Abstract
The determination of the composition of the microbial community in the oral cavity is usually based on cultivation methods; however, nearly half of the bacteria in the saliva and the dental plaque are not cultivable. In this study, we evaluated the difference in oral microbial diversity between children with severe early-childhood caries (S-ECC) and caries-free (CF) controls by means of a cultivation-independent approach called denaturing gradient gel electrophoresis (DGGE). Pooled dental plaque samples were collected from 20 children aged 2 to 8 years. Total microbial genomic DNA was isolated from those subjects, and a portion of the 16S rRNA gene locus was PCR amplified by using universal primers. We observed that the mean species richness of the bacterial population was greater in the CF children (n = 12) (42 ± 3.7) than in the S-ECC children (n = 8) (35 ± 4.3); the difference was statistically significant (P = 0.005). The overall diversity of plaque samples as measured by the Shannon index was 3.5 for the S-ECC group and 3.7 for the CF group (P = 0.004). Differences in DGGE profiles were distinguished on the basis of a cluster analysis. Sequence analysis of excised DGGE bands consisted of 2.7 phylotypes, on average. After adjusting for the number of observed bands, we estimated that the S-ECC group exhibited 94.5 total phylotypes and that the CF group exhibited 113.4. These results suggest that the microbial diversity and complexity of the microbial biota in dental plaque are significantly less in S-ECC children than in CF children.
Severe early-childhood caries (S-ECC) is an extremely destructive form of early-childhood caries involving multiple teeth, including the maxillary anterior teeth (14). The cause of S-ECC remains speculative. Formerly, S-ECC was thought to be associated with the prolonged use of a nursing bottle (19), but dietary intake alone may not account for the severe nature of this disease (10, 49). A compelling body of scientific evidence suggests that Streptococcus mutans is a major etiologic agent in the development of S-ECC (5, 20, 46); however, it remains to be determined whether S-ECC is caused by a single or specific consortium of bacteria (6, 46) or whether the biofilm as a whole undergoes a more complicated shift in multiple groups of bacteria, as suggested by several investigators (3, 4, 8, 45).
Quantitative and qualitative analyses of polymicrobial ecosystems such as plaque biofilms are complicated because they may consist of as many as hundreds of different bacterial species, many of which are not cultivable (1, 9, 22, 38). Previous studies by our group (24, 25) and others (17, 39, 40) demonstrated the value of a whole-microbiota survey method called denaturing gel gradient electrophoresis (DGGE) to depict the composition of the oral microbiota. With the DGGE approach, specific regions of the 16S rRNA gene locus are amplified by PCR, and the products are run on a denaturing gel that separates amplicons according to nucleotide composition. Different amplicons can then be displayed as bands with different migration distances to yield a distinctive fingerprint representing each of the various bacterial phylotypes or species on a single gradient gel. Microbial profiles of plaque or saliva from different individuals can be compared with a variety of measurement tools, and inferences about shifts in the ecological balance of the biofilm can be made. This approach has been widely used by environmental ecologists to survey entire bacterial communities without cultivation (15, 36, 37) and to analyze gastrointestinal microbial ecosystems (32, 43, 51). Specific bands of interest can be excised from gels, and these fragments of the 16S rRNA gene can be sequenced and compared with known sequences in an rRNA database (11).
The objective of this study was to characterize the microbial diversity in the complex dental plaque of children with S-ECC and in appropriate caries-free (CF) controls. We demonstrated that sufficient differences in DGGE profiles exist to distinguish the microbiota of S-ECC children from that of their CF counterparts.
(Part of this study was presented at the 83rd General Session of the International Association for Dental Research and the 34th Annual Meeting of the American Association for Dental Research, 2005.)
MATERIALS AND METHODS
Subjects.
Twenty children of Hispanic origin (10 boys and 10 girls; age range, 2.4 to 8.6 years) were included in this study. Eight of those subjects were CF children, and 12 were S-ECC children who had a score for decayed, missing, and filled teeth of 9.6 ± 3.6 (mean ± standard deviation [SD]) and a score for decayed, missing, and filled tooth surfaces of 17.9 ± 11.8 (mean ± SD) (Table 1). The S-ECC children were selected based on convenience from a list of children who were scheduled for extensive treatment under general anesthesia in the operating room at the Bellevue Hospital from April 2003 to April 2004. CF children comparable in age to the S-ECC cohort were selected from the pediatric dental clinic of the New York University College of Dentistry after having been diagnosed as being free of detectable caries. The study protocol was approved by the Institutional Review Board of New York University School of Medicine and the Bellevue Hospital for human subjects.
TABLE 1.
