Abstract
Severe Clostridium difficile associated disease is associated with outbreaks of the recently described BI/NAP1 epidemic clone. This clone is characterized by an 18-bp deletion in the tcdC gene and increased production of toxins A and B in vitro. TcdC is a putative negative regulator of toxin A&B production. We characterized tcdC genotypes from a collection of C. difficile isolates from a hospital that experienced an outbreak caused by the BI/NAP1 epidemic clone. Sequence analysis of tcdC was performed on DNA samples isolated from 199 toxigenic C. difficile isolates (31% BI/NAP1) from 2001 and 2005. Sequences obtained from 36 (18.6%) isolates predicted wild-type TcdC (232 amino acid residues), whereas 12 (6.1%) isolates had tcdC genotypes with previously described 18- or 39-bp deletions. The remaining isolates comprised 15 unique genotypes. Of these, 5 genotypes contain 18- or 36-bp deletions. Of these five genotypes, one is characterized by a single nucleotide deletion at position 117 resulting in a frameshift that introduces a stop codon at position 196, truncating the predicted TcdC to 65 amino acid residues. All 62 of the isolates in this collection comprising the epidemic clone are characterized by this genotype. This result suggests that severe truncation of TcdC is responsible for the increased toxin production observed in strains belonging to the BI/NAP1 clone and that the 18-bp deletion is probably irrelevant to TcdC function. Further investigations are required to determine the effect of this and other tcdC genotypes on toxin production and clinical disease.
Clostridium difficile is the etiologic agent responsible for human diseases ranging from mild diarrhea to severe pseudomembranous colitis, which are collectively referred to as C. difficile-associated disease (CDAD) (1, 2, 9). A hospital outbreak with increased rates of severe CDAD was noted at the University of Pittsburgh Medical Center-Presbyterian Hospital (UPMC-P) in 2000 to 2001, with 26 colectomies and 18 deaths in that period (3, 15). Similar outbreaks of severe CDAD have since been reported throughout the United States and Canada (8, 12). An epidemic C. difficile clone, designated BI by restriction endonuclease typing (REA), NAP-1 by pulsed field gel electrophoresis, and toxinotype III by the toxinotyping method of Rupnik et al. has emerged and is in part responsible for these outbreaks (12, 19-21). Whereas BI/NAP1-toxinotype III isolates comprised only 0.3% of >6,000 pre-2001 C. difficile isolates maintained in the reference collection at the Hines Veterans Affairs Hospital (HVA), 10 to 75% of post-2001 isolates in U.S. hospitals and 51% of 2001 UPMC-P hospital-acquired C. difficile isolates belong to the BI/NAP1 clone (12). Thus, the increased incidence of severe CDAD closely parallels the emergence of the BI/NAP1 clone.
Recent epidemic BI/NAP1 isolates have been found to hyperproduce toxins A and B in vitro, the organism's major virulence factors (24). These toxins are encoded on the 19.6-kb pathogenicity locus (PaLoc) by the tcdA and tcdB genes. In addition, three accessory genes tcdC, tcdR (formerly txeR or tcdD), and tcdE which encode proteins thought to be involved in toxin regulation and secretion are located on the PaLoc (5). In reference strain VPI10463, transcriptional analysis of the PaLoc demonstrates that tcdC is expressed during logarithmic growth phase in contrast to tcdA, tcdB, and tcdR, which are most highly expressed during the stationary phase (6, 7). These data suggest that tcdC functions as a negative regulator of toxin synthesis.
Spigaglia and Mastrantonio have described three tcdC genotypes from nonepidemic C. difficile isolates. Genotype tcdC-A is characterized by a nonsense mutation (C184T) that is predicted to result in severe truncation of the TcdC protein from 232 to 61 amino acid residues. In addition, this genotype contains a 39-bp deletion from nucleotides 341 to 379. Genotype tcdC-A was identified in C. difficile isolates of toxinotypes V and VI. In the same study, genotypes tcdC-B and tcdC-C were identified. These tcdC alleles were characterized by 18-bp deletions at nucleotides 330 to 347 (23). Of note, genotype tcdC-C was derived from a C. difficile isolate of an asymptomatic patient. On the other hand, similar 18-bp deletions have been detected by fragment size discrimination and sequencing of partial tcdC amplicons (G. Killgore, Centers for Disease Control and Prevention, unpublished data) among BI/NAP1 isolates and are proposed to result in a loss of toxin regulation accounting for the more severe disease attributed to the epidemic clone (12, 24). Six epidemic clone isolates from Canada and one reference strain from the United Kingdom have recently been shown to have tcdC genotypes containing both 18-bp deletions and a single nucleotide deletion at position 117 that introduces a frameshift mutation that truncates the predicted TcdC product to 65 amino acid residues (10).
