Abstract
Chromobox 7 (CBX7) is a chromobox family protein and a component of the Polycomb repressive complex 1 (PRC1) that extends the lifespan of cultured epithelial cells and can act independently of BMI-1 to repress the INK4a/ARF tumor suppressor locus. To determine whether CBX7 might be oncogenic, we examined its expression pattern in a range of normal human tissues and tumor samples. CBX7 was expressed at high levels in germinal center lymphocytes and germinal center-derived follicular lymphomas, where elevated expression correlated with high c-Myc expression and a more advanced tumor grade. By targeting Cbx7 expression to the lymphoid compartment in mice, we showed that Cbx7 can initiate T cell lymphomagenesis and cooperate with c-Myc to produce highly aggressive B cell lymphomas. Furthermore, Cbx7 repressed transcription from the Ink4a/Arf locus and acted epistatically to the Arf-p53 pathway during tumorigenesis. These data identify CBX7 as a chromobox protein causally linked to cancer development and may help explain the low frequency of INK4a/ARF mutations observed in human follicular lymphoma.
Keywords: Ink4a-ARF, oncogene, Polycomb, follicular lymphoma, p53
Polycomb (Pc) group (PcG) proteins are a class of epigenetic regulators that, despite being structurally unrelated, can be grouped functionally into two major multiprotein complexes referred to as Pc repressive complex 1 and 2 (PRC1 and PRC2). The PRC2 complex has histone deacetylase and histone methyl transferase activities (specific for K27 of histone H3) and establishes histone repressive marks on the chromatin (1). These marks are subsequently read by PRC1 complexes that alter higher chromatin organization, ultimately leading to transcriptional repression (2, 3). Although the precise composition of these two complexes may vary according to cellular context (4), the core components of the mammalian PRC1 complex are the homologs of Drosophila Pc, Posterior sex combs, Sex combs extra, and Polyhomeiotic. The PRC2 complex consists of Enhancer of Zeste (EZH2), Early Embryonic-Deficient (EED), Suppressor of Zeste (SUZ12), and other associated proteins.
PcG proteins regulate morphogenesis, chromosome X-inactivation, hematopoiesis, stem-cell self renewal, and cellular proliferation (reviewed in refs. 5 and 6). In addition, several PcG proteins have been linked to tumorigenesis (7, 8). BMI-1, one of the six mammalian orthologs of Drosophila Psc, is the PcG gene most strongly associated with cancer and was initially identified for its ability to cooperate with c-Myc in lymphomagenesis (9, 10). Furthermore, BMI-1 is overexpressed in mantle cell lymphoma and several other malignancies (11). Bmi-1 can extend proliferative lifespan, reduce apoptosis, and promote transformation of cultured cells and tumorigenesis in mice (12–14). These properties depend on its ability to repress transcription from the Ink4a/Arf tumor suppressor locus (13), which controls cellular senescence and apoptosis through regulating the Rb and p53 tumor suppressors (15). Other PcG proteins such as EZH2 and SUZ12 are also associated with tumorigenesis (7), but none of these has been validated as oncogenes in vivo.
The Pc homolog Chromobox 7 (CBX7) was identified by virtue of its ability to extend the lifespan of primary human prostate epithelial cells (16). CBX7 shares no homology with BMI-1 but belongs to a mammalian family of chromobox-containing proteins that comprises three homologs of HP1 and five homologs of Pc. Although CBX7 can also repress the INK4a/ARF locus, it can function independently of BMI-1 (16). Here, we demonstrate that CBX7 is up-regulated in follicular lymphoma and possesses oncogenic properties in the hematopoietic compartment, thus identifying CBX7 as a potential human oncogene.
Results
Elevated Expression of CBX7 in Human Follicular Lymphoma.