Comparisons of the diversity of DGGE profiles in children with different caries status
| Outcome variable | Value (mean ± SD) for group
|
P value | |
|---|---|---|---|
| S-ECC (n = 12) | CF (n = 8) | ||
| Age (yr) | 4.7 ± 1.3 | 6.1 ± 1.6 | 0.07a |
| Gender | |||
| No. (%) male | 7 (58) | 3 (38) | 0.65b |
| No. (%) female | 5 (42) | 5 (62) | |
| Caries statusd | |||
| dmft score | 9.6 ± 3.6 | 0 | |
| dmfs score | 17.9 ± 11.8 | 0 | |
| Microbial diversity | |||
| No. of DGGE bands | 35.4 ± 4.3 | 41.9 ± 3.7 | 0.005a |
| H′ | 3.5 ± 0.1 | 3.7 ± 0.1 | 0.004a |
| Similarity values (%) | |||
| Intergroup comparison | 91.3 ± 4.3 | 92.7 ± 3.6 | 0.35c |
| Intragroup comparison | 89.4 ± 4.7 | 0.001c | |
By nonparametric Mann-Whitney U test for independent samples.
By Fisher's exact test.
Results of ANOVA showed that the P value for the overall comparison was 0.001. Significant differences were also found between the S-ECC group and the intragroup comparison (91.3 versus 89.4; P = 0.02) and between the CF and the intragroup comparison (92.7 versus 89.4; P = 0.002).
dmft, decayed, missing, and filled teeth; dmfs, decayed, missing, and filled tooth surfaces.
Bacterial sample collection.
Bacterial samples from the 12 children with S-ECC were collected in the operating room of Bellevue Hospital, and samples from the 8 CF children were collected in a routine dental setting at the pediatric dental clinic of the New York University College of Dentistry. In brief, a sterile Gracey curette was used to collect a pooled plaque sample from the buccogingival surfaces and the accessible proximal surfaces of the molars and canines. The collected plaque sample was released from the curette by agitation in 1.0 ml of TE buffer (10 mM Tris-Cl [pH 7.5] and 1 mM EDTA). The plaque samples were immediately transported on ice to a microbiology laboratory at the New York University College of Dentistry. A portion of the sample was processed for DNA isolation.
DNA extraction.
The total plaque genomic DNA of the bacterial samples was isolated by means of a DNA purification kit (MasterPure; Epicenter, Madison, WI) with modifications. In brief, 300 μl of tissue and cell lysis solution (Epicenter) was added to the suspend pellet and sonicated for 30 seconds, and 10 μl of a proteinase K stock solution (QIAGEN Inc., Valencia, CA) of 10 mg/ml in TES buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, and 100 mM NaCl) and 2 μl of mutanolysin (Sigma-Aldrich, St. Louis, MO) (5,000 U/ml in phosphate-buffered saline) were added to 10 μl of lysozyme stock solution (100 mg/ml in TES buffer). That solution was incubated at 37°C for 1 h and was mixed gently at 15-min intervals. Proteinase K was inactivated via incubation at 65°C for 1 h. One microliter of 5-μg/μl RNase A was then added, and the mixture was incubated at 37°C for 30 min. The samples were then placed on ice for 3 to 5 min, and 400 μl of MPC protein precipitation reagent (Epicenter) was added and vortex mixed vigorously for 10 seconds; this was followed by a phenol-chloroform-isoamyl alcohol (25:24:1) extraction procedure and isopropanol precipitation. The DNA was then washed and dried. The quality and quantity of DNA samples were measured with a UV spectrophotometer at 260 nm and 280 nm (DU 640; Beckman, Hayward, CA). The final concentration of each DNA sample was adjusted to 10 ng/μl for all PCR applications.
PCR assay.
The complete ∼1,500-bp 16S rRNA gene locus was preamplified for all DNA extracts from S-ECC and CF plaques with a set of universal 16S rRNA gene sequence primers (23), followed by a second nested amplification of the V4-V5 hypervariable region (∼300 bp) of the 16S rRNA gene locus (46). In the first PCR, each reaction mixture (a total volume of 50 μl) contained a standardized 100 ng of the total genomic DNA, 200 μM of each deoxynucleoside triphosphate, 50 pmol of universal primers 16S-8f and 16S-1492r (23), 1.5 mM MgCl2, 5 μl of 10× PCR buffer II, and 2.5 U of Taq DNA polymerase (Applied Biosystems, Foster, CA).
In the second nested PCR, a second set of universal bacterial 16S rRNA gene primers (prbac1 and prbac2) (41) was used with a 40-nucleotide GC clamp, which was added to the 5′ end of prbac1 to prevent the dissociation of the 16S rRNA gene duplexes during denaturation electrophoresis (36, 42, 51). PCR conditions and reagents were as described elsewhere (24, 25). All PCR procedures were performed with the GeneAmp PCR system 9700 (Applied Biosystems). PCR products were evaluated by electrophoresis in 1.5% agarose gels that were run at 60 V for 100 min, and the sizes of all amplicons were confirmed according to a molecular size standard.