We describe tcdC genotypes from a collection of clinical C. difficile isolates from UPMC-P by sequence analysis of the entire tcdC gene to determine (i) the prevalence of previously reported and novel tcdC genotypes and (ii) the association between tcdC genotype and the epidemic BI/NAP1 clone.
MATERIALS AND METHODS
Setting.
All isolates of C. difficile in the present study originated from diagnostic specimens sent for C. difficile toxin testing for suspected CDAD from patients at UPMC-P, an 834-bed tertiary care teaching facility affiliated with the University of Pittsburgh Schools of the Health Sciences. This facility serves as a referral center for patients from institutions throughout western Pennsylvania, eastern Ohio, and northern West Virginia. Isolates from UPMC-P inpatients, affiliated outpatient clinics, and the UPMC-P emergency department and from non-hospital-acquired cases of CDAD were included.
Isolation of C. difficile.
Diagnostic testing for CDAD was carried out using a cell culture cytotoxicity assay for C. difficile toxins (CDT) (Diagnostic Hybrids; Athens, OH). Testing for CDAD was done at the discretion of the patients' healthcare providers. During the study period, all stools sent for CDAD testing were also cultured for C. difficile on Remel CCFA (cefoxitin, cycloserine, fructose agar) at 37°C after 4 h of anaerobic preincubation as previously described (13). Presumptive C. difficile isolates were identified by odor and colony morphology, subcultured on prereduced CDC anaerobe 5% sheep blood agar (Becton Dickinson, Franklin Lakes, NJ), and biochemically identified with a RapID ANA II panel (Remel, Lenexa, KS). C. difficile isolates were suspended in prereduced chopped meat broth (Anaerobe Systems, Morgan Hill, CA) and incubated for 48 h at 37°C. Then, 1.5 ml of the broth culture was centrifuged at 10,000 × g at room temperature. The supernatant was inoculated onto cell culture using the same CDT cytotoxicity assay used for CDT detection in stool specimens. CDT-positive isolates were stored in the remaining chopped meat broth at room temperature and were available for tcdC genotyping.
Selection of isolates.
A total of 199 UPMC-P toxigenic C. difficile isolates were genotyped with contributions from two time periods. The first group comprised 126 C. difficile isolates, a subset of CDT-positive stool specimens collected between 21 March and 19 December 2001. The second group comprised 73 C. difficile isolates, a subset of CDT-positive stool specimens collected between 2 April and 30 May 2005.
Sequencing of tcdC.
Isolates were subcultured onto prereduced TSA II with 5% sheep blood plates (Becton Dickinson, Franklin Lakes, NJ). DNA was extracted by QIAGEN DNeasy kit (Valencia, CA) according to the manufacturer's instructions. The tcdC gene was PCR amplified with primers C1 and C2 (IDT, Coralville, IA) as previously described (23). These primers amplify a 718-bp fragment of the PaLoc encompassing the entire tcdC gene (23). The reaction mixture contained AmpliTaq 10× PCR buffer, 125 nmol of MgCl2, 10 pmol of C1 and C2 primers, 10 nmol of each deoxynucleotide triphosphate, and 1.5 U of AmpliTaq gold (Applied Biosystems, Foster City, CA). The template was denatured at 95°C for 5 min, and DNA was amplified for 35 cycles consisting of 1 min at 95°C, 1 min at 50°C, and 1 min at 72°C. Sequencing was performed with the amplification (C1 and C2) primers with a BigDye terminator 3.1 kit (Applied Biosystems) according to the manufacturer's instructions. Capillary sequence analysis was performed with a 3730 DNA sequencer (Applied Biosystems) at the University of Pittsburgh's Genomics and Proteomics Core Laboratories facility. Sequences were analyzed and amino acid sequences deduced with the Lasergene 7.0.0 software package (DNAStar, Madison, WI) and compared to the published tcdC sequence for strain VPI10463. Isolates with sequences that differed from those previously published genotypes were named sequentially with the prefix sc (Fig. 1 and Table 1).
