functions, whereas serotype A Pka1A and serotype D Pka2D retain the original functions. Solid lines indicate branch points in evolution, and the numbers at the branch points indicate when the serotypes diverged.
Our model is consistent with the neofunctionalization model proposed by Ohno (21). In this model, after gene duplication, one gene retains the ancestral function while selection pressure on the other gene is reduced, allowing it to lose the original functions and possibly gain new ones. Although neither Pka2A nor Pka1D showed any obvious new functions, as is predicted by Ohno's model, this could be either because not enough time has elapsed since the gene duplication for these functions to have evolved or because we have not tested enough conditions to reveal novel functions. Alternatively, it is possible that instead of the subunits gaining functions, the Pka2A and Pka1D subunit genes may be in the process of becoming pseudogenes and will eventually be completely lost, leaving only one recognizable PKA catalytic subunit, Pka1A and Pka2D, in the respective serotypes.
The mechanism for the differential actions of Pka1 and Pka2 within each of the three serotypes is unclear. One possibility is that the Pka1 and Pka2 proteins interact differently with the regulatory subunit, Pkr1. Pkr1 is highly conserved among the serotypes (Pkr1B shares 89% amino acid identity with both Pkr1A and Pkr1D, and Pkr1A shares 95% amino acid identity with Pkr1D). Our yeast two-hybrid system data (Fig. 5) indicate that Pka1A, but not Pka2A, interacts strongly with Pkr1A. One explanation for this may lie in the amino acid residues in Pka1 and Pka2 that occur at positions in the catalytic subunit critical for binding to the regulatory subunit. Kim et al. (17) have identified several residues in the PKA catalytic subunit that are necessary for interactions with the regulatory subunit. A comparison of the PKA catalytic subunit protein sequence analyzed by Kim et al. and the protein sequences of the Pka1 and Pka2 catalytic subunits from serotypes A, B, and D reveals that at least four of the critical amino acids (at amino acid positions 379, 381, 396, and 398) are substantially different between the Pka1 subunit (isoleucine, tryptophan, glutamine, and lysine at the respective positions) and the Pka2 subunit (arginine, phenylalanine, leucine, and glutamine at the respective positions), although the residues are conserved within the Pka1 and Pka2 proteins of the respective serotypes. Another possibility is that Pka1 and Pka2 interact with different substrates or interact differentially with the same substrates. Finally, previous studies have indicated that the catalytic subunits of PKA have targets both in the cytoplasm and in the nucleus (9). Thus, it is possible that the Pka1 and Pka2 proteins are localized differently within the cell and may either act on different targets depending on their location or, alternatively, be functional only if they are localized in one organelle or another. Further studies will be necessary to address these and other models.
Studies have utilized comparative approaches for closely related species to examine gene duplication and to test models of gene duplication. This is especially true for the well-characterized yeast Saccharomyces cerevisiae, in which a whole-genome duplication occurred and was subsequently followed by extensive gene loss and gene specialization (15, 25, 31). The majority of these studies have focused on gene and protein structures (e.g., references 10 and 15), although some studies have advanced a functional approach (e.g., reference 28). In our study, we implemented a gene function approach rather than a gene structure approach by examining the roles of duplicated proteins in divergent, related species (C. gattii) to understand the origin of function in a more recently diverged pair of varieties (C. neoformans var. grubii and C. neoformans var. neoformans). Similarly, we utilized inter- and intraspecific complementation approaches, including the isolation and analysis of hybrid diploid strains, to decipher the functions of these signaling cascade genes. Both of these techniques may, in combination with the more commonly used gene structure comparison approach, have applications for determining gene function and examining the results of gene duplication events in other fungi, such as the sensu stricto strains of Saccharomyces.
Supplementary Material
Acknowledgments
We thank Alex Idnurm and Andrew Alspaugh for helpful suggestions in the preparation of the manuscript. We thank the Broad Institute for the serotype A and B genome sequences (http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans/Home.html and http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans_b/Home.html, respectively) and The Institute for Genomic Research for the serotype D genome sequence information (http://www.tigr.org/tdb/e2k1/cna1/).
This research was funded in part by NIH Interdisciplinary AIDS Training grant AI07392-14 (J.K.H.) and RO1 grant AI39115 (J.H.).