DGGE assay.
A standardized 20 μl of each PCR-amplified product was separated on gradient gels as previously described (24, 25). A 40% to 60% linear DNA denaturing gradient was formed in an 8% (wt/vol) polyacrylamide gel. PCR products were directly loaded in each lane and were run along with known species-specific DGGE reference markers (24). After electrophoresis, the gels were rinsed and stained for 15 min in a 0.5-μg/ml ethidium bromide solution, followed by 15 min of destaining in water. The DGGE images were digitally captured and recorded (Alpha Innotech Corporation, San Leandro, CA).
Analysis of microbial profiles.
All of the DGGE gel images were normalized first according to the known species-specific DGGE reference markers (24) by means of Fingerprinting II Informatix Software (Bio-Rad). The gel background was subtracted by use of mathematical algorithms according to the spectral analysis of overall densitometric curves. A minimal profiling setting (1.0%) was used for the band search for all DGGE gels. DGGE profiles were determined by measuring the migration distances and the intensities of the bands within each lane. The results were transferred into a microbial database that allowed us to cross-compare multiple DGGE profiles simultaneously. The percentage of similarity between fingerprinting profiles were calculated according to the Dice coefficient of pairwise comparisons (16, 29). The final parameters used to analyze the banding patterns included the numbers of the detected band, the band frequency distribution, the Shannon index (H′) for species richness (the number of different distinct bands in any individual lane) and evenness, and levels of pairwise similarity coefficients. Ward's algorithm was used to construct a dendrogram for cluster analysis (50).
Cloning and 16S rRNA gene sequencing.
Distinct amplicons from DGGE gels from both S-ECC and CF samples were excised from the gels, and DNA samples were eluted, purified, and reamplified with the same prbac1 and prbac2 primers but without the GC clamp. PCR amplicons were cloned with cloning kit (TOPO TA; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The transformed cells were then plated onto Luria-Bertani agar plates supplemented with kanamycin (50 μg/ml), and the plates were incubated overnight at 37°C. The clones were picked, and plasmid DNA was extracted. Purified plasmid DNA with 16S rRNA gene inserts was sequenced with the ABI Prism cycle sequencing kit (Applied Biosystems) and universal T3 primers. The sequences of 16S rRNA gene PCR amplicons obtained from the DGGE of the S-ECC or CF samples were analyzed against known sequences in the Ribosomal Database Project II (11).
Statistical analyses.
The differences in microbial diversity were assessed by comparing the DGGE profiles within and between the S-ECC and CF groups. The degree of correlation between the microbial diversity in the dental plaque and the caries status of the children was evaluated with analysis of variance (ANOVA), the nonparametric Mann-Whitney U test, the chi-square test, and Fisher's exact test. The analyses were performed with the SPSS software version 13.0 (Statistical Package for the Social Sciences; SSPS Inc, Chicago, IL). All P values of less than 0.05 were two tailed.
RESULTS
The mean age for the 12 S-ECC children was 4.7 years (±1.3), and that for the 8 CF children was 6.1 years (±1.6). This difference was not significant, as determined by the nonparametric Mann-Whitney U test for independent samples. The gender distribution between the two groups was not significantly different as determined by Fisher's exact test (Table 1).
DGGE banding pattern.
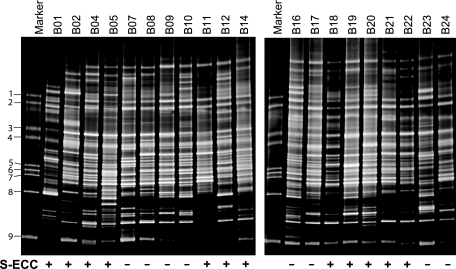
DGGE profiles were obtained from the plaque samples of the 20 subjects, among whom the caries status varied (Fig. 1). A total of 92 distinct amplicons were detected from the overall DGGE profiles after gel normalization. The number of distinct bands (amplicons) ranged from 29 to 46, with a mean ± SD of 37.9 ± 5.0 for each individual sample. On average, the total number of detectable bands was significantly higher in the CF group (41.9 ± 3.7) than in the S-ECC group (35.4 ± 4.3), and that difference was statistically significant (P = 0.005 by the Mann-Whitney U test) (Table 1). Also shown in Table 1 is the comparison of the Shannon index, which represents a measure of the richness and evenness of microbial diversity in a given sample. The diversity of bacteria in the CF group (H′ = 3.7 ± 0.1) was greater than that in S-ECC group (H′ = 3.5 ± 0.1). That difference was also statistically significant (P = 0.004 by the Mann-Whitney U test). Intragroup and intergroup comparisons of the similarity values of the microbial profiles revealed that the mean similarity values of the DGGE profiles were 91.3% ± 4.3% within the S-ECC samples and 92.7% ± 3.6% within the CF samples, but it decreased significantly to 89.4% ± 4.7% (P = 0.001 by ANOVA) in a comparison between the two groups (Table 1).
