Abstract
Sequence-based epidemiological typing of methicillin-resistant Staphylococcus aureus (MRSA) has recently been promoted because it results in unambiguous data sets that can be organized in local and global databases. The replacement of previous typing methods, such as the highly discriminatory pulsed-field gel electrophoresis (PFGE), has been attempted with various markers and typing schemes, including spa typing and multilocus sequence typing. However, despite a number of advantages, none of these methods showed convincing evidence for performance in epidemiological typing comparable to that of PFGE. By using three sets of 48 MRSA strains comprising isolates that were (i) genetically highly diverse, (ii) genetically related, and (iii) obtained from long-term carriers, we analyzed the performance of the four highly polymorphic S. aureus markers: clfA, clfB, fnbA, and spa. Typeability, discriminatory power, in vivo stability, and evolution of these markers were compared to those of PFGE. Clearly, none of the markers alone could match the discriminatory power of PFGE (63 genotypes; index of discrimination of 0.96). Instead, this could be achieved by combining markers in pairs. We showed that by using only 3′ partial sequences of approximately 500 bp, the majority of each marker's discriminatory power was displayed, and using the partial sequences, the best performance was obtained with the combination of clfB and spa (57 genotypes; index of discrimination of 0.94). Genetic changes were not observed for any of the sequence markers over a period of 3 years and in the case of partial sequences for a period of more than 4 years. This is in contrast to PFGE where changes occurred after several months. The genetic differences found between isolate pairs of long-term carriers and among highly related isolates indicated clonal evolution. A typing scheme based on 500-bp 3′ partial sequences of clfB and spa is proposed.
The quality and use of a typing method are crucial for epidemiological analyses. The epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) has in the past been analyzed by an array of different typing methods ranging from phenotypic analyses, such as phage typing, to genetic analyses, e.g., pulsed-field gel electrophoresis (PFGE) of DNA macrorestriction products. Recently, new DNA sequence-based techniques have been developed and promoted. These new techniques allowed the creation of unambiguous data sets that can repeatedly be created in different laboratories, readily be exchanged between laboratories, and be organized in global databases.
Two different DNA sequence-based S. aureus typing methods have been introduced for which global databases are available. One, spa typing, is based on the highly variable repeat region (X region) of the spa gene (10). The other, multilocus sequence typing (MLST), is based on 450- to 500-bp DNA fragments of seven moderately variable housekeeping genes (5).
Epidemiological investigations range from local applications, such as transmission from patient to patient, to global applications, such as the study of internationally circulating “clones.” Local investigations require a method based on highly variable markers in order to ensure the best discrimination. A number of studies suggested that spa typing is useful in local investigations because of its great discriminatory power (10, 12, 15), while an earlier study showed that spa typing was less discriminatory than PFGE (22). A recent study suggested that discriminatory power could be increased when the spa gene was used in combination with clfB, which contains a highly variable serine-aspartate (SD) repeat region (16).
Investigations at a global level require a method based on genetic markers which are more stable, such as housekeeping genes, in order to be able to recognize clones. MLST is thus more prone for global investigations, and in addition, it can also be used to analyze the population structure of a bacterial species. Analysis of global S. aureus MLST data sets showed that a limited number of clonal groups of strains, also called clonal complexes (CCs), existed in the population with only a few differences within groups (6). It also showed that the population structure of S. aureus is predominantly clonal (7), which confirmed the results of a previous report based on multilocus enzyme electrophoresis data sets (19). A way to increase the limited resolution within CCs had been to add seven polymorphic cell surface genes (sas genes) to the MLST typing schemes (21). However, none of the currently available typing methods satisfactorily covered the complete range from local to global applications; thus, depending on the goal of the investigator, different methods have to be applied that cannot easily be compared.
Although DNA sequence-based methods are generally considered superior to other methods as stated above, one problem is that time and cost of analysis increase with the number and size of gene fragments used in a typing scheme. Both constraints hugely influence the choice of a typing method. Therefore, affordable sequence-based typing schemes will have to rely on a small number of loci and their efficient use.
This study was designed to set up a typing scheme of S. aureus with the following goals: (i) to investigate outbreaks, (ii) to study endemic situations, and (iii) to follow the dynamics of predominant strains at a local and, if possible, at an international level. So far, PFGE is the current state-of-the-art method for such investigations. However, PFGE is a comparative typing method (24), the data are not unambiguous (2, 26), and interlaboratory reproducibility studies highlight the problems of standardization (18). A sequence-based typing method is definitive and does not suffer from these problems (1). As a prerequisite for future epidemiological analyses, a new method should perform as well as PFGE to discriminate between isolates, while time and cost of analysis should, in the best case, not be higher than for PFGE. In light of these constraints, a typing scheme based on spa seems to offer a clear advantage, because only one marker gene fragment would have to be sequenced. However, there is confusion about the discriminatory power and thus about its performance in epidemiological typing.