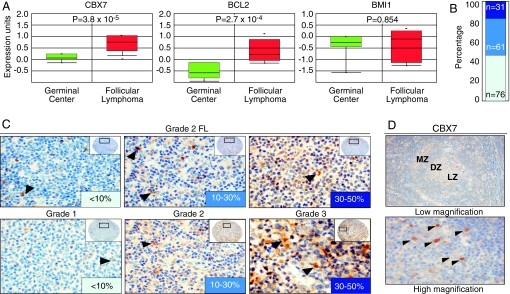
To explore the involvement of CBX7 in human cancer, we examined its expression in the ONCOMINE database (17). Analyzing data from a multicancer study (18), we noted that CBX7 was expressed at significantly higher levels in follicular lymphoma relative to normal germinal center cells, from which follicular lymphoma arises (P = 3.8 × 10−5). The association between CBX7 expression and follicular lymphoma (Q = 5.3 × 10−3) was even more significant than for BCL-2 expression (Q = 1.6 × 10−2), a striking result given that t(14; 18) translocations involving BCL-2 are a hallmark of this disease, and >90% of indolent follicular lymphomas express high levels of BCL-2 (19). In accordance with previous reports (11), we did not observe an association between BMI-1 expression and follicular lymphoma (Fig. 1A). Furthermore, no other PcG gene analyzed was significantly increased in this data set [supporting information (SI) Table 1].
Fig. 1.
Aberrant expression of CBX7 in human follicular lymphomas. (A) Expression of CBX7, BCL-2, and BMI-1 in germinal center (GC, green) and follicular lymphoma (FL, red), as reported in ref. 18. (B) FL samples were stained by IHC by using anti-CBX7 antibodies and the CBX7 levels evaluated and quantified as explained in Materials and Methods. (C) Representative pictures of FL samples stained with CBX7 antibodies are shown. (D) CBX7 expression in secondary GC of the tonsil. Arrows indicate high CBX7 expression in germinal center histiocytes.
To extend these observations, we analyzed CBX7 levels by immunohistochemistry (IHC) using tissue microarrays that contained 168 cases of follicular lymphoma. To this end, we used a previously described polyclonal antibody specific for CBX7 (16) and validated for IHC (20) (see also SI Fig. 5 for further analyses). A subset of samples (54%) displayed strong or moderate levels of CBX7 staining (Fig. 1 B and C). Using Fisher's exact test, a statistically significant correlation was observed between CBX7 expression and Ki67 (P = 0.015) and between CBX7 expression and higher-grade follicular lymphoma (P = 0.0004) (SI Table 2). Consistent with the microarray data, a subset of lymphomas with high CBX7 levels displayed overexpression of CBX7 transcripts as assessed by quantitative RT-PCR analysis of paraffin shavings. These same samples showed no evidence of increased copy number of the CBX7 gene or translocation at the CBX7 locus (22q13), suggesting that high CBX7 levels arise at least in part from increased transcription or mRNA stability (SI Fig. 6 and data not shown).
Elevated CBX7 expression also correlated with the presence of t(14;18) translocations (P = 0.045) involving BCL-2, a hallmark of follicular lymphoma, and positive immunostaining for ZBTB7 (also known as Pokemon) (P = 0.007) (21) and c-MYC (P = 0.007), both markers of poor prognosis in follicular lymphoma (22, 23). However, only one of eight cases coexpressing high CBX7 and c-MYC protein was associated with a translocation of the c-MYC gene. Apart from follicular lymphoma, no evidence for consistent up-regulation of CBX7 was observed in a limited survey of other solid tumors and lymphoma subtypes, including mantle cell lymphoma, with the possible exception of a subset of diffuse large B cell lymphomas (data not shown).
CBX7 Expression in the Germinal Center.
The germinal center contains a complex microenvironment that provides a niche for B cell differentiation. Different PcG proteins present distinct patterns of expression in germinal centers and play a key role in the generation and maturation of B and T cells (5). We therefore analyzed BMI-1 and CBX7 expression in the germinal centers of normal tonsils. Consistent with previous reports (5), BMI-1 was expressed predominantly in mantle zone (MZ) lymphocytes and to a lesser extent in centrocytes and scattered centroblasts in the dark zone (DZ) and the light zone within the germinal center (SI Fig. 7). In contrast, CBX7 was rarely expressed in MZ lymphocytes but accumulated in the germinal center with high expression in histiocytes and lower expression in centrocytes and scattered centroblasts, most predominantly in the DZ (Fig. 1D). Although the CBX7 levels observed in CBX7-positive follicular lymphomas were higher than in normal germinal center cells, from expression data alone, it is difficult to judge whether CBX7 contributes directly to tumorigenesis or represents a marker of the cell of origin of this malignancy. A similar situation applies to BMI-1 (11), which is expressed at high levels in mantle cell lymphomas (derived from the MZ) but not in follicular lymphoma (derived from germinal center cells).
CBX7 Promotes Lymphomagenesis.