FIG. 1.
DGGE profiles of PCR-amplified bacterial 16S rRNA gene segments. The DGGE gel images were obtained from the total genomic DNAs of the pooled dental plaque samples of 12 children with S-ECC and 8 CF children. For each individual lane, the number of detected amplicons ranged from 29 to 46, with a mean ± SD of 37.9 ± 5.0. The oral microbial diversity was greater (P = 0.005 by the nonparametric Mann-Whitney U test) in the CF group than in the S-ECC group. The marker is the DGGE reference marker corresponding to 16S rRNA gene fragments from specific bacterial species: 1, Fusobacterium nucleatum subsp. vincenti (ATCC 49256); 2, F. nucleatum subsp. nucleatum (ATCC 25586); 3. Streptococcus sanguinis (ATCC 10556); 4, Streptococcus oralis (ATCC 35037); 5, Streptococcus salivarius (ATCC 7073); 6, Streptococcus mutans (ATCC 700610); 7, Lactobacillus paracasei subsp. paracasei (ATCC 25598); 8, Porphyromonas gingivalis (ATCC 33277); 9, Actinomyces naeslundii genospecies 1 (ATCC 12104).
Clustering of DGGE profiles based on caries status.
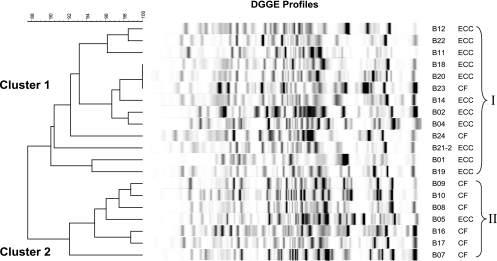
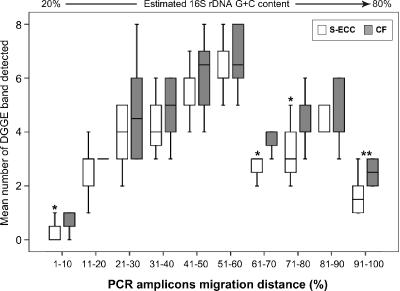
Figure 2 depicts the results of Ward's analysis in which the Dice coefficient for measuring similarity algorithm in banding patterns was applied. The S-ECC and CF groups displayed a statistically significant clustering of profiles, cluster I (S-ECC) and cluster II (CF) (P = 0.004 by Fisher's exact test). The analysis of specific regions of the profiles, which were determined by partitioning the migration distances into 10 segments, revealed that the CF group had more detected bands in specific segments 1 (P = 0.05), 7 (P = 0.034), 8 (P = 0.046), and 10 (P = 0.008) (Fig. 3.). The overall differences in the band distribution between the S-ECC and CF groups were statistically significant (P = 0.007 by the chi-square test). However, only four bands (4.2%) were present in more than 85% of the samples.
FIG. 2.
Cluster analysis. The difference in microbial diversity was clearly distinguished by cluster analysis with Ward's algorithm based on the Dice coefficient. A distinct cluster from the S-ECC group was observed, and 11 of 13 S-ECC profiles were grouped into one dendrogram branch (P = 0.004 by Fisher's exact test). The DGGE profiles of the CF children were differentiated from those of the S-ECC children in a separate cluster. The difference in the mean similarity values (89.4% ± 4.7%; P = 0.001 by ANOVA) between the two groups was statistically significant.
FIG. 3.
Distribution of the PCR amplicons. The frequency distribution of the PCR amplicons in DGGE gels is shown. The x axis of the graph is the migration distance divided into 10 segment groups that correspond to the percent GC content of the 16S rRNA gene amplicons. The y axis is the mean number of bands detected on the DGGE gels. A comparison of the mean in each segment showed that more DGGE bands were detected in the CF group in 4 of the 10 category groups according to their migration distances. Segment 1, 1% to 10% (P = 0.05); 7, 61% to 70% (P = 0.034); 8, 71% to 80% (P < 0.046); 10, 91% to 100% (P = 0.003).
Sequence analysis of excised bands.
