Abstract
At various times after orthopedic operations (more than a few weeks, with an average of 29.9 days), 11 patients had a sudden onset of high temperature (average 38.9°C) and local cellulitis at different sites on the operated sides. The wounds had completely healed, without complicated infections, when the cellulitis occurred. The clinical picture of cellulitis in all patients was atypical: diffuse salmon-pink skin color, local heat, swelling, spontaneous pain, and tenderness but no eruptions. No patient had any underlying immunocompromising conditions or had been given immunosuppressive agents. Gram-negative spiral bacteria were isolated from blood cultures and were identified as Helicobacter cinaedi on the basis of 16S rRNA gene sequencing and DNA-DNA hybridization using standard strains. By means of phylogenetic analysis, we divided these clinical isolates into two clones. The H. cinaedi strain isolated via fecal cultures from two patients without intestinal symptoms was the same clone as the blood isolate. All isolates were quite susceptible to various antibiotics, and clinical and inflammatory symptoms of bacteremia and cellulitis improved after treatment with penicillins and cephalosporins. A relatively high incidence of recurrence of the same disease was observed, however. Almost all patients responded immunologically to the infection, as evidenced by the production of serum antibody against H. cinaedi. We thus suggest that H. cinaedi should not be regarded as simply an opportunistic pathogen but that it may be a pathogen in immunocompetent hosts and may cause infections together with bacteremia and cellulitis.
Helicobacter cinaedi, first identified as a Campylobacter species but now known to be an enterohepatic Helicobacter species, is usually found in the intestinal tract and/or liver of humans and other mammals (2, 8, 28, 30, 32, 33, 35-37). The last two decades have seen increasing numbers of reports of H. cinaedi infections, mainly in humans and particularly in individuals with underlying immunosuppressive conditions such as AIDS, malignant diseases, and chronic alcoholism (3, 11, 18, 20, 23). Vandamme et al. previously documented a few cases of infection with H. cinaedi isolated from feces and blood from an apparently nonimmunocompromised child and adult (36). Other groups have also described invasive infections caused by H. cinaedi and other Campylobacter-like organisms (Helicobacter fennelliae) in patients without underlying diseases (15, 26). However, despite these reports, the pathogenicity and etiological properties of this spiral bacterium are only poorly understood. Also, the molecular epidemiology of various strains isolated from humans has not yet been systematically analyzed.
We recently observed 11 cases of H. cinaedi bacteremia and cellulitis that occurred consecutively during a particular period in the same hospital. The clinical and epidemiological features of the H. cinaedi infections were investigated in the present study, which may thus provide important insights into the pathogenesis and epidemiology of this emerging pathogen.
MATERIALS AND METHODS
Isolation and culture of bacteria from samples of blood and feces.
An aliquot of 10 ml of patients' venous or arterial blood was cultured by using the BD BACTEC Plus Aerobic/F system according to the manufacturer's protocols (Becton Dickinson and Company, Franklin Lakes, NJ). After the culture-positive notification, a part of the culture sample was inoculated onto Campylobacter agar (Becton Dickinson) and incubated at 37°C under microaerobic conditions (CampyPak microaerophilic system; Becton Dickinson) with high humidity. After Gram staining of the colony-forming bacteria, bacterial growth was examined microscopically. A fecal culture was performed similarly to the blood culture by inoculating and cultivating the samples of feces on Campylobacter agar (Becton Dickinson). The spiral bacteria isolated were used for further investigation as described below.
Identification of the species of the clinical isolates.
DNA from type strains of H. cinaedi (CCUG 18818T) and Helicobacter canis (NCTC 12379T) and representative strains of clinical isolates (isolate 377 from case 2 and isolate 717 from case 5) were prepared according to a standard procedure (21). DNA from each strain was labeled with photobiotin (Vector Laboratories, Inc., Burlingame, CA), and microplate quantitative DNA-DNA hybridization was performed according to previously described methods (6) to determine the taxonomic species name.
Phylogenetic analysis by using 16S rRNA and hsp60 gene sequences.
16S rRNA and hsp60 genes of all isolates were amplified by PCR as previously described (7, 16, 22). The sequences were determined by using an automatic sequencer (model 3100; Applied Biosystems, Foster City, CA) and a dye terminator reaction kit (Applied Biosystems). About 1,430 bp of the 16S rRNA gene sequence and 530 bp of the hsp60 gene sequence were determined for each strain. To detect closely related species, each sequence discovered was analyzed by means of the FASTA search system (29) found at the DNA DataBank of Japan (DDBJ) website (http://www.ddbj.nig.ac.jp). Sequences of the 16S rRNA and hsp60 genes of closely related species of the genus Helicobacter were taken from the DDBJ, GenBank, and European Molecular Biology Laboratory (EMBL) databases. CLUSTAL-X software, originally described by Thompson et al. (34), was then used to ascertain the phylogenetic relationships for each isolate. The phylogenetic tree was drawn by using TreeView software (27, 29).
Typing via PFGE.