Next to clfB and spa, some other highly polymorphic genes belonging to the same class of cell surface adhesion genes have previously been used for the analysis of MRSA and were proposed as potentially useful markers for typing approaches (11, 17). For the present study, we selected four polymorphic genes, clfA, clfB, fnbA, and spa, for comparison in performance to PFGE and, for a limited number of strains, also to MLST. All four genes contain variable repeat sequences. They are distributed over the entire chromosome, all code for cell wall-associated adhesin proteins (8), and their nonrepeat regions followed a clonal evolution (17).
We analyzed three different collections of isolates: (i) a diverse set of MRSA isolates to evaluate the global diversity of markers and typeability; (ii) a clonally related set of MRSA isolates to evaluate the discriminatory power of markers; (iii) 24 pairs of isolates, obtained more than 2 years apart from the same patients, to evaluate the in vivo stability of markers at a local level and to study their in vivo evolution.
MATERIALS AND METHODS
Bacterial isolates. (i) Diverse MRSA.
Forty-eight isolates from a globally diverse collection were previously analyzed by PFGE, MLST, spa typing, and also using the nonrepeat region of nine adhesion genes (17). It included 9 isolates of all 6 CCs which are known to comprise MRSA strains, 23 isolates of 12 epidemic clones, and 14 sporadic strains.
(ii) Related MRSA.
Molecular epidemiological surveillance of MRSA in our area allowed the identification of a group of isolates that all differed by no more than six bands in PFGE restriction patterns (genetically related). However, a predominant pattern has never been observed, as it is the case for epidemic strains spreading in our area. We selected 48 isolates of this group from 51 isolates that were collected in a 2-year period of time (2003 to 2004).
(iii) LTC MRSA.
During the MRSA surveillance in our hospital over the last 10 years, we collected isolates from over 2,000 patients. From these patients, we identified those for whom at least two isolates were available and selected 24 showing the longest period of time between the first and the last isolate (ranging from 214 to 1,722 days). Only pairs of isolates that differed by no more than six bands in their PFGE restriction profiles were included and considered long-term carriers (LTCs). Previously, it had been shown that isolate pairs of LTCs with more than six band differences should be considered two genetically different strains (3).
Primer design for PCR amplification.
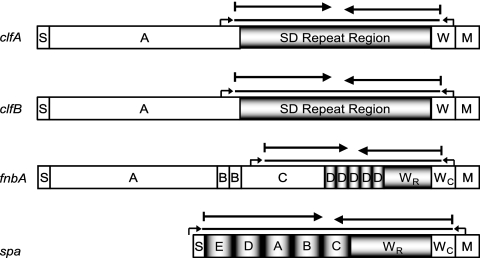
The repeat-containing regions of genes clfA, clfB, fnbA, and spa were chosen for molecular typing (see Fig. 1). These regions are located in the 3′ half for all genes (see Fig. 1). On the basis of DNA sequence alignments from seven publicly available S. aureus genome sequences (strains Mu50, N315, MW2, MSSA476, COL, MRSA252, and NCTC8325), we targeted primer binding sites at conserved regions flanking the repeat-containing region. Primer sequences, their positions on database sequences, lengths, and specific melting temperatures are summarized in Table 1.
FIG. 1.
Schematic representation of clfA, clfB, fnbA, and spa, constructed on the basis of data from a previous report (8). Boxes indicate segments of each gene with signal sequences (S), wall-spanning regions (W), repeated and conserved wall-spanning regions (WR and WC, respectively), and membrane-spanning regions (M). Boxes labeled A to E are domains that were previously assigned. Repeated segments are indicated with vertical black and white shadings (each segment represents one repeat motif), whereas regions with small repeat motifs are indicated with horizontal black and white shading. Small arrows indicate primer annealing sites located in conserved regions flanking the repeat-containing regions. The different sequences of regions used for analyses are indicated by lines above the boxes. Continuous black lines represent the whole amplified repeat-containing regions (clfA, clfB, fnbA, and spa), long arrows indicate 500-bp partial sequences of the 3′ or 5′ region of the genes (3p500 or 5p500, respectively) which were taken for analysis. PCR amplification products varied in size mostly due to variation in repeat number; thus, it is possible that partial 5′ and 3′ sequences overlap.