That CBX7 appeared to be aberrantly expressed in human lymphoma prompted us to test whether it could promote lymphomagenesis in mice. To this end, we produced transgenic chimeric mice by introducing genes into hematopoietic stem cells (HSCs) or early progenitor cells using recombinant retroviruses followed by adoptive transfer into irradiated recipients (SI Fig. 8A). By using different retroviruses or target cell populations, it is possible to assess the impact of genes and gene combinations in a manner that is much more rapid than traditional germ-line transgenic methods. Moreover, the expression of each gene is strictly confined to the hematopoietic compartment, and the coexpressed GFP reporter allows identification of transgene-expressing cells and tumor imaging. In contrast to germ-line transgenic mice, gene function is studied in a chimeric setting, where developing tumor cells are surrounded by their normal counterparts.
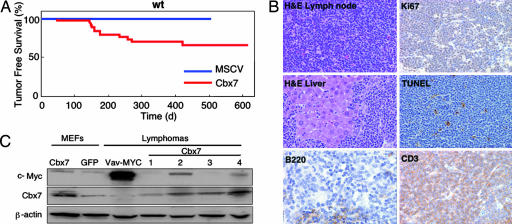
We produced a murine stem cell virus (MSCV) vector that coexpressed murine Cbx7 with GFP and efficiently expressed the Cbx7 protein. Moreover, enforced expression of Cbx7 from this vector was capable of overcoming Ras-induced senescence and c-Myc-induced apoptosis of rodent fibroblasts, leading to an increase in their oncogenic transformation (SI Fig. 9). A significant proportion of mice reconstituted with Cbx7-expressing HSCs derived from C57BL/6 WT mice (11/30) developed tumors within 1 year of adoptive transfer (Fig. 2A). The affected animals invariably displayed an enlarged thymus and splenomegaly (data not shown). In contrast, none of the animals reconstituted with HSCs harboring the control vector showed signs of tumor development (Fig. 2A and SI Fig. 8B), excluding insertional mutagenesis as the cause of the observed pathologies. Cbx7-expressing tumors were disseminated T cell lymphomas with a high proliferative index (Ki67) and a low apoptotic rate (as assessed by TUNEL) (Fig. 2B). Characterization of Cbx7-expressing lymphomas by FACS showed they were invariably GFP-positive and a mature CD4- or CD8-positive T cell type (data not shown). Interestingly, Eμ-Bmi-1 transgenic mice develop both B and T cell lymphomas with a similar latency to that observed upon Cbx7 expression, although these tumors were immature in nature (24). T (and B) cell lymphomas were also observed in Ink4a/Arf−/− mice (25).
Fig. 2.
Expression of Cbx7 in the murine lymphoid compartment causes mature T cell lymphomas. (A) Kaplan–Meier curve showing tumor-free survival. Mice reconstituted with HSCs infected as indicated were monitored for lymphoma onset and illness until they reached a terminal stage and were killed. The data are presented in a Kaplan–Meier format, showing the percentage of mouse survival at various times after reconstitution. (B) Characterization of Cbx7-overexpressing lymphomas. Representative pictures are shown. (C) Western blot of Cbx7 overexpressing lymphomas showing exogenously expressed Cbx7 and endogenous c-Myc levels. β-Actin is used as a loading control. MEFs of the indicated genotypes and a lymphoma arising in a vav-MYC transgenic mouse were used as controls.
Cbx7 Cooperates with Myc to Produce Aggressive Lymphomas.