Patterns of large restriction fragments of genomic DNA were obtained by pulsed-field gel electrophoresis (PFGE) as described elsewhere previously (25). Organisms cast into plugs were lysed with lysozyme, sodium dodecyl sulfate, and proteinase K. The genomic DNA embedded in the plugs was digested with SpeI restriction endonuclease and was separated in a 1% agarose gel under conditions of 6 V/cm for 20 h at 14°C and pulse times of 5.3 to 34.9 s, ramped linearly using the CHEF-DR III system (Bio-Rad Laboratories, Hercules, CA). A lambda DNA ladder (New England BioLabs, Hitchin, United Kingdom) was used as a molecular size marker. Gels were stained with ethidium bromide and photographed under UV light. Banding patterns were compared by visual inspection.
Analysis by RAPD.
Random amplified polymorphism DNA (RAPD) analysis was performed according to previously described methods (1, 4). After preliminary experiments, we selected two primers (primer 1281 [5′-AAC GCG CAA-3′] and primer 1283 [5′-GCG ATC CCC A-3′]) for RAPD typing of the H. cinaedi strains. PCR was carried out in 50 μl of a mixture containing 1 μl of extracted DNA (∼20 ng), 5 μl of 10× PCR buffer containing 20 mM Mg2+, 2.5 μl of deoxynucleoside triphosphate mix (2.5 mM each), 5 μl of primer (4 μM), and 0.5 μl of hot-start EX-Taq (5 U/μl; Takara Bio Inc., Shiga, Japan). The cycling program was 1 cycle each at 94°C for 2 min, 37°C for 1 min, and 72°C for 4 min and 29 cycles each at 94°C for 2 min, 37°C for 3 min, and 72°C for 7 min. After PCR, 1.0% agarose gel electrophoresis with ethidium bromide staining was performed, and the DNA fingerprint of each strain was evaluated.
ELISA for determination of antibody response to the infection.
For the enzyme-linked immunosorbent assay (ELISA) for the determination of the antibody response to the infection, H. cinaedi whole-cell antigen was first prepared by sonication of bacterial cells of clinical isolates that had been cultured on and collected from Campylobacter agar according to a method reported previously (14). We first tested the antibody titer by use of whole-cell antigen prepared from several different clinical isolates (e.g., strains 923 and 1035 as representatives) (Table 1) of two different clones that we identified in the present genomic analysis. Because the ELISA showed almost similar responses for the sera from different patients, we used strain 1035 to prepare the antigen throughout the study. The whole-cell extract was then treated with Triton X114 to remove lipopolysaccharide as described previously (19), and a fraction obtained via ultrafiltration (<100 kDa; Millipore, Billerica, MA) was used as an H. cinaedi antigen in the ELISA. Each well of a 96-well microtiter plate was coated with 50 μl of H. cinaedi extract (0.0625 μg of protein/well) in 0.1 M carbonate buffer (pH 9.6), blocked with 0.5% gelatin, and washed three times with phosphate-buffered saline containing 0.05% Tween 20 (washing buffer). Samples (400-fold-diluted human serum) in the wells were incubated for 1 h at room temperature. Wells were then washed with washing buffer and reacted with horseradish peroxidase-conjugated anti-human immunoglobulin G antibody (Sigma-Aldrich Corporation, St. Louis, MO), followed by a reaction with 1,2-phenylenediamine dihydrochloride. The reaction was terminated by the addition of 50 μl of 2.0 mol/liter sulfuric acid, and absorbance at 490 nm was read by means of a micro-ELISA plate reader. To examine the host immune response against H. cinaedi infection, serum levels of antibody against H. cinaedi antigen of H. cinaedi-infected groups and those of three different control groups, Helicobacter pylori-infected subjects, age- and sex-matched controls without apparent H. cinaedi infection, and infant subjects younger than 1 year, were compared.
TABLE 1.