TABLE 1.
Details of primers used in PCR mixtures to amplify the repeat regions of four adhesion genes
| Gene | Supposed or known function | Accession no. of EMBL database sequence | Primer name | Sequence (5′-3′) | Optimal annealing temp (°C)a | Position on database sequence |
|---|---|---|---|---|---|---|
| clfA | Clumping factor A; fibrinogen receptor | Z18852 | SA_clfA_1902.for | TTCTGGTGACGGTATCGATAAACC | 58 | 1902 |
| SA_clfA_2983.rev | TTCAGAACCTGTATCTGGTAATGG | 58 | 2983 | |||
| clfB | Clumping factor B; fibrinogen receptor | AJ224764 | SA_clfB_1586.for | TCGGTTGGAATAATGAGAATGTTG | 58 | 1586 |
| SA_clfB_2658.rev | TTTGTGTTTTCGCTCTTATCTCCT | 58 | 2686 | |||
| fnbA | Fibronectin-binding protein A | J04151 | SA_fnbA_1780.for | ATTACCACACAGCTATAGATGGTG | 58 | 1780 |
| SA_fnb_3064.rev | TTGATTCTTCTCCACCTGTTTCAG | 58 | 3064 | |||
| spa | Protein A | M18264 | SA_spa_729.for | ACTAGGTGTAGGTATTGCATCTGT | 54 | 729 |
| SA_spa_1984.rev | TCCAGCTAATAACGCTGCACCTAA | 54 | 1984 |
Optimal PCR and sequencing annealing temperatures were obtained by testing for the largest amplicon amounts in PCR mixtures with different temperatures covering a range from 6°C below and above the melting temperature of the oligonucleotide.
Molecular procedures.
Bacterial isolates were grown overnight in 1 ml of brain heart infusion in 96-well plates. After lysis of 100 μl precipitated bacteria in 200 μl lysis buffer (1× Tris-EDTA buffer, 0.35 M NaCl, and 0.05 mg/ml lysostaphin), DNA was obtained as described previously (4). PCR and sequencing were performed as described previously (17) with the following differences. For PCR and sequencing reactions, the annealing temperatures were adjusted according to each primer pair's optimal annealing temperature. During PCR, cycling extension time (72°C) was 2 min in all cases. Purification of PCR mixtures and sequencing reaction mixtures was carried out using Montage PCR μ96 kit (Millipore) and Montage SEQ 96 kit (Millipore), respectively, following the manufacturer's instructions. Purified sequencing reaction mixtures were analyzed using an ABI 3730 sequencer (Applied Biosystems) with standard conditions. Sequence read errors were removed by visual inspection of electropherograms. Contigs of each gene sequence were assembled with ContigExpress of the software package Vector NTI Advance 10 (Invitrogen).
Resolution of the markers.
Discriminatory power was evaluated by the number of types (variant alleles) and by calculation of the index of discrimination (ID) (13). Variant alleles were taken into account beginning from the smallest possible genetic difference, i.e., a point mutation. In general, we consider that discriminatory power positively correlates with the number of types found.
The index of discrimination was calculated from the distribution of types with the Discriminatory Power Calculator (http://biophp.org/stats/discriminatory_power/demo.php). It describes the probability that two unrelated isolates drawn at random from a given population will be placed into different typing groups. An ID value of 1 would indicate that the typing method was able to distinguish each isolate from all others. Conversely, an index of 0 would indicate that all isolates were of an identical type.
Marker stability and microevolution.
Short-term in vivo genetic changes of clfA, clfB, and spa and genetic differences between highly related isolates were analyzed to study the stability of markers and their microevolution. We compared sequences of isolates that were obtained from LTC patients at two different moments in time. The length of the period of time without genetic changes was taken as a measure of stability. Genetic differences between the first and second isolates could be due to (i) polymerase slippage during replication leading to duplication or loss of DNA repeat(s), (ii) misincorporation of nucleotides leading to point mutations, or (iii) acquisition or replacement of repeats by foreign repeats. Changes following the definition of the first two points were considered clonal events, while those of the third point were considered events of recombination. By scoring the number of events for either type of event, the genetic evolution of spa and clfB during a short period of time was investigated.