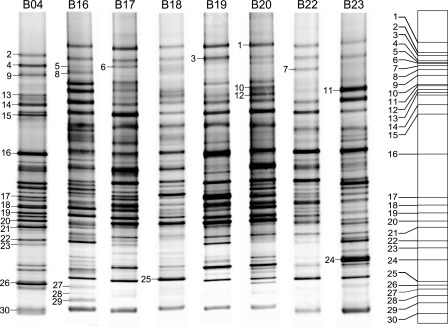
Of the 92 discrete PCR amplicons detected in the DGGE profiles, 30 bands (22 from the S-ECC group and 8 from the CF group) were excised from the gels (Fig. 4), after which the DNA was extracted, reamplified, and cloned. An average of 11 clones per unique band (for a total of 396 clones) were picked and sequenced. The phylogenetic affiliation of each sequence was estimated by comparison with the RDP-II database (11). From the 396 sequences examined, 26 genera were identified (Table 2), 46% of which were gram negative, 27% gram positive, and 19% unknown. On average, 2.7 genera per band sequenced were found. Bands 8, 25, and 29 were significantly less present among the S-ECC group samples, and band 21 was more predominant in the S-ECC samples than in the CF samples (P = 0.006 by the chi-square test).
FIG. 4.
Illustration of the DGGE bands cut for sequence analysis. A total of 30 discrete PCR amplicons (22 from S-ECC children and 8 from CF children) were excised from the DGGE gels, after which the DNA was extracted, reamplified, and cloned. Three hundred ninety-six clones were picked and sequenced, and 24 genera were identified. Bands 8, 25, and 29 were significantly less present among the S-ECC samples, and band 21 was more predominant in the S-ECC samples than in the CF samples.
TABLE 2.
Sequence analysis of genera identified in the study subjects
| Band no. | Genus(era) | Distribution (%)
|
|
|---|---|---|---|
| S-ECC | CF | ||
| 1 | Bacteroidetes, Capnocytophaga, Fusobacterium | 66.7 | 50.0 |
| 2 | Bacteroidetes | 33.3 | 75.0 |
| 3 | Bacteroidetes, Capnocytophaga, Leptotrichia | 41.7 | 25.0 |
| 4 | Bacteroidetes, Capnocytophaga, Fusobacterium | 41.7 | 50.0 |
| 5 | Bacteroidetes, Capnocytophaga, Fusobacterium, TM7 phylum | 33.3 | 37.5 |
| 6 | Capnocytophaga, Fusobacterium, Sarcina, uncultured bacterium | 41.7 | 50.0 |
| 7 | Bacteroidetes, Fusobacterium, Neisseria | 41.7 | 50.0 |
| 8 | Bacteroidetes, Fusobacterium, Filifactor, Leptotrichia | 16.7 | 62.5 |
| 9 | Fusobacterium | 25.0 | 12.5 |
| 10 | Fusobacterium, Bacteroidetes, uncultured bacterium | 33.3 | 50.0 |
| 11 | Fusobacterium, uncultured bacterium X112 | 66.7 | 37.5 |
| 12 | Fusobacterium | 41.7 | 25.0 |
| 13 | Capnocytophaga, Fusobacterium, uncultured bacterium | 41.7 | 62.5 |
| 14 | Fusobacterium | 50.0 | 37.5 |
| 15 | Bacteroides, Fusobacterium, Leptotrichia | 66.7 | 25.0 |
| 16 | Kingella, Leptotrichia, Streptococcus | 66.7 | 75.0 |
| 17 | Corynebacterium, Neisseria, Rothia | 66.7 | 87.5 |
| 18 | Cardiobacterium, Leptotrichia, Streptococcus | 66.7 | 87.5 |
| 19 | Gemella, Kingella, Leptotrichia, Streptococcus, Veillonella | 75.0 | 100 |
| 20 | Granulicatella, Leptotrichia, Neisseria | 66.7 | 87.5 |
| 21 | Leptotrichia, Neisseria, Streptococcus | 66.7 | 12.5 |
| 22 | Bacteroides, Leptotrichia | 58.3 | 50.0 |
| 23 | Eubacterium, Leptotrichia, Prevotella, uncultured bacterium | 33.3 | 37.5 |
| 24 | Lautropia, Leptotrichia, Prevotella | 58.3 | 75.0 |
| 25 | Corynebacterium, Prevotella, Lautropia | 8.3 | 37.5 |
| 26 | Bacteroidetes | 50.0 | 37.5 |
| 27 | Actinomyces, Treponema, uncultured human oral bacterium A11 | 50.0 | 37.5 |
| 28 | Actinomyces, Neisseria | 25.0 | 50.0 |
| 29 | Treponema | 8.3 | 37.5 |
| 30 | Campylobacter, Leptotrichia, Veillonella | 41.7 | 62.5 |
DISCUSSION
DGGE profiling of the bacterial populations in the plaque of children with S-ECC displayed significantly less diversity than that for the CF controls. This finding suggests that the caries-associated microbiota becomes less diverse, perhaps because certain groups of microbes supplant or dominate the plaque biofilm as caries progresses. The results of this study are consistent with our previous study with adults (24). Although not a focus of the present study, one possible explanation for the lesser microbial diversity in S-ECC plaque is that caries plaques contain higher proportions of acidogenic and aciduric bacteria than caries-free plaques, as some research papers have reported (4, 18, 26, 48). For S-ECC, several studies suggest that this form of caries results predominantly from a mutans streptococcus infection (3, 5, 27). Other, nonmutans streptococcus bacterial groups capable of tolerating low pH have also been found associated with dental caries (47, 48). A recent study that used a DNA-based reverse capture checkerboard assay indicated that more than 10 bacterial species were overabundant in the oral cavities of caries-active children (12). Those bacteria have previously been associated with diseased subjects but not healthy subjects (38). Each individual bacterial species may collectively contribute to the overall cariogenicity of the microbial biocommunity of the dental plaque associated with dental caries (13, 21). Another explanation for the decrease in diversity is that caries lesions create more retentive niches for cariogenic microorganisms, which increase their total numbers of cariogenic bacteria but subsequently decrease the overall richness of the plaque community. This notion was supported by several studies (2, 3, 47). For example, Becker, et al. (3) showed that bacterial levels were significantly increased at each of the caries sites, including white spot, cavitated lesions, and excavated carious dentin, compared to those found on intact enamel surfaces. They also reported that a number of other bacterial species may also be associated with caries initiation and development. Clearly, the concept that dental caries is a polymicrobial infectious disease was well supported by the published literature (7, 8, 28, 33). Further studies, however, are needed to determine whether caries-associated plaques are more or less diverse overall and to elucidate the determinant associated with the microbial composition.