Clinical characteristics of patients with H. cinaedi infectionsa
| Case | Age (yr) | Sex | Isolated strain(s) | Ward floor | Orthopedic diagnosis | Date of onset of cellulitis | Duration after operation (days) | Location of cellulitis | WBC count (cells/mm3) | CRP (mg/dl) | Recurrence | Stool culture | Diarrhea | Antibiotic treatment | Date of serum sampling (ELISA) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 65 | F | 923 | 7th | L HF, RF | 19 June 2004 | 18 | L forearm and chest | 13,160 | 12.3 | + | − | + | SBTPC | 23 February 2005 |
| 2 | 71 | F | 377 | 7th | LH | 1 July 2004 | 8 | Back | 12,600 | 13.5 | − | ND | − | IPM-CS | ND |
| 3 | 68 | F | 944 | 7th | L HF | 21 July 2004 | 21 | L upper arm | 18,420 | 12.3 | + | − | − | CTM | 4 March 2005 |
| 4 | 22 | M | 409 | 5th | R FeF, TF, FiF | 9 August 2004 | 18 | R leg | 18,880 | 13.7 | − | ND | − | SBTPC | ND |
| 5 | 76 | F | —b | 7th | R OA | 1 July 2004 | 12 | R lateral thigh | 14,830 | 19.3 | − | ND | − | IPM-CS | 1 July 2005 |
| 6th | L OA | 15 September 2004 | 10 | L medial thigh | 15,120 | 22.0 | − | ND | − | CTM | ND | ||||
| 6 | 58 | M | 717 | 6th | L HF | 28 October 2004 | 28 | L upper arm | 10,470 | 14.9 | + | − | − | CTM | 25 February 2005 |
| —b | 6th | L CF | 28 October 2004 | 42 | L leg | CTM | ND | ||||||||
| 7 | 79 | F | 1035, 14 | 6th | R HF | 24 December 2004 | 113 | R upper arm | 13,300 | 5.6 | + | − | − | SBTPC | 9 February 2005 |
| 896 | 6th | R CF | 20 January 2005 | 42 | R leg | 5,540 | 0.6 | − | ND | − | SBTPC | ND | |||
| 8 | 70 | F | 643 | 6th | L TF, FiF | 17 January 2005 | 56 | L leg | 7,250 | 6.0 | − | ND | − | CTM | ND |
| 9 | 72 | F | 56, 361 | 6th | R TF, FiF | 1 February 2005 | 8 | R leg | 10,230 | 10.2 | − | + | − | CTM | 21 February 2005 |
| 10 | 62 | F | 1007 | 6th | R OA | 18 February 2005 | 11 | R thigh | 6,140 | 18.7 | − | − | − | SBTPC | 8 March 2005 |
| 11 | 77 | F | 545, 892 | 6th | R OA | 10 March 2005 | 31 | R thigh | 15,790 | 19.2 | − | + | − | CTM | 17 March 2005 |
Abbreviations: CF, calcaneous fracture; FeF, femoral fracture; FiF, fibular fracture; HF, humeral fracture; RF, radial fracture; TF, tibial fracture; LH, lumbar herniation; OA, osteoarthritis, R, right; L, left; WBC, white blood cell; CRP, C-reactive protein; ND, not done; SBTPC, sultamicillin tosylate; CTM, cefotiam hydrochloride; IPM-CS, imipenem-cilastatin sodium; F, female; M, male.
Bacteria had first been isolated and identified, but these isolates became extinct during successive cultures.
Procedures for obtaining human serum samples used in this study were in accordance with those recommended by the Regional Ethical Committee on Human Experimentation of Kumamoto University and Kumamoto Orthopedic Hospital. Written informed consent was obtained from each subject.
Statistical analysis.
Statistical significance between groups was determined by the two-tailed unpaired Student's t test. A P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
All sequence data determined in this work were deposited in the DDBJ/GenBank/EMBL databases under the accession numbers listed in Fig. 2.
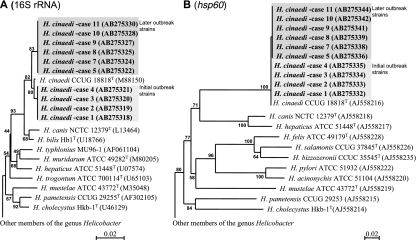
FIG. 2.
Genetic analysis of clinical isolates of H. cinaedi. 16S rRNA (A) and hsp60 (B) genes were analyzed by means of the FASTA search system. Phylogenetic relationships for these clinical isolates and representative members of the genus Helicobacter were analyzed on the basis of the 16S rRNA gene sequence (1,430-bp area) and hsp60 gene sequence (530-bp area) obtained from the DDBJ, GenBank, and EMBL databases. The accession numbers for the 16S rRNA and hsp60 gene sequences of each strain are shown in parentheses.
RESULTS
Case reports.
During 9 months from July 2004 to March 2005, 11 patients who had been admitted to Kumamoto Orthopedic Hospital (204 beds) in Kumamoto, Japan, for orthopedic surgery suffered from bacteremia and cellulitis caused by H. cinaedi. Table 1 summarizes the clinical characteristics of the patients (two men and nine women). These patients, whose ages ranged from 22 to 79 years (average, 65.5 years), had a sudden onset of cellulitis accompanied by high temperature. The clinical symptoms appeared an average of 29.9 days (range, 8 to 113 days) after the orthopedic operations. At the time that the patients showed these symptoms, wounds at operative sites had completely healed. H. cinaedi was isolated from cultures of blood obtained from all patients: four isolates from venous blood and seven isolates from arterial blood. In the past, three patients (79-, 76-, and 65-year-old women) had had hypertension, but it was well controlled when they were infected, and patients' histories contained no records of autoimmune or malignant diseases or their treatment. Also, the patients did not have any underlying disorders that would cause an impairment of antimicrobial host defense, nor did they have abnormal findings from physical and laboratory examinations except for the orthopedic diseases, i.e., bone fractures and osteoarthritis (Table 1), which necessitated the orthopedic surgeries. Operations included six operations for bone fractures (a 22-year-old man had an open fracture), four for artificial knee joint replacement for osteoarthritis, and one for lumbar herniation. All patients had cellulitis on their operated side together with fever (38.5°C to 39.5°C; highest mean temperature, 38.9°C); at that time, H. cinaedi organisms were isolated from blood. One patient (case 1) had a symptom of enteritis (diarrhea); however, H. cinaedi was not isolated from this patient by fecal culture. H. cinaedi organisms were isolated from two other patients without intestinal symptoms via fecal cultures (cases 9 and 11). Two patients (cases 5 and 7) underwent two operations at different times, and each time, cellulitis occurred in the operated side, with isolation of bacteria from blood culture.