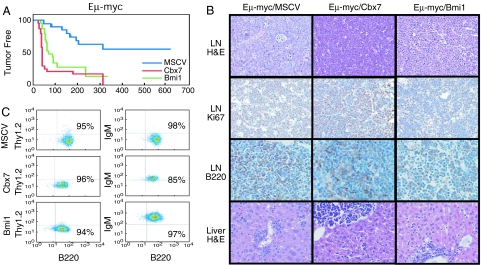
The long latency and incomplete penetrance observed after overexpression of Cbx7 suggest that aberrant Cbx7 expression is not sufficient for lymphomagenesis. Our immunohistochemical analysis indicated that human follicular lymphomas with elevated CBX7 expression often overexpressed c-MYC (see SI Table 2), and immunoblotting of Cbx7 induced lymphomas indicated that two of four also up-regulated c-Myc through an unknown mechanism (Fig. 2C; see Cbx7 lymphomas 2 and 4). To test whether Cbx7 can cooperate with c-Myc, we performed lymphomagenesis experiments as described above, using fetal liver cells derived from Eμ-myc transgenic animals, which produce B cell lymphomas within 3–6 months of adoptive transfer (26, 27). Expression of Cbx7 or Bmi-1 resulted in lymphomas in the majority of recipient mice by 4–6 weeks (Fig. 3A, median tumor-free survival 43 days for Eμ-myc/Cbx7 and 74 days for Eμ-myc/Bmi-1 respectively; P < 0.0001 for Eμ-myc/MSCV vs. Eμ-myc/Cbx7 or Eμ-myc/MSCV vs. Eμ-myc/Bmi-1; n = 20, 26 and 23), and overall survival was similarly affected (SI Fig. 8C). Importantly, all Eμ-myc/Cbx7 and Eμ-myc/Bmi-1 lymphomas analyzed were GFP-positive and overexpressed either Cbx7 or Bmi-1 (data not shown).
Fig. 3.
Cbx7 accelerates the onset of lymphomas triggered by c-Myc. (A) Kaplan–Meier curve showing tumor-free survival in recipient animals reconstituted with Eμ-myc fetal liver cells infected as indicated. (B) Characterization of Eμ-myc-expressing lymphomas. Representative pictures are shown. (C) Immunophenotyping by flow cytometry.
Whole-body imaging (SI Fig. 8D) and pathological examination revealed that the Eμ-myc/Cbx7 lymphomas were aggressive, involving all major lymph node groups and causing splenomegaly and thymic enlargement. Lymphomas disseminated to the liver, lungs, kidneys, and occasionally the heart (Fig. 3B and data not shown). All Eμ-myc-derived lymphomas displayed a high proliferative index as assessed by Ki67 staining (Fig. 3B). Immunological analysis showed that all of the Eμ-myc/Cbx7 lymphomas were of B cell origin (B220 positive), similar to that observed for Eμ-myc/GFP and all Eμ-myc/Bmi-1 lymphomas. However, in contrast with Eμ-myc/GFP, all Eμ-myc/Cbx7 and Eμ-myc/Bmi-1 lymphomas tested presented with a mature B cell phenotype (Thy1.2− B220+ IgM+; Fig. 3C), similar to the immunophenotype of Eμ-myc/Ink4a/Arf+/− lymphomas (12). These results suggest that Cbx7 can both initiate and accelerate tumorigenesis.
The Ink4a/Arf Locus Is a Target of Cbx7 in Lymphomagenesis.
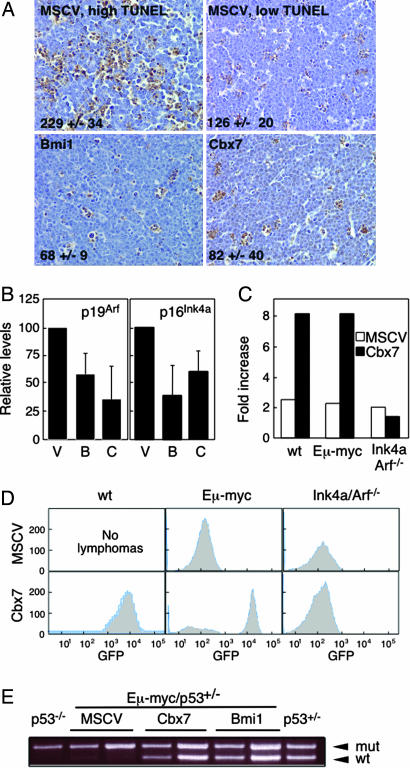
In the Eμ-myc model, loss of Ink4a/Arf accelerates lymphomagenesis mainly by disabling the Arf-p53 circuit and impacting p53-dependent apoptosis (12, 28, 29). Because Cbx7 can repress Ink4a/Arf transcription (16), we analyzed expression of p16Ink4a, p19Arf and apoptosis in a panel of Eμ-myc/MSCV, Eμ-myc/Cbx7, and Eμ-myc/Bmi-1 lymphomas by quantitative RT-PCR and TUNEL analysis of lymphoma sections. Control lymphomas displayed a variable range in the expression of both Ink4a/Arf transcripts and of apoptosis (see Fig. 4A for representative TUNEL staining; 8/12 Eμ-myc/MSCV lymphomas showed high TUNEL staining, and four showed low levels of apoptosis). However, lymphomas overexpressing either Cbx7 or Bmi-1 expressed low levels of p16Ink4a and p19Arf compared with MSCV (Fig. 4B), and their levels of apoptosis were consistently low (Fig. 4A). Similarly, splenic lymphomas produced with Cbx7 as the only exogenous transgene, expressed low levels of p16Ink4a and p19Arf transcripts and p19Arf protein compared with normal spleen, low levels of apoptosis (39+/−17 TUNEL-positive cells per 400× field as compared with Fig. 4A) and low levels of p53 by IHC (SI Fig. 10). In addition, PCR performed using genomic DNA from the two Cbx7-overexpressing lymphomas where DNA was available showed no deletions of the Ink4a/Arf locus (data not shown). Although it is difficult to infer a mechanism from end-stage tumors that may have uncharacterized secondary mutations, these results are consistent with our in vitro data (see SI Fig. 9) and suggest that Cbx7 can suppress the Ink4a/Arf locus and reduce apoptosis in vivo.