FIG. 1.
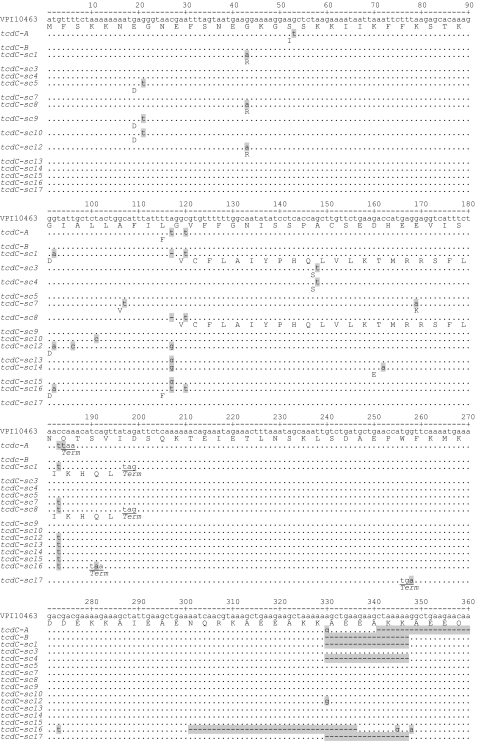
Comparison of TcdC nucleotide and amino acid sequences to the published sequence for the reference strain VPI10463. Dots and dashes indicate identical bases and deletions, respectively. Only amino acid changes are depicted. Stop codons are underlined. Genotype tcdC-sc2 is not depicted but is identical to tcdC-sc1 with the exception of an inserted nucleotide (t) in the untranslated region at position 212.
TABLE 1.
Characteristics of tcdC genotypes of isolates from 199 cases of CDAD at UPMC 2001 and 2005
| tcdC genotypea | No. of cases (%) | Predicted truncating mutations | Nucleotide deletions (>1 bp) | Deletion (>1 bp) beyond stop codon? | Predicted TcdC length (amino acid residues) | No. of residue changes in TcdC (no. deleted) | REA type(s) within group (no. typed) |
|---|---|---|---|---|---|---|---|
| Wild type (VPI10463) | 36 (18.1) | 232 | 0 (0) | K (1) | |||
| tcdC-A | 3 (1.5) | C184T | 39 | Yes | 61 | 2 (171) | |
| tcdC-B | 9 (4.5) | None | 18 | No | 226 | 0 (6) | |
| tcdC-sc1 | 91 (45.7) | Δ117A | 18 | Yes | 65 | 27 (167) | BI 9 (3), BI10 (2) |
| tcdC-sc2* | 1 (0.5) | Δ117A | 18 | Yes | 65 | 27 (167) | BR2 (1) |
| tcdC-sc3 | 23 (11.9) | None | None | 232 | 1 (0) | J9, 28-31 (6) | |
| tcdC-sc4 | 1 (0.5) | None | 18 | No | 226 | 1 (6) | |
| tcdC-sc5 | 5 (2.5) | None | None | 232 | 2 (0) | ||
| tcdC-sc7 | 6 (3.0) | None | None | 232 | 2 (0) | ||
| tcdC-sc8 | 1 (0.5) | Δ117A | None | 65 | 26 (167) | ||
| tcdC-sc9 | 9 (4.5) | None | None | 232 | 1 (0) | CL (1) | |
| tcdC-sc10 | 1 (0.5) | None | None | 232 | 2 (0) | ||
| tcdC-sc12 | 2 (1.0) | None | None | 232 | 2 (0) | ||
| tcdC-sc13 | 1 (0.5) | None | None | 232 | 0 (0) | ||
| tcdC-sc14 | 3 (1.5) | None | None | 232 | 1 (0) | BM (1) | |
| tcdC-sc15 | 5 (2.5) | None | None | 232 | 0 (0) | ||
| tcdC-sc16 | 1 (0.5) | C191A | 36 | Yes | 63 | 2 (169) | |
| tcdC-sc17 | 1 (0.5) | G258A | 18 | Yes | 85 | 0 (147) |
*, Genotype tcdC-sc2 is identical to genotype tcdC-sc1 except for the insertion of a single nucleotide (t) at position 212.