All patients showed atypical clinical symptoms of cellulitis, including diffuse, pale, salmon-pink skin color; local heat; swelling; spontaneous pain; tenderness; and no eruptions. Figure 1 provides a representative picture of the appearance and magnetic resonance image of cellulitis. Cellulitis occurred in the left lower leg, the same side as the operation but not near the surgical incision and wound, 42 days after the surgery. The lesion of this patient was accompanied by swelling, salmon-pink skin color, local heat, spontaneous pain, and tenderness. Coronal and axial T2-weighted magnetic resonance images of the left lower leg confirmed subcutaneous edematous and inflammatory changes, which are typical findings of cellulites. In all cases, healing of the surgical wounds had been completed by the time of the onset of cellulitis, and there were no apparent infection-related findings such as redness, pain, swelling, and purulent discharge in the wound tissues. Hematology and blood chemistry studies indicated leukocytosis and elevated C-reactive protein and erythrocyte sedimentation rate values when patients had cellulitis and fever. All these symptoms of inflammation were thought to be unrelated to a direct effect of the operations, because they appeared more than 1 week after the date of surgery, when the surgical wounds had already healed. Antibiotic chemotherapy was effective, so cellulitis and fever along with other inflammatory findings improved within a few days after starting treatment. However, 4 of 11 patients had recurrences of cellulitis in the same area, although the degree of severity of the cellulitis diminished gradually with each recurrence. For example, one patient (case 6, a 58-year-old man) had five recurrences of cellulitis during follow-up at an outpatient clinic, and for the later recurrences, the cellulitis spontaneously disappeared without any specific treatment.
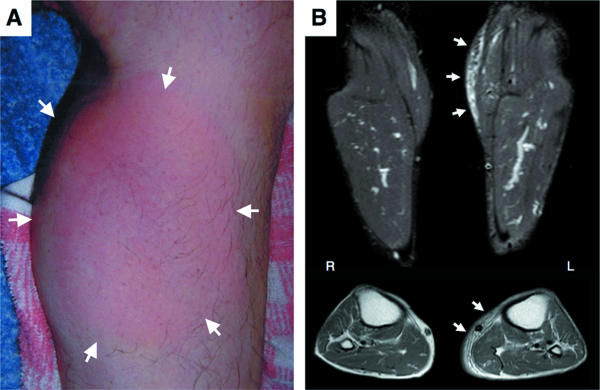
FIG. 1.
A typical case of postoperative cellulitis caused by H. cinaedi (case 6). (A) A 58-year-old man with a left calcaneous fracture resulting from a traffic accident underwent orthopedic surgery in September 2004. Cellulitis occurred in the left lower leg. Note the salmon-pink color and swelling (arrows). (B) Magnetic resonance images of the left lower leg. Coronal and axial T2-weighted images showed a diffuse, widespread, high-intensity area, indicating inflammatory lesions in the subcutaneous tissues (arrows).
Culture and isolation of bacteria.
Culture of blood from all 11 patients via the BACTEC Plus system produced positive results after 4 to 7 days of incubation of cultures. Microscopic examination of bacterial organisms grown in this system revealed that all strains were a gram-negative spiral-shaped bacterium. Subculture from the culture bottle on Campylobacter agar resulted in growth under microaerobic conditions, but the organisms were very fastidious with regard to growth conditions such as nutrient and gas composition and humidity. For example, on a Columbia blood agar plate (bioMérieux, Marcy l'Etoile, France) or other media under the same culture conditions, isolates manifested no growth. A CO2 incubator for tissue cultures (supplied with 5% CO2 gas) with humidification or an anaerobic incubator (N2 base gas with 10% CO2 and 10% H2) also resulted in no effective growth of this organism. The fecal culture for the two patients who had no apparent enteritis symptoms grew the same gram-negative spiral bacterial organisms as those grown via blood culture.
Identification of bacterial strains.
All isolates showed very high 16S rRNA gene sequence similarity to the type strains of H. cinaedi (more than 99.3%), whereas less than 98.5% similarity to other species of the genus Helicobacter was found (Fig. 2). The genomic DNA-DNA hybridization result revealed that the isolates (two representative strains) belonged to the species H. cinaedi, because they showed more than 82% homology with the type strain of H. cinaedi (Table 2).
TABLE 2.
DNA-DNA hybridization similarity of two clinical H. cinaedi isolates
| Isolate | % Homology for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
H. cinaedi type strain
|
H. cinaedi, case 2
|
H. cinaedi, case 5
|
H. canis type strain
|
|||||
| Optimal conditions | Stringent conditions | Optimal conditions | Stringent conditions | Optimal conditions | Stringent conditions | Optimal conditions | Stringent conditions | |
| H. cinaedi CCUG 18818T | 100.0 | 100.0 | 108.0 | 106.1 | 94.0 | 89.9 | 7.4 | 3.7 |
| H. cinaedi, case 2 | 82.9 | 81.8 | 100.0 | 100.0 | 82.3 | 76.4 | 5.7 | 2.7 |
| H. cinaedi, case 5 | 101.6 | 99.9 | 103.4 | 100.6 | 100.0 | 100.0 | 7.6 | 3.8 |
| H. canis NCTC 12379T | 9.5 | 4.4 | 10.7 | 5.4 | 9.1 | 4.3 | 100.0 | 100.0 |
| Salmon DNA | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
Epidemiological features of the isolates.