Isolates with no difference in PFGE patterns are generally considered highly related. Given the close relationship of isolates within one PFGE group, we hypothesized that isolates with a single allelic difference have evolved from isolates with predominant (most frequent) alleles. Therefore, first, groups of isolates with identical PFGE patterns were formed. Within these groups, we then identified the predominant and variant alleles for clfA, clfB, and spa. We analyzed the genetic differences found in isolates that differed in one marker from the predominant types. These were classified according to the above criteria.
Genetic relatedness.
The question that is often asked when two isolates are found to be different by a typing method is are they related or unrelated? For PFGE in outbreak investigation, it has been arbitrarily decided that isolates with one to six band differences should be considered genetically related (25). On the basis of PFGE data, we grouped isolates according to these criteria and genetically related isolates were marked with the same letter. Within these groups, isolates were marked with a numerical identifier. Using gene sequences as markers, relatedness could be established using evolutionary tools. However, for repeat regions of genes, these methods fail to work, as deletion or duplication of a repeat will not be considered a single genetic event. We aimed to overcome this problem by analyzing regions of DNA within the repeat regions that are putatively more conserved. Alignments of alleles, obtained from highly related isolates, revealed that gain and loss of repeats mainly occurred in the middle of the repeat regions. This led us to analyze only the first two, three, or five repeats and the DNA sequence flanking the repeat region. Grouping of isolates using this approach was compared to PFGE and MLST data (when available).
Nucleotide sequence accession numbers.
DNA sequences have been submitted to the EMBL database (accession numbers AM406818 to AM407390).
RESULTS
Repeat-containing regions of clfA, clfB, fnbA, and spa.
DNA sequences of the repeat-containing regions of clfA, clfB, fnbA, and spa were obtained. The length of DNA sequences was found to be variable between isolates, mostly due to variation in the number of repeats. The fnbA and spa genes contained two different repeat motifs, and variation between isolates was observed for both (Fig. 1). For fnbA, the first motif (D) had a length of 114 bp and was repeated three to five times. The second motif had a length of 42 bp and was repeated between three and seven times (comprised within region WR). For spa, the first motif had a length of 174 bp and was repeated four to five times (Fig. 1, spa, boxes A to E). The second repeat motif, which is used for spa typing, had a length of 24 bp and was found to vary between 6 and 16 times (comprised within region WR). The clfA and clfB genes contained only one repeat region (SD repeat region). For clfB, the repeat motif was previously determined and comprised 18 bp (16). According to the previously defined search algorithm, in this study, the repeat motif was repeated between 12 and 53 times. Similar to clfB, the repeat motif for clfA was found to comprise 18 bp. The number of repeats varied between 21 and 50 times.
Typeability.
For clfA, clfB, and spa, amplification products were obtained for all 144 tested strains. For fnbA, no amplification product was obtained for three strains. Additional analyses using a primer set designed for amplification of the first half of the gene showed that fnbA was also present in these strains (data not shown). Most likely, sequence variation in primer binding sites accounted for the lack of amplification.
Resolution of markers.
The performance of each marker (except for MLST) was determined for three data sets, all 144 isolates, genetically diverse MRSA, and clonally related MRSA (Table 2). In all data sets, PFGE showed the best performance with either parameter, number of types (63, 17, and 41, respectively) and ID (0.96, 0.86, and 0.99). None of the four markers alone could match this resolution, and this is most evident when looking at the analysis of clonally related MRSA. Of all sequence-based markers, clfB performed best in the number of types (54, 12, and 31, respectively) and spa alone performed best in ID (0.90, 0.59, and 0.97, respectively).
TABLE 2.