C. difficile genotyping methods.
All isolates tested had previously undergone molecular genotyping by MLVA (for multilocus, variable number of tandem repeats analysis) (11). Briefly, C. difficile isolate genomic DNA was PCR amplified and sequenced at seven tandem repeat loci. Tandem repeat copy numbers were manually counted and concatenated to generate an MLVA type for each isolate. Minimum spanning tree (MST) analysis of MLVA types was performed to determine the genetic distance between isolates using the summed tandem repeat difference (STRD) as a coefficient of genetic distance in the BioNumerics software (v3.0; Applied Maths, Austin, TX). This method was validated against HVA reference strains including isolates belonging to the epidemic BI/NAP1 clone (11). Fifteen isolates in the present study had been REA typed at the HVA reference lab as part of a previous study (11).
In the present study, MST analysis of MLVA data was used to attribute tcdC genotypes to genetically related isolates. Clonal complexes were defined by an STRD of ≤2 (shaded areas in Fig. 2). Based on this definition, isolates were considered to be part of the BI/NAP1 epidemic population if they clustered with REA-typed HVA BI reference strains.
FIG. 2.
Minimum-spanning tree of MLVA data depicting genetic relatedness of 199 isolates of C. difficile from cases of CDAD at UPMC-P in 2001 and 2005. Each circle depicts one MLVA genotype, with the STRD between adjacent isolates displayed in gray. Where no numeral is shown, the STRD equals 1. STRDs of ≥11 are depicted with dashed lines. Clouds of gray surround isolates with an STRD of ≤2; these complexes represent closely related isolates. White circles depict genotypes represented by one isolate; light gray circles with black numerals, dark gray circles with white numerals, and black circles with white numerals depict genotypes represented by 2, 3, and ≥5 isolates, respectively. The circles are labeled with the tcdC genotype described in Fig. 1 (wild-type tcdC = 0, tcdC-A = A, tcdC-B = B, tcdC-sc1 = 1, tcdC-sc2 = 2, tcdC-sc3 = 3, etc.); available REA types are displayed below the tcdC genotype. The tree has been redrawn for ease of viewing and is not to scale.
Nucleotide sequence accession numbers.
The unique nucleotide sequences of the tcdC genes of representative C. difficile strains for genotypes tcdC-sc1, tcdC-sc2, tcdC-sc3, tcdC-sc4, tcdC-sc5, tcdC-sc7, tcdC-sc8, tcdC-sc9, tcdC-sc10, tcdC-sc12, tcdC-sc13, tcdC-sc14, tcdC-sc15, tcdC-sc16, and tcdC-sc17 were assigned GenBank accession numbers DQ861412, DQ861414, DQ861415, DQ861416, DQ861417, DQ861418, DQ861419, DQ861420, DQ861421, DQ861422, DQ861423, DQ861424, DQ861425, DQ861426, and DQ861413, respectively.
RESULTS
Frequency of tcdC genotypes and new genotypes identified.
The frequencies of the genotypes within this collection of 199 C. difficile isolates are summarized in Table 1. Wild-type tcdC sequences corresponding to that of toxigenic strain VPI10463 comprised 36 of 199 (18.1%) isolates. Genotypes previously described, tcdC-A and tcdC-B, were present in 3 of 199 (1.5%) and 9 of 199 (4.5%) isolates, respectively.