Molecular epidemiology of the clinical isolates was studied by means of phylogenetic analysis. On the phylogenetic tree, the type strain of H. cinaedi and all isolates formed one cluster (Fig. 2) that comprised two subclusters, the first composed of four strains, which were isolated at the time of the initial H. cinaedi outbreak (cases 1 to 4), and the second containing the other six isolates from the later outbreak (cases 5 and 7 to 11). The type strain of H. cinaedi was located at a position intermediate between the subclusters. The DNA sequences of strains within each subgroup were identical. Five and nine base substitutions were found for the type strain versus isolates of the first subcluster and the second subcluster, respectively; 12 base substitutions were found between the isolates of the first and second subclusters.
Almost the same result was observed with hsp60 gene sequence analysis (Fig. 2). All isolates and the type strain of H. cinaedi clearly made one cluster, with two subclusters appearing within it. Members of each subcluster were exactly the same as those found via 16S rRNA gene phylogenetic analysis.
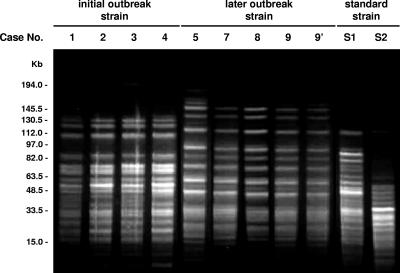
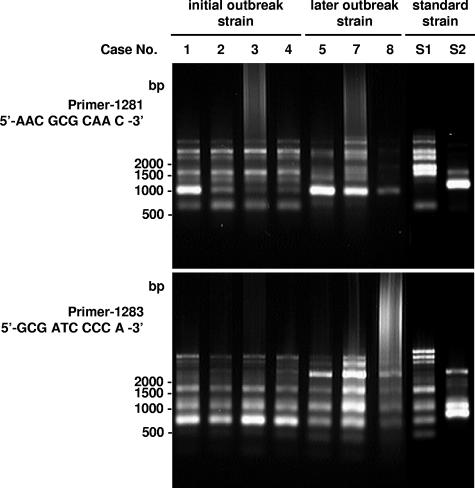
As shown in Fig. 3, PFGE revealed that the type strain of H. cinaedi and clinical strains were completely different: some large bands (more than 112.0 kb) were missing, and some extra intermediate-sized bands (48.5 to 112.0 kb) existed only in the type strain. Also, four isolates from the initial infections (cases 1 to 4) and four from the later outbreak (cases 5, 7, 8, and 9) showed slightly different PFGE patterns. RAPD analysis provided similarly different profiles for the genome of the H. cinaedi isolates (Fig. 4).
FIG. 3.
PFGE DNA fingerprints for H. cinaedi isolates digested with SpeI. PFGE patterns for the clinical strains differed from those for the type strains of H. cinaedi CCUG 18818T (S1) and H. canis NCTC 12379T (S2). Strains of clinical isolates were obtained from blood samples, except for sample 9′, which was isolated from a fecal sample. Case numbers are identical to those in Table 1.
FIG. 4.
RAPD analysis of the genome of H. cinaedi clinical isolates. Primers used in this analysis are indicated in the figure. See the text for details. Case numbers are identical to those in Table 1. S1, H. cinaedi CCUG 18818T; S2, H. canis NCTC 12379T.
Antibody titers for H. cinaedi in patients and control subjects.
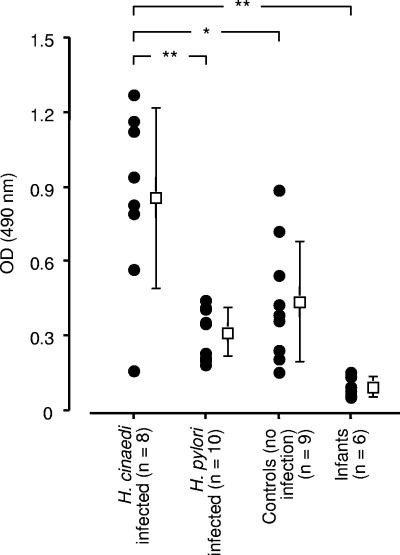
The specific host immune response against H. cinaedi infection was examined by determining the antibody titers for H. cinaedi raised in patients whose blood was used to isolate and culture H. cinaedi organisms. Figure 5 illustrates that the patients' antibody titers were significantly higher than those of control groups, including age- and sex-matched subjects, H. pylori-infected patients, and infants.
FIG. 5.
Determination of antibody response to H. cinaedi infection by ELISA. Serum antibody levels against the H. cinaedi antigen of the H. cinaedi-infected patients were compared with those of control groups: H. pylori-infected subjects, age- and sex-matched controls without apparent H. cinaedi infection, and infants younger than 1 year of age. *, P < 0.05; **, P < 0.01.