Discriminatory power of repeat-containing regions of four genes and their partial sequences in comparison to other typing methods
| Typing method, gene, or partial gene sequencea | Discriminatory power of gene or partial sequence for:
|
|||||
|---|---|---|---|---|---|---|
| All isolates (n = 144)
|
Related MRSA (n = 48)
|
Diverse MRSA (n = 48)
|
||||
| No. of genotypes | IDb | No. of genotypes | ID | No. of genotypes | ID | |
| PFGE | 63 | 0.96 | 17 | 0.86 | 41 | 0.99 |
| clfB_spa_3p500 | 57 | 0.94 | 17 | 0.77 | 35 | 0.98 |
| clfA_spa_3p500 | 48 | 0.94 | 13 | 0.83 | 30 | 0.97 |
| clfA_clfB_3p500 | 48 | 0.92 | 14 | 0.67 | 29 | 0.97 |
| spa | 42 | 0.90 | 8 | 0.59 | 30 | 0.97 |
| spa_3p500 | 39 | 0.90 | 8 | 0.59 | 29 | 0.97 |
| clfB | 54 | 0.89 | 12 | 0.47 | 31 | 0.97 |
| clfA | 35 | 0.87 | 9 | 0.51 | 22 | 0.93 |
| clfB_3p500 | 41 | 0.86 | 11 | 0.38 | 27 | 0.96 |
| clfA_3p500 | 27 | 0.83 | 8 | 0.51 | 17 | 0.90 |
| clfB_5p500 | 31 | 0.83 | 6 | 0.24 | 19 | 0.94 |
| clfA_5p500 | 26 | 0.79 | 3 | 0.08 | 19 | 0.91 |
| fnbA | 20 | 0.76 | 6 | 0.48 | 15 | 0.82 |
| spa_5p500 | 14 | 0.73 | 2 | 0.04 | 12 | 0.85 |
| fnbA_3p500 | 16 | 0.72 | 3 | 0.29 | 12 | 0.80 |
| fnbA_5p500 | 14 | 0.65 | 3 | 0.19 | 11 | 0.76 |
| MLST | NDc | ND | ND | ND | 21 | 0.93 |
Partial gene sequences (500 bp) of the 3′ or 5′ region of the genes are indicated by 3p500 or 5p500, respectively.
The index of discrimination (ID) was calculated by the method of Hunter (13).
ND, not determined.
In order to later reduce the workload and cost, we wanted to know how much resolution could be obtained with either 3′ or 5′ partial sequences of the repeat region. Beginning from either end, these sequences comprised a short part of the conserved flanking sequence plus a given number of repeats to obtain a length of approximately 500 bp.
As expected, 3′ and 5′ partial sequences showed lower resolution than the entire regions did. Yet, for all markers we observed better resolution with 3′ partial sequences than with 5′ partial sequences, and we observed that the resolution obtained with 3′ partial sequences did not greatly differ from the resolution with the entire repeat regions. The majority of the resolution can be found within the 3′ partial sequences.
For clfA, clfB, and spa, the lengths of the selected 3′ partial sequences were 498 bp, 508 bp, and 501 bp, respectively, comprising 27, 25 and 13 repeats, respectively. For spa, this meant that in 131 isolates of all 144 isolates (91%), the entire X-repeat region (as it is used for spa typing) was comprised because only 13 or less repeats were present.
To further increase resolution, we formed and tested pairwise combinations of markers using 3′ partial sequences of spa, clfA, and clfB. Due to the generally low resolution obtained with fnbA, no combination was tested with this marker, and it was excluded from further analyses. We found that all three combinations performed better than any single entire repeat-containing marker sequence. Considering the number of types and the ID, the best performance in resolution was achieved with the 3′ combination of clfB and spa, which was similar to that obtained with PFGE (Table 2). This combination was taken for further analysis. For naming purposes, we kept the type numbers for alleles of both markers and called the combination a double-locus sequence type (DLST) for future typing approaches.
Our analysis of the collection of highly diverse MRSA showed that the majority of the studied markers had an ID value of >0.90, ranging from 0.76 to 0.99. However, for the collection of clonally related MRSA, we observed a much wider ID range from 0.04 to 0.86. This marked difference between collections showed that ID values obtained from collections including highly diverse MRSA were skewed toward 1. As a result, it appeared that the two worst performing markers among related MRSA, 5′ spa and 5′ clfA (ID values of 0.04 and 0.08, respectively) showed reasonably good resolution among genetically highly diverse MRSA (ID values of 0.85 and 0.91, respectively). Thus, the collection of isolates taken for analysis had a great influence on the evaluation of a marker's performance.
Genetic relatedness.
A comparison of 57 DLSTs and 63 pulsotypes is summarized in Table 3. We observed differences in the groups formed on the basis of either PFGE or DLST data, in that identical isolates of either group are found to vary with the other method. Examples were DLSTs 1-1, 2-2, 4-4, 3-3, 3-34, and 8-5, which all comprised at least two different PFGE types. Conversely, we observed PFGE types A, A1, C, C1, C10, and C12 that were present in at least two different DLSTs.
TABLE 3.
Genetic relationship between strains with identical DLST and DLST-R
The combination of the type numbers for the alleles of the markers clfB_3p500 and spa_3p500.
The type numbers for the alleles of the markers clfB_3p500 and spa_3p500 considering only five and three repeats, respectively.