The remaining 151 isolates belonged to one of 15 unique tcdC genotypes (Fig. 1). Of these, genotype tcdC-sc1 represented the greatest proportion (91 of 199 isolates [45.7%]), followed by tcdC-sc3 (23 isolates [11.6%]). Three of these tcdC genotypes (tcdC-sc1, tcdC-sc2, and tcdC-sc8) are characterized by a single nucleotide deletion at nucleotide position 117 (Δ117). This deletion results in a frameshift that introduces a nonsense mutation at position 196 and a radical alteration in the preceding 26 deduced amino acid residues. The predicted tcdC gene product is truncated from 232 to 65 amino acid residues. Both tcdC-sc1 and tcdC-sc2 contain 18-bp deletions at nucleotides 330 to 347, which are identical to the previously described 18-bp deletions characteristic of the BI/NAP1 clone (11, 12, 24). Because the 18-bp deletion is located downstream of the mutated nucleotide at position 117 this region of tcdC is predicted to be untranslated. Unlike tcdC-sc1 and tcdC-sc2, tcdC-sc8 lacks the downstream 18-bp deletion. tcdC-sc2 contains an insertion of a single nucleotide (t) at position 212, i.e., in an untranslated region.
Two genotypes, tcdC-sc16 and tcdC-sc17, were characterized by single-nucleotide nonsense mutations (C191A and G258A, respectively) that are predicted to truncate the TcdC protein to 63 and 85 amino acid residues, respectively. In addition, these genotypes were remarkable in that a 36-bp deletion was observed at nucleotides 301 to 336 in the tcdC-sc16 genotype and an 18-bp deletion was observed at nucleotides 330 to 347 in the tcdC-sc17genotype, the latter similar to the tcdC-B and -C alleles described by Spigaglia and Mastrantonio (23). In each case, the 18- and 36-bp deletions in tcdC-sc16 and tcdC-sc17 reside downstream of the nonsense mutation in a region of the tcdC gene that is predicted to be untranslated. Neither genotype was associated with more than one case of CDAD (Table 1), nor was either genotype genetically related to epidemic clone isolates by MLVA (Fig. 2). Genotype tcdC-sc16 was most closely related to isolates with wild-type tcdC genotypes, whereas genotype tcdC-sc17 was closely related to isolates with tcdC-B, a tcdC genotype characterized by an 18-bp deletion only.
Genotype tcdC-sc4 has an 18-bp deletion that, like the previously described tcdC-B allele, is in frame and predicted to result in a 6-amino-acid residue deletion. Unlike tcdC-sc17, the tcdC-sc4 genotype does not harbor upstream mutations that disrupt the reading frame and result in truncation of TcdC. A single, polar amino acid substitution (A50S) caused by the single-nucleotide mutation G148T distinguishes this genotype from tcdC-B.
Ten tcdC genotypes had silent mutations that are predicted to generate either wild-type TcdC (tcdC-sc13 and tcdC-sc15) or TcdC containing minor residue changes (Fig. 1). The salient features of all genotypes occurring in this collection are summarized in Table 1. There were no statistically significant differences in the proportions of the genotypes from 2001 to 2005 with the exception of two genotypes, tcdC-sc3 and wild-type tcdC. TcdC-sc3 occurred in 22 of 126 (17.5%) 2001 isolates and 1 of 73 (1.4%) 2005 isolates (P = 0.001). Wild-type isolates occurred in 12 of 126 (9.5%) 2001 isolates and 24 of 73 (32.9%) 2005 isolates (P = 0.001).
Relationship between tcdC genotype, the epidemic clone, and other REA/MLVA types.
MST analysis of MLVA genotypes from the study isolates and their corresponding tcdC genotypes are shown in Fig. 2. In general, tcdC genotypes correlated with the genetic relationships depicted by MLVA genotyping even though this genotyping method relies on VNTRs in loci distinct from the tcdC gene. Genetically related isolates had identical tcdC genotypes. For example, most isolates bearing the tcdC-sc1 genotype are highly related by MLVA. Similarly, isolates bearing the tcdC-sc3 genotype belong to another genetically related population, and isolates with the wild-type (labeled “0” in Fig. 2) tend to cluster together by MST of MLVA genotypes. There was only one example of isolates with identical MLVA genotypes containing more than one tcdC genotype (Fig. 2). In this instance, an isolate characterized by wild-type tcdC was identical by MLVA to an isolate with the previously identified genotype, tcdC-B.