DISCUSSION
Here, we describe 11 cases of H. cinaedi bacteremia associated with cellulitis but without apparent immunocompromised conditions, particularly human immunodeficiency virus infection. Previously published epidemiological studies and case reports indicated a strong association of the onset of H. cinaedi infection with an immunocompromised state and/or human immunodeficiency virus infection or a specific condition, e.g., homosexuality (3, 5, 8, 11, 24, 31, 32). We believe, however, that the cases of H. cinaedi infections that we detail here all occurred in apparently immunocompetent subjects, although they may have been prone to develop H. cinaedi infections, as discussed below.
One unique feature of the present H. cinaedi infections is that these 11 cases occurred as a cluster at the same hospital, all after orthopedic operations. These cases may thus represent a nosocomial outbreak. Epidemiological and phylogenetic analysis showed that the clinical isolates could be classified into two groups with several base substitutions. One group of H. cinaedi isolates was obtained from patients who stayed in the hospital ward, mainly on the seventh floor, from June to September 2004, and the other group was derived from patients who stayed on the sixth floor of the same ward from October 2004 to March 2005 (Table 1). Therefore, each clone seems to have spread in a relatively limited environment in the hospital and in a serial fashion during several months.
Because of this epidemiological evidence and because all cases occurred after orthopedic surgery, the present outbreak may have an iatrogenic cause or may have originated from physical areas and people (other patients and hospital staff) that were contaminated with H. cinaedi organisms. A prospective surveillance study was therefore initiated to clarify the epidemiology of H. cinaedi, and infection control measures were reinforced according to recommendations of the Hospital Infection Control Practice Advisory Committee. Because H. cinaedi is thought to have an oral route of infection, water from a feed tank was cultured for the presence of bacteria. Also, swab cultures of surgical instruments and materials used for nursing care of patients in hospital rooms, e.g., wet towels, were taken. More importantly, to identify any H. cinaedi carriers in the hospital, stool samples for culture were obtained from patients without any symptoms of H. cinaedi infection and medical staff members, including physicians, nurses, and physiotherapists, who had close contact with the infected patients. However, all these surveillance measures produced negative results for the presence of H. cinaedi organisms.
Therefore, although the apparent source and route of this outbreak remain unclear, because H. cinaedi was isolated via fecal culture from 2 of 11 patients, the H. cinaedi-infected patients themselves may be the source of the outbreak, which may have spread via an oral route. We also found that one physiotherapist who had no clinical symptoms and negative fecal culture results showed a high serum antibody titer for H. cinaedi by the ELISA, which was performed for hospital medical staff members who had worked closely with the H. cinaedi-infected patients. These data may thus support the hypothesis that some H. cinaedi-infected patients may have served as carriers of the infection in the hospital.
With regard to the pathogenetic mechanism of cellulitis, one characteristic feature of cellulitis in the present cases is that the skin lesions were always found on the operated side (Fig. 1). This feature may be due to impaired regional blood flow in the operated limbs, which may facilitate bacterial adhesion to and intrusion onto the vascular endothelium. In addition, because all lesions occurred at locations that were somewhat remote from the surgical wounds, and because they appeared after the wounds had completely healed, the cellulitis is thought not to be directly caused by an infection at the surgical wound. Rather, observations showed a strong trend for the appearance of cellulitis at the area of the skin where cold compresses had been applied. This finding suggests that cold-induced stress causes impaired local blood flow, which in turn may promote H. cinaedi invasion of skin tissue via the vasculature. This idea is supported by the fact that after the application of cold compresses to the operated area in patients who underwent orthopedic surgery was stopped, no additional cases of H. cinaedi cellulitis occurred in the hospital.
Because of the fastidious and slowly growing nature of H. cinaedi on conventional bacterial culture medium, an appreciable number of cases may be overlooked during routine processing for bacterial isolation and identification at clinical microbiological laboratories. In addition to the 11 patients reported here, we found several patients with atypical cellulitis and high temperature during these outbreaks at the hospital, but H. cinaedi organisms were not isolated from blood and stool samples from these patients. These patients showed significantly high serum antibody titers for H. cinaedi by ELISA (data not shown). More efficient H. cinaedi isolation and diagnostic techniques, which enable clarification of the epidemiology and pathogenesis of H. cinaedi infection, are thus required.
During treatment of the H. cinaedi infections reported here, all patients responded well to antimicrobial agents including broad-spectrum penicillins and cephalosporins (Table 1). However, a high incidence (40%) of recurrence of H. cinaedi cellulitis (bacteremia) was observed. which is consistent with previous case reports of immunocompromised patients (17), even though the patients described here were apparently not so compromised. Recurrence is thus thought to be a typical feature of H. cinaedi infection. In this context, our most interesting cases were the patients (cases 5 and 7) (Table 1) who underwent two operations because of osteoarthritis and bone fracture, respectively, with repeated H. cinaedi-related cellulitis and bacteremia. We suspect that H. cinaedi may have the potential to be a latent pathogen in some tissues yet unidentified, even after appropriate antimicrobial treatment. The recurrence of the H. cinaedi infections is not due to antibiotic resistance of the bacteria acquired after antibiotic treatment, because in case 7, isolates obtained before and after the recurrence of the infection showed similarly good susceptibilities to various antimicrobial agents. Otherwise, this organism could invade the vascular and blood circulation repeatedly from the intestinal tract, where it exists among normal flora as a resident microbiota (32).