The results of our study also demonstrate that the DGGE profiles of each caries type formed significant group-specific clusters (Fig. 2). The delineation of two distinct clusters was reflected by the number of bands detected (richness), the intensity, and the migration distribution of the PCR 16S amplicons. This finding is noteworthy, considering that the S-ECC and CF profiles were generated on multiple gels. Moreover, the overall S-ECC and CF profiles were more similar within each group than between the groups, which suggests the presence of common phylotypes associated with diseased or healthy status. These results demonstrated that caries group can be predicted with reasonable accuracy based on DGGE banding patterns. Even though the molecular fingerprinting profile does not provide immediate discrimination among bacterial species, it does enable the simultaneous analysis of multiple samples and thus facilitates the direct comparison of microbial communities from different samples of interest (35, 36). Additionally, DGGE-generated molecular fingerprinting also allows the study of changes in individual microbial communities over time (25, 30, 31, 44).
One feature of DGGE profiling is that bands (amplicons) of interest can be excised from the gels and sequenced to obtain a better approximation of their taxonomic identity (35). By doing so, we were able to obtain valuable information in three areas: the distribution of gram-positive versus gram-negative microbes, the identification of each amplicon to at least the genus level, and the number of phylotypes represented in each band. Interestingly, we found that on average, each band represented not just one, but 2.7 phylotypes. If we assume that each band present on the DGGE gels from the plaque samples represented 2.7 different phylotypes, it would mean that the overall species richness of the samples for the 20 children as a group is 248.4 (92 × 2.7) and that the group-specific species richness is 94.5 (35 × 2.7) for the S-ECC children and 113.4 (42.2 × 2.7) for the CF children. Since a known limitation of DGGE in most instances is that the identification is based on partial 16S rRNA gene sequences, which may not be sufficient for species-level assignment, this adjusted value could be an overestimation of the actual numbers of phylotypes in the supragingival plaque samples.
With a different population (adults) and using a different sampling method, Munson et al. (34) found an average of 32 phylotypes present within caries lesions in which gram-positive species dominated the sample, and they noted that lactobacilli were the most prevalent of all species present. Kroes and coworkers (22) reported an overall diversity of 59 phylotypes from subgingival plaque samples collected from a healthy adult. Paster and colleagues (38) found 215 novel phylotypes in subgingival samples, and on average healthy subjects harbored 72 of the 215 phylotypes of bacteria. They estimated that this number represented only 84% of the total number present based on coverage estimation. An adjusted value of 99 (72 identified plus 27 expected) would be closer to our estimation of 113.4 for caries-free individuals. Becker et al. (3) reported that only 11 phylotypes were present in the tooth surface plaque from a caries-free child. More recently, Aas et al. (1) reported that in five healthy volunteers, 52 species were detected on sound tooth surfaces. This discrepancy in the number of phylotypes among the different studies may be due to differences in sampling methods, sampling sites, the number of clones that were sequenced, and the number of subjects that were included in different studies. In our study, greater species richness was found than in previous studies. Possible reasons could be that we gathered pooled supragingival samples from each child to get better consistency in the plaque sample and that DGGE profiles were obtained and systematically analyzed for all 20 children, i.e., 12 highly caries-active children and 8 caries-free children. Moreover, the bacterial lysis method we employed appears to have yielded a better representation of gram-positive and gram-negative organisms as evident on DGGE profiles (Fig. 1). Because pooled supragingival plaque is dominated by gram-positive streptococci and that group of bacteria is difficult to lyse, all of the bacterial samples were treated with a cocktail lysis buffer containing mutanolysin, proteinase K, and a lysozyme treatment, which yielded a high quality and concentration of total bacterial genomic DNA and a wide range in the 16S rRNA gene diversity. Sequencing of excised amplicons from DGGE gels further demonstrated that the distribution of the amplicons ranged from low GC content to high GC content (Table 2). Since the goal of this study was to evaluate the overall variation in oral microbial diversity in children of different caries status by means of the DGGE technique, we did not conduct extensive cloning or sequence entire sets of bands from each subject to identify all members of the microbial population. If more precise taxonomical identification is required, then the full-length 16S rRNA pool from which the nested DGGE amplicons were generated can be cloned and sequenced.