Significantly, all 62 (100%) isolates that were identified as belonging to the epidemic clone (BI/NAP1) by MLVA (sc1 isolates contained in the shaded area in Fig. 2) have the tcdC-sc1 genotype characterized by the nucleotide 117 deletion predicted to truncate the TcdC protein. Isolates bearing tcdC-A or tcdC-B genotypes were not genetically related to the epidemic (BI/NAP1) clone by MLVA despite containing 39- and 18-bp deletions in tcdC.
Genotype tcdC-sc1 was found in 91 of 199 (45.7%) of C. difficile isolates examined. Of these isolates, 62 were from 2001 (62 of 126 [49.2%]) and 29 were from 2005 (29 of 73 [39.7%]). Thirty nonepidemic clone isolates were genotype tcdC-sc1 but were nonetheless closely related to the BI/NAP1 epidemic clone (Fig. 2). Of these 30 isolates, 11 (36.7%) are from 2001 and 19 (63.3%) are from 2005, and the 3 isolates most distantly related to the epidemic clone depicted in Fig. 2 by STRD of 24 are themselves typed as BI 11 by the HVA REA genotyping system.
Genotype tcdC-sc8, which shares the deletion of nucleotide 117 found in tcdC-sc1 (Table 1), is related to the epidemic clone isolates cluster by MLVA (Fig. 2). Genotype tcdC-sc2, however, which also shares the deletion of nucleotide 117, is more distantly related to the epidemic clone by MLVA, suggesting that potential de novo mutations or lateral transfers of tcdC in this strain may have occurred.
Association of 18-, 36-, and 39-bp deletions in tcdC genotypes with the epidemic clone.
Two of the previously described tcdC genotypes and five of the genotypes described in the present study contain deletions of 18, 36 or 39 bp, but in only two of these genotypes (tcdC-B and tcdC-sc4) are these deletions predicted to alter the predicted TcdC protein in the form of deletions of three amino acid residues. In the other six genotypes (tcdC-A, tcdC-sc1, sc2, sc16, and sc17), upstream mutations predict truncations of 147 to 171 amino acid residues of the TcdC protein in addition to radically altering 2 to 27 amino acid residues prior to the truncations due to frameshifts (Table 1).
If one defines a highly mutated TcdC protein as one having >6 amino acid deletions, as is the case with tcdC genotypes tcdC-A, tcdC-sc1, tcdC-sc2, tcdC-sc8, tcdC-sc16, and tcdC-sc17, 18-, 36-, or 39-bp deletions accompany 97 of 98 (99.0%) isolates with tcdC genotypes predicted to have highly mutated TcdC proteins. Using 18-, 36-, or 39-bp deletion analysis as a predictor of such high degrees of tcdC mutation is therefore 99.0% sensitive and 90.1% specific for genotypes resulting in highly mutated TcdC proteins. The only false negative in this regard is genotype tcdC-sc8, which lacks an 18-bp deletion and yet has a highly mutated TcdC protein predicted by its single nucleotide deletion, Δ117A.
DISCUSSION
The key finding of the present study was the identification of a genotype, tcdC-sc1, that was present in all of our epidemic BI/NAP1 isolates and related isolates by MLVA. This genotype is characterized by an upstream deletion at nucleotide 117 that causes severe truncation of the predicted TcdC protein. A similar genotype, tcdC-sc2, shared this deletion but was more distantly related by MLVA to the BI/NAP1 epidemic clone isolates. Genotypes tcdC-sc1 and tcdC-sc2 share the Δ117A mutation together with 18-bp deletions, features identified by MacCannell et al. in six epidemic strains of C. difficile from sites in Canada and one reference strain from the United Kingdom (10).
The emergence of a strain with dysfunctional suppression of log-phase toxin elaboration may explain recently reported CDAD outbreaks with fulminant presentations and increased severity of disease. These strains have been demonstrated to produce increased levels of toxins A and B during both phases of growth (logarithmic and stationary) (24). Therefore, the apparent decreased responsiveness to appropriate antimicrobial therapies may be due to an increased C. difficile toxin burden prior to CDAD recognition (14, 16-18).