Many isolates of H. cinaedi have been discovered, via rectal swab and stool culture, in asymptomatic subjects and individuals with clinical signs of enteritis and proctocolitis. The same species has also been isolated from pet and farm animals, e.g., dogs, cats, and hamsters, and is a potential causative agent of intestinal infections and bacteremia (9, 10, 12, 13, 17, 32). However, all the patients described here did not have a history of pet care before the onset of H. cinaedi infection.
Also, our patients had no background of homosexuality or underlying diseases that are risk factors for H. cinaedi infection. Although the transmission route of the infection is yet to be identified, the present outbreak may have been mediated by an H. cinaedi carrier who, having been admitted to the hospital before the outbreak, may have spread the bacteria in a limited area in the hospital.
It is of great interest that the first and second clones of H. cinaedi that were isolated from our patients affected patients on the seventh and sixth floors, respectively, although there was some overlap in the admittance floor (Table 1). In other words, the present outbreak was not caused by just a single clone of H. cinaedi, and one H. cinaedi clone appears to have been replaced by the other clone during the outbreak period. The mechanism of substitution of one clone for the other is unclear. It may have resulted from a genetic mutation that occurred during the spread of bacteria, or two coexisting clones may have infected and/or colonized a particular individual or the hospital or patient community environment. Also, because the same clone was recovered from both blood and fecal samples of a few patients during the outbreak, H. cinaedi organisms might be transmitted via an oral route among human subjects, colonize the intestinal tract, and then, under particular conditions such as orthopedic surgery, disseminate via the vascular system to cause bacteremia and cellulitis.
In conclusion, we report here a unique outbreak of H. cinaedi infections caused by two different bacterial clones. Although the epidemiology of the infection is not fully understood, these patients did not have the particular risk factors or underlying conditions, except for prior surgical operation, that might lead to this infection. Because of the lack of highly sensitive bacterial identification techniques, other immunocompetent subjects infected with H. cinaedi may have been overlooked in the past. Our present findings now warrant further investigation of the pathogenesis, etiology, and epidemiology of this emerging disease.
Acknowledgments
We are very grateful to Judith B. Gandy for her excellent editing of the manuscript. We thank Tatsuya Kawaguchi, Chief of Infection Control Team at Kumamoto University Hospital, and Tadashi Nakamura and Shuuichi Higashi at Kumamoto Orthopedic Hospital and Keita Yamakawa, a former member of the Infection Control Team at the same hospital, for their helpful discussion and cooperation in the surveillance study. Thanks are also due to Akira Nishizono, Oita University, for providing us with serum from patients infected with H. pylori and neonate serum and with technical support for the ELISA.
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, and Science (MEXT) and by a grant for the future science promotion program from Kumamoto University.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Akopyanz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, L. P. 2001. New Helicobacter species in humans. Dig. Dis. 19:112-115. [DOI] [PubMed] [Google Scholar]
- 3.Burman, W. J., D. L. Cohn, R. R. Reves, and M. L. Wilson. 1995. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin. Infect. Dis. 20:564-570. [DOI] [PubMed] [Google Scholar]
- 4.Burucoa, C., V. Lhomme, and J. L. Fauchere. 1999. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis. J. Clin. Microbiol. 37:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimolai, N., M. J. Gill, A. Jones, B. Flores, W. E. Stamm, W. Laurie, B. Madden, and M. S. Shahrabadi. 1987. “Campylobacter cinaedi” bacteremia: case report and laboratory findings. J. Clin. Microbiol. 25:942-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 7.Ezaki, T., N. Li, Y. Hashimoto, H. Miura, and H. Yamamoto. 1994. 16S ribosomal DNA sequences of anaerobic cocci and proposal of Ruminococcus hansenii comb. nov. and Ruminococcus productus comb. nov. Int. J. Syst. Bacteriol. 44:130-136. [DOI] [PubMed] [Google Scholar]