In summary, we observed a significant variation in the DGGE profiles between the S-ECC and CF groups. The microbial diversity and the complexity of the microbial biota in plaque were less in children with S-ECC than in CF children. The results of our study also demonstrated that PCR-based 16S rRNA gene DGGE is a sufficiently valuable tool for differentiating the microbial composition of the oral plaque in S-ECC children from that of CF children. Moreover, DGGE may be further developed as a pattern recognition tool with which to identify specific group of bacteria predominantly colonized in children of various caries status. The information, therefore, can be used as a basis for targeted caries intervention and prevention.
Acknowledgments
We acknowledge Peter Catapano and Untray Brown from the pediatric clinic at Bellevue Hospital in New York City, New York, and Michele Kaider and Josh Abene of the New York University College of Dentistry in New York City for their clinical assistance.
This study was supported by the NIH/NIDCR grant DE13937.
Footnotes
Published ahead of print on 1 November 2006.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babaahmady, K. G., S. J. Challacombe, P. D. Marsh, and H. N. Newman. 1998. Ecological study of Streptococcus mutans, Streptococcus sobrinus and Lactobacillus spp. at sub-sites from approximal dental plaque from children. Caries Res. 32:51-58. [DOI] [PubMed] [Google Scholar]
- 3.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, et al. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beighton, D. 2005. The complex oral microflora of high-risk individuals and groups and its role in the caries process. Community Dent. Oral Epidemiol. 33:248-255. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz, R. 1996. Etiology of nursing caries: a microbiologic perspective. J. Public Health Dent. 56:51-54. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz, R. J., J. Turner, and C. Hughes. 1984. Microbial characteristics of the human dental caries associated with prolonged bottle-feeding. Arch. Oral Biol. 29:949-951. [DOI] [PubMed] [Google Scholar]
- 7.Boue, D., E. Armau, and G. Tiraby. 1987. A bacteriological study of rampant caries in children. J. Dent. Res. 66:23-28. [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw, D. J., and P. D. Marsh. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res. 32:456-462. [DOI] [PubMed] [Google Scholar]
- 9.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleaton-Jones, P., B. D. Richardson, L. Granath, L. P. Fatti, R. Sinwell, A. R. Walker, et al. 2000. Nutritional status and dental caries in a large sample of 4- and 5-year-old South African children. S. Afr. Med. J. 90:631-635. [PubMed] [Google Scholar]
- 11.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, et al. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corby, P. M., J. Lyons-Weiler, W. A. Bretz, T. C. Hart, J. A. Aas, T. Boumenna, et al. 2005. Microbial risk indicators of early childhood caries. J. Clin. Microbiol. 43:5753-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton, J. W. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137-140. [DOI] [PubMed] [Google Scholar]
- 14.Drury, T. F., A. M. Horowitz, A. I. Ismail, M. P. Maertens, R. G. Rozier, and R. H. Selwitz. 1999. Diagnosing and reporting early childhood caries for research purposes. J. Public Health Dent. 59:192-197. [DOI] [PubMed] [Google Scholar]
- 15.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, et al. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto, C., H. Maeda, S. Kokeguchi, S. Takashiba, F. Nishimura, H. Arai, et al. 2003. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 38:440-445. [DOI] [PubMed] [Google Scholar]
- 18.Hayes, M. L., E. C. Carter, and S. J. Griffiths. 1983. The acidogenic microbial composition of dental plaque from caries-free and caries-prone people. Arch. Oral Biol. 28:381-386. [DOI] [PubMed] [Google Scholar]
- 19.Ismail, A. I., and W. Sohn. 1999. A systematic review of clinical diagnostic criteria of early childhood caries. J. Public Health Dent. 59:171-191. [DOI] [PubMed] [Google Scholar]
- 20.Kohler, B., I. Andreen, and B. Jonsson. 1988. The earlier the colonization by mutans streptococci, the higher the caries prevalence at 4 years of age. Oral Microbiol. Immunol. 3:14-17. [DOI] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E., N. Ganeshkumar, F. J. Cassels, and C. V. Hughes. 1993. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 7:406-413. [DOI] [PubMed] [Google Scholar]
- 22.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-147. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., West Sussex, England.