Fig. 4.
The Ink4a/Arf locus and p53 mediate Cbx7 effects in lymphomagenesis. (A) TUNEL analysis. Number of TUNEL-positive cells per 400× field is shown. (B) Quantitative RT-PCR analysis of transcript levels for p19Arf and p16Ink4a for lymphomas of the indicated genotypes. (C) Expression of Cbx7 confers a growth advantage on fetal liver cells obtained from WT or Eμ-myc embryos but not on those derived from Ink4a/Arf−/− embryos, as measured by the increase in GFP-positive cells after 7 days of in vitro culture. (D) Fetal liver cells derived from embryos of genotypes as indicated were infected with the named constructs coexpressing GFP and injected into irradiated mice. Resulting lymphomas were analyzed for GFP expression. (E) Eμ-myc/p53+/− fetal liver cells were infected with the indicated constructs coexpressing GFP and injected into irradiated mice. Loss of heterozygosity of p53 was analyzed by allele-specific PCR in the resulting lymphomas.
To directly test the genetic interactions between Cbx7, Ink4a/Arf, and p53, we examined whether Cbx7 would confer a proliferative advantage to cells already lacking Ink4a/Arf and could diminish the selective pressure to disable the p53 pathway during lymphomagenesis. Cbx7 and control retroviral vectors (MSCV and Bmi-1) were introduced into enriched HSC cultures from fetal livers of different genotypes (C57BL/6 WT, Eμ-myc, or Ink4a/Arf−/−), and the coexpressed GFP reporter was used to monitor Cbx7 effects during proliferation and tumorigenesis. Expression of Cbx7 conferred a growth advantage to fetal liver cells derived from WT or Eμ-myc embryos, as evidenced by a notable in vitro expansion of GFP-positive cells (8-fold increase within 1 week) that eventually overgrew their noninfected (GFP-negative) counterparts (Fig. 4C and data not shown). However, Cbx7 expression did not confer a growth advantage to Ink4a/Arf−/− fetal liver cells in vitro. Similarly, whereas all of the Cbx7 and Bmi-1 lymphomas obtained after adoptive transfer of WT or Eμ-myc HSCs into recipient animals were GFP-positive, none of the lymphomas arising from Ink4a/Arf-deficient HSC populations expressed GFP (Fig. 4D; 3/3 Cbx7 and 2/2 Bmi-1 lymphomas). Finally, lymphomas arising from Eμ-myc/p53+/− HSCs expressing Cbx7 or Bmi-1 invariably retained the WT p53 allele, whereas those arising from MSCV-infected controls were p53-null, because they lost this allele (Fig. 4E). Sequence analysis of the retained p53 allele in the Cbx7 lymphomas confirmed it to be WT (data not shown). Thus, Cbx7 requires Ink4a/Arf to confer a selective advantage in vitro and in vivo, and the presence of Cbx7 can compensate for p53 loss during the process of lymphomagenesis.