- 8.Fennell, C. L., P. A. Totten, T. C. Quinn, D. L. Patton, K. K. Holmes, and W. E. Stamm. 1984. Characterization of Campylobacter-like organisms isolated from homosexual men. J. Infect. Dis. 149:58-66. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez, K. R., L. M. Hansen, P. Vandamme, B. L. Beaman, and J. V. Solnick. 2002. Captive rhesus monkeys (Macaca mulatta) are commonly infected with Helicobacter cinaedi. J. Clin. Microbiol. 40:1908-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores, B. M., C. L. Fennell, L. Kuller, M. A. Bronsdon, W. R. Morton, and W. E. Stamm. 1990. Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect. Immun. 58:3947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox, J. G. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, J. G., L. Handt, B. J. Sheppard, S. Xu, F. E. Dewhirst, S. Motzel, and H. Klein. 2001. Isolation of Helicobacter cinaedi from the colon, liver, and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J. Clin. Microbiol. 39:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhart, C. J., C. L. Fennell, M. P. Murtaugh, and W. E. Stamm. 1989. Campylobacter cinaedi is normal intestinal flora in hamsters. J. Clin. Microbiol. 27:1692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto, T., A. Nishizono, T. Fujioka, J. Ikewaki, K. Mifune, and M. Nasu. 1999. Local secretory immunoglobulin A and postimmunization gastritis correlate with protection against Helicobacter pylori infection after oral vaccination of mice. Infect. Immun. 67:2531-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsueh, P. R., L. J. Teng, C. C. Hung, Y. C. Chen, P. C. Yang, S. W. Ho, and K. T. Luh. 1999. Septic shock due to Helicobacter fennelliae in a non-human immunodeficiency virus-infected heterosexual patient. J. Clin. Microbiol. 37:2084-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 17.Kiehlbauch, J. A., D. J. Brenner, D. N. Cameron, A. G. Steigerwalt, J. M. Makowski, C. N. Baker, C. M. Patton, and I. K. Wachsmuth. 1995. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J. Clin. Microbiol. 33:2940-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiehlbauch, J. A., R. V. Tauxe, C. N. Baker, and I. K. Wachsmuth. 1994. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann. Intern. Med. 121:90-93. [DOI] [PubMed] [Google Scholar]
- 19.Liu, S., R. Tobias, S. McClure, G. Styba, Q. Shi, and G. Jackowski. 1997. Removal of endotoxin from recombinant protein preparations. Clin. Biochem. 30:455-463. [DOI] [PubMed] [Google Scholar]
- 20.Mammen, M. P., Jr., N. E. Aronson, W. J. Edenfield, and T. P. Endy. 1995. Recurrent Helicobacter cinaedi bacteremia in a patient infected with human immunodeficiency virus: case report. Clin. Infect. Dis. 21:1055. [DOI] [PubMed] [Google Scholar]
- 21.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 22.Mikkonen, T. P., R. I. Kärenlampi, and M. L. Hänninen. 2004. Phylogenetic analysis of gastric and enterohepatic Helicobacter species based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 54:753-758. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, H., M. Goto, E. Ono, E. Sawabe, M. Iwata, K. Okuzumi, K. Yamaguchi, and T. Takahashi. 2003. Isolation of Helicobacter cinaedi from blood of an immunocompromised patient in Japan. J. Infect. Chemother. 9:344-347. [DOI] [PubMed] [Google Scholar]
- 24.Ng, V. L., W. K. Hadley, C. L. Fennell, B. M. Flores, and W. E. Stamm. 1987. Successive bacteremias with “Campylobacter cinaedi” and “Campylobacter fennelliae” in a bisexual male. J. Clin. Microbiol. 25:2008-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkusu, K., L. E. Bermudez, K. A. Nash, R. R. MacGregor, and C. B. Inderlied. 2004. Differential virulence of Mycobacterium avium strains isolated from HIV-infected patients with disseminated M. avium complex disease. J. Infect. Dis. 190:1347-1354. [DOI] [PubMed] [Google Scholar]
- 26.Orlicek, S. L., D. F. Welch, and T. L. Kuhls. 1993. Septicemia and meningitis caused by Helicobacter cinaedi in a neonate. J. Clin. Microbiol. 31:569-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 28.Pasternak, J., R. Bolivar, R. L. Hopfer, V. Fainstein, K. Mills, A. Rios, G. P. Bodey, C. L. Fennell, P. A. Totten, and W. E. Stamm. 1984. Bacteremia caused by Campylobacter-like organisms in two male homosexuals. Ann. Intern. Med. 101:339-341. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinn, T. C., S. E. Goodell, C. Fennell, S. P. Wang, M. D. Schuffler, K. K. Holmes, and W. E. Stamm. 1984. Infections with Campylobacter jejuni and Campylobacter-like organisms in homosexual men. Ann. Intern. Med. 101:187-192. [DOI] [PubMed] [Google Scholar]
- 31.Sacks, L. V., A. M. Labriola, V. J. Gill, and F. M. Gordin. 1991. Use of ciprofloxacin for successful eradication of bacteremia due to Campylobacter cinaedi in a human immunodeficiency virus-infected person. Rev. Infect. Dis. 13:1066-1068. [DOI] [PubMed] [Google Scholar]
- 32.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14:59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tee, W., B. N. Anderson, B. C. Ross, and B. Dwyer. 1987. Atypical campylobacters associated with gastroenteritis. J. Clin. Microbiol. 25:1248-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totten, P. A., C. L. Fennell, F. C. Tenover, J. M. Wezenberg, P. L. Perine, W. E. Stamm, and K. K. Holmes. 1985. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J. Infect. Dis. 151:131-139. [DOI] [PubMed] [Google Scholar]
- 36.Vandamme, P., E. Falsen, B. Pot, K. Kersters, and J. De Ley. 1990. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J. Clin. Microbiol. 28:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, and J. De Ley. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88-103. [DOI] [PubMed] [Google Scholar]