- 24.Li, Y., C. Y. Ku, J. Xu, D. Saxena, and P. W. Caufield. 2005. Survey of oral microbial diversity using PCR-based denaturing gradient gel electrophoresis. J Dent. Res. 84:559-564. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., D. Saxena, V. Barnes, H. Trivedi, Y. Ge, and T. Xu. 2006. PCR-based denaturing gradient gel electrophoresis in the evaluation of oral microbiota. Oral Microbiol. Immunol. 21:333-339. [DOI] [PubMed] [Google Scholar]
- 26.Lingstrom, P., F. O. van Ruyven, J. van Houte, and R. Kent. 2000. The pH of dental plaque in its relation to early enamel caries and dental plaque flora in humans. J. Dent. Res. 79:770-777. [DOI] [PubMed] [Google Scholar]
- 27.Loesche, W. J. 1979. Clinical and microbiological aspects of chemotherapeutic agents used according to the specific plaque hypothesis. J. Dent. Res. 58:2404-2412. [DOI] [PubMed] [Google Scholar]
- 28.Marchant, S., S. R. Brailsford, A. C. Twomey, G. J. Roberts, and D. Beighton. 2001. The predominant microflora of nursing caries lesions. Caries Res. 35:397-406. [DOI] [PubMed] [Google Scholar]
- 29.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCracken, V. J., J. M. Simpson, R. I. Mackie, and H. R. Gaskins. 2001. Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota. J. Nutr. 131:1862-1870. [DOI] [PubMed] [Google Scholar]
- 31.Meroth, C. B., J. Walter, C. Hertel, M. J. Brandt, and W. P. Hammes. 2003. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millar, M. R., C. J. Linton, A. Cade, D. Glancy, M. Hall, and H. Jalal. 1996. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J. Clin. Microbiol. 34:2506-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milnes, A. R., and G. H. Bowden. 1985. The microflora associated with developing lesions of nursing caries. Caries Res. 19:289-297. [DOI] [PubMed] [Google Scholar]
- 34.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer, G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317-322. [DOI] [PubMed] [Google Scholar]
- 36.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nubel, U., F. Garcia-Pichel, M. Kuhl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, et al. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasiah, I. A., L. Wong, S. A. Anderson, and C. H. Sissons. 2005. Variation in bacterial DGGE patterns from human saliva: over time, between individuals and in corresponding dental plaque microcosms. Arch. Oral Biol. 50:779-787. [DOI] [PubMed] [Google Scholar]
- 40.Rocas, I. N., J. F. Siqueira, Jr., M. C. Aboim, and A. S. Rosado. 2004. Denaturing gradient gel electrophoresis analysis of bacterial communities associated with failed endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 98:741-749. [DOI] [PubMed] [Google Scholar]
- 41.Rupf, S., K. Merte, and K. Eschrich. 1999. Quantification of bacteria in oral samples by competitive polymerase chain reaction. J. Dent. Res. 78:850-856. [DOI] [PubMed] [Google Scholar]
- 42.Sheffield, V. C., D. R. Cox, L. S. Lerman, and R. M. Myers. 1989. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc. Natl. Acad. Sci. USA 86:232-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 44.Suchodolski, J. S., C. G. Ruaux, J. M. Steiner, K. Fetz, and D. A. Williams. 2004. Application of molecular fingerprinting for qualitative assessment of small-intestinal bacterial diversity in dogs. J. Clin. Microbiol. 42:4702-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Houte, J. 1994. Role of micro-organisms in caries etiology. J. Dent. Res. 73:672-681. [DOI] [PubMed] [Google Scholar]
- 46.van Houte, J., G. Gibbs, and C. Butera. 1982. Oral flora of children with “nursing bottle caries.” J. Dent. Res. 61:382-385. [DOI] [PubMed] [Google Scholar]
- 47.van Houte, J., J. Lopman, and R. Kent. 1996. The final pH of bacteria comprising the predominant flora on sound and carious human root and enamel surfaces. J. Dent. Res. 75:1008-1014. [DOI] [PubMed] [Google Scholar]
- 48.van Houte, J., C. Sansone, K. Joshipura, and R. Kent. 1991. Mutans streptococci and non-mutans streptococci acidogenic at low pH, and in vitro acidogenic potential of dental plaque in two different areas of the human dentition. J. Dent. Res. 70:1503-1507. [DOI] [PubMed] [Google Scholar]
- 49.Walker, A. R., J. Gregory, G. Bradnock, J. Nunn, and D. White. 2000. National Diet and Nutrition Survey: young people aged 4 to 18 years. The Stationery Office, London, United Kingdom.
- 50.Ward, J. H. 1963. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58:236-244. [Google Scholar]
- 51.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]