Discussion
In this investigation, we used a bioinformatics prescreening analysis followed by histopathology to narrow down the potential relevant target cell type for CBX7-mediated oncogenesis to lymphocytes and demonstrated that this PcG gene can transform the lymphoid compartment in mice. Although the potential contribution of other PcG genes to tumorigenesis has been widely assumed (5, 7, 30), Cbx7 is a previously uncharacterized example of a chromobox protein with potent oncogenic potential in vivo. In follicular lymphoma, CBX7 up-regulation occurs predominantly at the RNA level, although we did not identify a mutational mode of oncogene activation in this study. Bmi-1 is the only other PcG protein whose oncogenic properties have been investigated and validated to a similar extent (10, 31). Of note, although ≈10% of mantle cell lymphomas (MCLs) amplify 10p12–13, where BMI-1 resides, many MCLs also up-regulate BMI-1 transcript in the absence of genomic amplification (11).
Despite their structural (and presumably biochemical) differences, both BMI-1 and CBX7 are subunits of PRC1 complexes repressing the transcription of the INK4a/ARF locus, and both promote lymphomagenesis. However, BMI-1 and CBX7 appear to exist in distinct complexes and can exert their effects independently of each other (16). In keeping with these observations, Bmi-1 protein levels were undetectable in Eμ-myc/Cbx7 lymphomas, and conversely Cbx7 was not detected in Eμ-myc/Bmi-1 lymphomas (data not shown). Importantly, as reported here, BMI-1 and CBX7 also have different patterns of expression in normal germinal centers and are associated with distinct lymphoma subtypes. Together, these results demonstrate that PcG genes other than BMI-1 can be overtly oncogenic and could contribute to various human pathologies.
In our experiments, hematopoietic progenitor cells transduced with Cbx7 were subsequently used to reconstitute the hematopoietic compartment of irradiated recipient mice. In the absence of any other exogenously supplied oncogenic lesion, Cbx7 promotes T cell lymphomagenesis. However, when a sensitizing oncogene is directed to the B cell compartment, Cbx7 promotes aggressive B cell lymphomagenesis with a penetrance and latency similar to Bmi-1. Although the resulting lymphomas are of a mature B cell type, they are distinct from human follicular lymphoma in that they do not reflect a postgerminal center origin (data not shown). Nevertheless, that Cbx7 cooperates with c-Myc in murine lymphomagenesis and is coexpressed with c-MYC in human follicular lymphomas reinforces the relevance of our models to human disease.
Although CBX7 probably affects the transcription of multiple genes, repression of the INK4a/ARF locus appears important for its tumorigenic potential. Thus, like Ink4a/Arf loss, Cbx7 cooperates with c-Myc during lymphomagenesis. Furthermore, Cbx7 expression mimics Ink4a/Arf loss in murine fibroblasts and is unable to further enhance the proliferative potential of Ink4a/Arf−/− hematopoietic progenitors. In these assays, the effects of Cbx7 can be explained by its impact on Arf expression alone, but in other contexts, suppression of p16Ink4a may also be important (16). Similar mechanisms may be operating during the development of human follicular lymphoma, where CBX7 levels are often high but INK4a/ARF deletions and P53 mutations are rare (32, 33). In human follicular lymphoma, where transformation to a fatal, aggressive form can involve various lesions (34), CBX7 may act in conjunction with BCL-2 to restrict INK4a/ARF and P53 activities, during the more indolent stages of this disease and perhaps also during the process of transformation to diffuse large B cell lymphomas. Consistent with this possibility, preliminary studies suggest that coexpression of Bcl-2 with Cbx7 accelerates lymphomagenesis compared with expression of Cbx7 alone, although at this stage our data are not yet statistically significant (P = 0.06, unpublished observations).
Together with previous work, our studies reinforce the importance of disabling the INK4a/ARF tumor surveillance during tumorigenesis. For example, ZBTB7 (Pokemon) has also been identified as a potent oncogene that can promote lymphomagenesis in mice and is up-regulated in human follicular lymphoma; moreover, like CBX7, ZBTB7 is a negative regulator of ARF (21). As shown here, CBX7 is often cooverexpressed with ZBTB7 in human follicular lymphomas, raising the possibility that these proteins act together to control expression from the INK4a/ARF locus. However, based on its functional similarity to BMI-1, it is also possible that CBX7 contributes to lymphomagenesis by enhancing stem cell self-renewal and/or increasing the replicative potential of cancer stem cells. In either case, CBX7 could be a target for control of follicular lymphoma in the clinic.
Materials and Methods
Histopathology of Human Tumors and Tissues.