Other evidence that tcdC mutations might be the causal link to severe disease is also tantalizing. Strain 8864, which contains a rearrangement of its PaLoc that leaves it with no functional tcdA or tcdC but which is nonetheless TcdB positive, is known to be exceptionally more cytotoxic in vitro than strain VPI10463 and more pathogenic in animal models (22). Recently, TcdC has been demonstrated to be membrane associated, leading to speculation that if it does perform a role in transcriptional regulation, it may also serve as a sensor for metabolite deprivation or have some other signaling function (4). Ultimately, determination of causality for tcdC mutations and severe CDAD will require development of the necessary molecular tools for genetic manipulation of Clostridium spp.
The present findings should be interpreted with caution, however, as there is as yet no definitive evidence that TcdC functions as a transcriptional regulator. Whether the predicted shortened TcdC's are responsible for the increase in CDAD incidence and severity or are simply a marker of increased virulence cannot be ascertained from the present study. Even if TcdC is a transcriptional regulator of tcdA and tcdB, the impact of the various predicted changes in the protein that we observed are not known. Ten tcdC genotypes with silent or conservative predicted mutations were also identified in the present study, and the effects of the latter on tcdC phenotype are unknown. Mutations such as the A50S amino acid substitution present in the predicted TcdC in the modest outbreak of tcdC-sc3/REA type J isolates in 2001 are of unknown significance and merit further investigations of toxin phenotypes in vitro. Moreover, the existence of genotypes with severe TcdC truncation (tcdC-sc2, tcdC-sc16, and sc17) falling far outside the BI/NAP1 epidemic clone are intriguing. Further studies are required to determine whether these tcdC genotypes are associated with either increased toxin production and more severe disease or disease in low-risk populations.
Regardless of its impact on C. difficile pathogenesis, tcdC genotyping in conjunction with other methods such as MLVA could provide additional information regarding the clonality and pathogenic potential of C. difficile populations. The finding that tcdC genotypes were reasonably concordant with MLVA and REA types (Fig. 2) supports this view.
The presence of 30 isolates with genotype tcdC-sc1 outside of the main epidemic clone outlined by MLVA but within the BI designation by REA as depicted in Fig. 2 suggests clonal expansion and evolution of the epidemic clone rather than de novo generation of or lateral gene transfer to a different strain. Alternatively, these 30 isolates may represent a clonal population that is closely related yet distinct from BI/NAP1.
We found that the 18- and 39-bp deletions previously described as an indicator of tcdC mutation are for the most part silent deletions except in the case of tcdC-B and tcdC-sc4 (12). Thus, detection of these deletions in tcdC by conventional PCR is only 90% specific for severe TcdC truncation. Future molecular epidemiologic studies of CDAD should therefore focus on sequencing the entire tcdC gene to establish the presence or absence of single-nucleotide nonsense and frameshift mutations rather than strictly upon 18-, 36-, or 39-bp deletions. Although tcdC-sc8 has only been found in one isolate thus far, detection of similar genotypes would further reduce the sensitivity of an assay for severe tcdC mutation based solely on PCR amplicon size.
The present study demonstrates that clinical isolates of C. difficile frequently harbor tcdC genetic variants. Furthermore, the identification of a single mutant tcdC genotype strongly associated with the epidemic BI/NAP1 clone supports the hypothesis first proposed by Spigaglia and Mastrantonio that TcdC variants may produce different levels of toxin that results in C. difficile strains of unique pathogenic potential. This inference can be tested by studying the association of various tcdC genotypes with CDAD severity.
Acknowledgments
This study was supported partially by an NIH training grant (T32 AI007333 [S.R.C.]) and by a National Institute of Allergy and Infectious Diseases research career award (K24 AI52788 [L.H.H.]).
We are grateful to Dale Gerding and Stuart Johnson for performing the REA typing and Eilleen Driscoll for isolation of the C. difficile from stool and mixed cultures.
Footnotes
Published ahead of print on 11 October 2006.