Footnotes
Published ahead of print on 22 December 2006.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahn, Y.-S., G. M. Cox, J. R. Perfect, and J. Heitman. 2005. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 15:2013-2020. [DOI] [PubMed] [Google Scholar]
- 3.Bahn, Y.-S., J. K. Hicks, S. S. Giles, G. M. Cox, and J. Heitman. 2004. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot. Cell 3:1476-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahn, Y.-S., K. Kojima, G. M. Cox, and J. Heitman. 2005. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol. Biol. Cell 16:2285-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 6.Fraser, J. A., S. S. Giles, E. C. Wenink, S. G. Geunes-Boyer, J. R. Wright, S. Diezmann, A. Allen, J. E. Stajich, F. S. Dietrich, J. R. Perfect, and J. Heitman. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360-1364. [DOI] [PubMed] [Google Scholar]
- 7.Fraser, J. A., R. L. Subaran, C. B. Nichols, and J. Heitman. 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island. Eukaryot. Cell 2:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granger, D. L., J. R. Perfect, and D. T. Durack. 1985. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Investig. 76:508-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harootunian, A. T., S. R. Adams, W. Wen, J. L. Meinkoth, S. S. Taylor, and R. Y. Tsien. 1993. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Mol. Biol. Cell 4:993-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, X., and J. Zhang. 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks, J. K., C. A. D'Souza, G. M. Cox, and J. Heitman. 2004. Cyclic-AMP dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 3:14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hull, C. M., M.-J. Boily, and J. Heitman. 2005. Sex-specific homeodomain proteins Sxi1α and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot. Cell 4:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idnurm, A., Y.-S. Bahn, K. Nielsen, X. Lin, J. Fraser, and J. Heitman. 2005. Deciphering the model pathoegenic fungus Cryptococcus neoformans. Nat. Rev. Microbiol. 3:753-764. [DOI] [PubMed] [Google Scholar]
- 14.Idnurm, A., J. L. Reedy, J. C. Nussbaum, and J. Heitman. 2004. Cryptococcus neoformans virulence gene discovery through insertional mutagenesis. Eukaryot. Cell 3:420-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellis, M., B. W. Birren, and E. S. Lander. 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428:617-624. [DOI] [PubMed] [Google Scholar]
- 16.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. MacDougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, C., X. Nguyen-Huu, and S. S. Taylor. 2005. Crystal structure of a complex between the catalytic and regulatory (RIα) subunits of PKA. Science 307:690-696. [DOI] [PubMed] [Google Scholar]
- 18.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. M. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore, T. D. E., and J. C. Edman. 1993. The α-mating type locus of Cryptococcus neoformans contains a peptide pheromone gene. Mol. Cell. Biol. 13:1962-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen, K., G. M. Cox, P. Wang, D. L. Toffaletti, J. R. Perfect, and J. Heitman. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect. Immun. 71:4831-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, New York, NY.
- 22.Pan, X., and J. Heitman. 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal growth. Mol. Cell. Biol. 22:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pukkila-Worley, R., Q. D. Gerrald, P. R. Kraus, M.-J. Boily, M. J. Davis, S. S. Giles, G. M. Cox, J. Heitman, and A. Alspaugh. 2005. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 4:190-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson, L. S., and G. R. Fink. 1998. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc. Natl. Acad. Sci. USA 95:13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scannell, D. R., K. P. Byrne, J. L. Gordon, S. Wong, and K. H. Wolfe. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440:341-345. [DOI] [PubMed] [Google Scholar]
- 26.Sherman, F. 1991. Getting started with yeast, p. 3-21. In C. Guthrie and G. R. Fink (ed.), Methods in enzymology, vol. 194. Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 27.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Hoof, A. 2005. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171:1455-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, P., M. E. Cardenas, G. M. Cox, J. R. Perfect, and J. Heitman. 2001. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waugh, M. S., C. B. Nichols, C. M. DeCesare, G. M. Cox, J. Heitman, and J. A. Alspaugh. 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148:191-201. [DOI] [PubMed] [Google Scholar]
- 31.Wolfe, K. H., and D. C. Shields. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708-713. [DOI] [PubMed] [Google Scholar]
- 32.Xu, J., R. J. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.