We used the ONCOMINE database, version 1.0 (www.oncomine.org; ref. 35) to analyze the expression of CBX7 in publicly available microarray data, comparing normal tissue with cancer samples, and established a P < 0.05 as the cutoff limit for considering the data significant. Graphics and statistic values from this analysis were generated by using ONCOMINE. For IHC analysis, reactive tonsils and tissue microarrays of follicular lymphoma and diffuse large B cell lymphomas were stained with anti-BMI-1 (mouse monoclonal; citric acid, pH 6.0, 1:50 dilution) or anti-CBX7 (rabbit polyclonal; citric acid, pH 6.0, 1:2,000 dilution) antibodies. Antibodies were detected with a biotinylated anti-mouse or -rabbit antibody and detected with a streptavidin detection system or the PowerVision+ Detection system (ImmunoVision, Springdale, AR). Criteria for scoring and analyzing CBX7 expression included approximate quantitation of percentage of tumor cells that showed nuclear localization, which ranged from <5% to >50%. Criteria for scoring BCL-2, CD10, Ki67, BCL-6, and ZBTB7 expression was as published (21). Criteria for scoring c-MYC included approximate quantization of percent tumor cells showing nuclear localization which ranged from 0% (rare scattered) to >5% of tumor cells. Statistical analyses for correlations between various markers were performed by using Fisher's exact test.
Generation of Lymphomas.
Cell populations enriched for HSCs or early progenitor cells of various genotypes were derived from fetal livers at embryonic days 13.5–15.5 and infected with retroviruses expressing various genes. Infected cells were used to reconstitute the hematopoietic compartment of C57BL/6 mice irradiated with a single 8-Gy-dose of total body γ-irradiation (Cesium source; 0.8 Gy/min) (29). The retroviral vectors used in this study were MSCV IRES GFP (referred as MSCV), MSCV Cbx7 IRES GFP (referred as Cbx7), and MSCV Bmi-1 IRES GFP (referred as Bmi-1). Mice were observed for lymphoma onset with periodic palpation of peripheral lymph nodes, blood smears, overall morbidity, and by whole-body fluorescence imaging. After the appearance of well palpable lymphomas, tumors were harvested and either fixed for histological evaluation or rendered as single-cell suspensions and used for different assays or stored frozen in 10% DMSO. All mouse experiments were performed according to protocols approved by Cold Spring Harbor Laboratory. Statistical analysis was performed with a one-way ANOVA test by using PRISM (Version 3.0, GraphPad, San Diego, CA).
Histopathology and Immunophenotyping of Murine Tumors.
Tissue samples were fixed in 4% normal buffered formalin, embedded in paraffin, sectioned into 5-μm slices, and stained with hematoxylin/eosin. For IHC, the following antibodies were used: Ki67 (rabbit ab, Novocastra, Norwell, MA), p53 (rabbit ab, CM5, Novocastra), CD3 (rabbit ab, DAKO Cytomation), and B220 (rat Ab, clone RA3–6B2, BD Biosciences PharMingen, Franklin Lakes, NJ). Representative sections were deparaffinized, rehydrated in graded alcohols, and processed by using the avidin-biotin immunoperoxidase method. Sections were then subjected to antigen retrieval by using microwave heating on citric buffer (except for the B220 Ab). Appropriate biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA) were used. Diaminobenzidine was used as the chromogen, and hematoxylin was used to counterstain nuclei. The apoptotic rate was analyzed by TUNEL assay according to published protocols (29). For immunophenotyping, samples were incubated with phycoerythrin or APC-Cy7-conjugated antibodies directed against Thy1.2, B220, IgM, CD4, or CD3 (Pharmigen, San Diego, CA), and data were collected on a LSRII flow cytometer equipped with 488- and 635-nm lines (BD, San Jose, CA). Debris and dead cells were gated out during analysis.
Western Blotting and Real-Time PCR.
Both were performed by using standard approaches, as described in SI Text.
Loss of Heterozygosity (LOH) Analysis of p53.
Eμ-myc/p53+/− fetal liver cells were obtained from embryos of crosses between Eμ-myc transgenic mice and p53+/− mice, all on the C57BL/6 genetic background. Lymphomas were resected from tumor-bearing mice and DNA prepared from FACS-sorted GFP-positive B220-positive cells. LOH at the p53 locus was detected by allele-specific PCR (29).