REFERENCES
- 1.Bartlett, J. G., T. W. Chang, M. Gurwith, S. L. Gorbach, and A. B. Onderdonk. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531-534. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778-782. [PubMed] [Google Scholar]
- 3.Dallal, R. M., B. G. Harbrecht, A. J. Boujoukas, C. A. Sirio, L. M. Farkas, K. K. Lee, and R. L. Simmons. 2002. Fulminant Clostridium difficile: an underappreciated and increasing cause of death and complications. Ann. Surg. 235:363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Govind, R., G. Vediyappan, R. D. Rolfe, and J. A. Fralick. 2006. Evidence that Clostridium difficile TcdC is a membrane-associated protein. J. Bacteriol. 188:3716-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond, G. A., and J. L. Johnson. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 6.Hammond, G. A., D. M. Lyerly, and J. L. Johnson. 1997. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb. Pathog. 22:143-154. [DOI] [PubMed] [Google Scholar]
- 7.Hundsberger, T., V. Braun, M. Weidmann, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735-742. [DOI] [PubMed] [Google Scholar]
- 8.Loo, V. G., L. Poirier, M. A. Miller, M. Oughton, M. D. Libman, S. Michaud, A. M. Bourgault, T. Nguyen, C. Frenette, M. Kelly, A. Vibien, P. Brassard, S. Fenn, K. Dewar, T. J. Hudson, R. Horn, P. Rene, Y. Monczak, and A. Dascal. 2005. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N. Engl. J. Med. 353:2442-2449. [DOI] [PubMed] [Google Scholar]
- 9.Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacCannell, D. R., T. J. Louie, D. B. Gregson, M. Laverdiere, A. C. Labbe, F. Laing, and S. Henwick. 2006. Molecular analysis of Clostridium difficile PCR ribotype 027 isolates from Eastern and Western Canada. J. Clin. Microbiol. 44:2147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh, J. W., M. O'Leary, M., K. A. Shutt, A. W. Pasculle, S. Johnson, D. N. Gerding, C. A. Muto, and L. H. Harrison. 2006. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J. Clin. Microbiol. 44:2558-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 13.Mundy, L. S., C. J. Shanholtzer, K. E. Willard, D. N. Gerding, and L. R. Peterson. 1995. Laboratory detection of Clostridium difficile: a comparison of media and incubation systems. Am. J. Clin. Pathol. 103:52-56. [DOI] [PubMed] [Google Scholar]
- 14.Musher, D. M., S. Aslam, N. Logan, S. Nallacheru, I. Bhaila, F. Borchert, and R. J. Hamill. 2005. Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole. Clin. Infect. Dis. 40:1586-1590. [DOI] [PubMed] [Google Scholar]
- 15.Muto, C. A., M. Pokrywka, K. Shutt, A. B. Mendelsohn, K. Nouri, K. Posey, T. Roberts, K. Croyle, S. Krystofiak, S. Patel-Brown, A. W. Pasculle, D. L. Paterson, M. Saul, and L. H. Harrison. 2005. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect. Control Hosp. Epidemiol. 26:273-280. [DOI] [PubMed] [Google Scholar]
- 16.Pepin, J., M. E. Alary, L. Valiquette, E. Raiche, J. Ruel, K. Fulop, D. Godin, and C. Bourassa. 2005. Increasing risk of relapse after treatment of Clostridium difficile colitis in Quebec, Canada. Clin. Infect. Dis. 40:1591-1597. [DOI] [PubMed] [Google Scholar]
- 17.Pepin, J., L. Valiquette, M. E. Alary, P. Villemure, A. Pelletier, K. Forget, K. Pepin, and D. Chouinard. 2004. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepin, J., L. Valiquette, and B. Cossette. 2005. Mortality attributable to nosocomial Clostridium difficile-associated disease during an epidemic caused by a hypervirulent strain in Quebec. CMAJ 173:1037-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microbiol. Infect. 7:417-420. [DOI] [PubMed] [Google Scholar]
- 20.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 22.Soehn, F., A. Wagenknecht-Wiesner, P. Leukel, M. Kohl, M. Weidmann, C. von Eichel-Streiber, and V. Braun. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864: implications for transcription, expression, and enzymatic activity of toxins A and B. Mol. Gen. Genet. 258:222-232. [DOI] [PubMed] [Google Scholar]
- 23.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]