Supplementary Material
Acknowledgments
We thank M. Roussel (St. Jude Children's Research Hospital, Memphis, TN) for the p19Arf antibody and J. C. Acosta, M. Asher, I. Linkov, T. Matos, M. E. Dudas, D. Pant, E. Jansen, K. Campbell, R. Sachidanandam, M. Serrano, S. Powers, M. McCurrach, and M. Collado for assistance and helpful advice. C.L.S. was a Seligson Clinical Fellow and a Special Fellow of the Leukemia and Lymphoma Society of America and is currently supported by the Australian National Health and Medical Research Council (CDA 406675). D.M. was supported in part by a grant from Joan's Legacy Foundation. This work is supported by the Don Monti Foundation, the Wellcome Trust (D.B.), the Medical Research Council (D.B. and J.G.), Cancer Research UK (G.P. and J.G.), the National Cancer Institute (C.C.-C. and S.W.L.), and the Leukemia and Lymphoma Society of America (S.W.L.).
Abbreviations
- Pc
Polycomb
- PcG
Pc group
- PRC
Pc repressive complex
- CBX7
Chromobox 7
- MSCV
murine stem cell virus
- HSC
hematopoietic stem cells
- IHC
immunohistochemistry
- IRES
internal ribosomal entry site.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608721104/DC1.
References
- 1.Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 4.Otte AP, Kwaks TH. Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 5.Raaphorst FM. Hum Mol Genet 14 Spec No. 2005;1:R93–R100. doi: 10.1093/hmg/ddi111. [DOI] [PubMed] [Google Scholar]
- 6.Lessard J, Sauvageau G. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 7.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Gil J, Bernard D, Peters G. DNA Cell Biol. 2005;24:117–125. doi: 10.1089/dna.2005.24.117. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 10.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 11.Bea S, Tort F, Pinyol M, Puig X, Hernandez L, Hernandez S, Fernandez PL, van Lohuizen M, Colomer D, Campo E. Cancer Res. 2001;61:2409–2412. [PubMed] [Google Scholar]
- 12.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 14.Haupt Y, Bath ML, Harris AW, Adams JM. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- 15.Sherr CJ. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 16.Gil J, Bernard D, Martinez D, Beach D. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, et al. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong D. J Clin Oncol. 2005;23:6358–6363. doi: 10.1200/JCO.2005.26.856. [DOI] [PubMed] [Google Scholar]
- 20.Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J. Oncogene. 2005;24:5543–5551. doi: 10.1038/sj.onc.1208735. [DOI] [PubMed] [Google Scholar]
- 21.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 22.Lossos IS, Alizadeh AA, Diehn M, Warnke R, Thorstenson Y, Oefner PJ, Brown PO, Botstein D, Levy R. Proc Natl Acad Sci USA. 2002;99:8886–8891. doi: 10.1073/pnas.132253599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Climent JA, Alizadeh AA, Segraves R, Blesa D, Rubio-Moscardo F, Albertson DG, Garcia-Conde J, Dyer MJ, Levy R, Pinkel D, Lossos IS. Blood. 2003;101:3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- 24.Alkema MJ, Jacobs H, van Lohuizen M, Berns A. Oncogene. 1997;15:899–910. doi: 10.1038/sj.onc.1201262. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 26.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 27.Schmitt CA, Fridman JS, Yang M, Lee S, Baranov E, Hoffman RM, Lowe SW. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 28.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tokimasa S, Ohta H, Sawada A, Matsuda Y, Kim JY, Nishiguchi S, Hara J, Takihara Y. Exp Hematol. 2001;29:93–103. doi: 10.1016/s0301-472x(00)00620-2. [DOI] [PubMed] [Google Scholar]
- 31.Alkema MJ, van der Lugt NM, Bobeldijk RC, Berns A, van Lohuizen M. Nature. 1995;374:724–727. doi: 10.1038/374724a0. [DOI] [PubMed] [Google Scholar]
- 32.Elenitoba-Johnson KS, Gascoyne RD, Lim MS, Chhanabai M, Jaffe ES, Raffeld M. Blood. 1998;91:4677–4685. [PubMed] [Google Scholar]
- 33.Lo Coco F, Gaidano G, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. Blood. 1993;82:2289–2295. [PubMed] [Google Scholar]
- 34.Lossos IS. Leukemia. 2005;19:1331–1333. doi: 10.1038/sj.leu.2403801. [DOI] [PubMed] [Google Scholar]
- 35.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.