For the analysis of genetic relationships with putative conserved repeat regions, we observed that the 3′ flanking sequence plus three spa repeats and five clfB repeats performed best and that some congruence was apparent in comparison to PFGE and MLST data. Similar to DLST, numerical identifiers were given to each type. To distinguish types obtained this way from DLST, we called them DLST-R types, because they were restricted to a limited number of repeats. Six groups of isolates with identical DLST-R type were found. Compared to MLST data, we noted that none of the DLST-R types was found in different CCs, meaning that no evidence for potential homoplasy was found. We also observed that different STs were included within DLST-R groups (Table 3).
Marker stability.
Analysis of 24 LTC isolates showed that no genetic changes were found for the entire repeat region of clfA during a period of 2 years and in clfB, fnbA, and spa during a period of 3 years. For the proposed 3′ partial sequences, we found no changes for clfA and fnbA, while for clfB and spa, we found changes only after more than 4 years. Overall, six LTC pairs comprised genetic differences (Table 4). Of these, only one pair comprised differences within 3′ partial sequences. We observed more genetic differences in PFGE patterns (6 of 24 related isolates) than in any of the sequence-based markers (0 to 3 of 24 related isolates). In general, we noticed that the greater the time interval between the first and second isolates, the more observed genetic changes. No such correlation was found with PFGE types, where the differences appeared independently of time (Table 4).
TABLE 4.
In vivo stability of PFGE patterns clfA, clfB, fnbA, and spa genes over time in long-term carriers of MRSA
| Patient | Δ Daysa | Stability of PFGE patternsb | Stability of sequence-based markersc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| clfA | clfA_3p500 | clfB | clfB_3p500 | fnbA | fnbA_3p500 | spa | spa_3p500 | |||
| 1 | 1,722 | —d | — | 1x12 du | 1x12 du | 1x24 d, 1x48 d | 1x48 d | |||
| 2 | 1,548 | — | 1 m | 1x174 d | ||||||
| 3 | 1,420 | — | — | |||||||
| 4 | 1,293 | — | — | 1x54 d | ||||||
| 5 | 1,162 | — | — | 1 m | ||||||
| 6 | 1,124 | 6 | — | 2x18 d | ||||||
| 7 | 1,089 | — | — | |||||||
| 8 | 1,053 | — | — | |||||||
| 9 | 992 | — | — | |||||||
| 10 | 927 | 5 | — | |||||||
| 11 | 891 | — | 1 m | |||||||
| 12 | 800 | 1 | — | |||||||
| 13 | 767 | — | — | |||||||
| 14 | 539 | — | — | |||||||
| 15 | 517 | 2 | — | |||||||
| 16 | 432 | — | — | |||||||
| 17 | 371 | — | — | |||||||
| 18 | 347 | — | — | |||||||
| 19 | 330 | — | — | |||||||
| 20 | 287 | 2 | — | |||||||
| 21 | 285 | — | — | |||||||
| 22 | 229 | — | — | |||||||
| 23 | 222 | — | — | |||||||
| 24 | 214 | 6 | — | |||||||
Δ Days is the number of days between the two isolates collected from long-term carriers of MRSA.
For PFGE, the results are expressed as the number of band differences in restriction profiles.
For sequence-based analyses, the results are expressed as the number and type of event (point mutation [m], deletion [d], and duplication [du]), plus the size of insertions and deletions (number of base pairs). 1 m, one point mutation; 1x12 du, one 12-bp duplication.
—, no difference between the two isolates of a long-term carrier.
Microevolution of the markers.
Differences in microevolutionary patterns between markers were studied by observing in vivo genetic changes of 24 pairs of LTC isolates and by studying differences between isolates that shared the same PFGE type. For eight pairs of LTC isolates, the following 10 genetic changes were recorded (Table 4). For clfA, two pairs comprised one point mutation. For clfB, one pair showed one partial repeat duplication (12 bp), one pair showed a deletion of three repeats (54 bp), and one pair showed two deletions of one repeat (18 bp). For spa, one pair showed one point mutation, one pair showed a deletion of the first repeat motif (174 bp), and one pair showed two deletions, one comprising one repeat (24 bp) and the other comprising two repeats (48 bp). All events were concordant with genetic changes due to clonal evolution.
Among the 144 studied isolates, we found five groups of isolates with indistinguishable PFGE patterns (A, A16, C, C10, and C1) (Table 5). Within these groups, seven variant alleles were found, three for spa, two for clfA, and two for clfB. Two variant alleles (spa type 34 and clfA type 22) were found in two different PFGE patterns, subtypes C and C1. Genetic differences were due to five deletions, one point mutation, and one duplication. All genetic differences were concordant with genetic changes in clonal evolution (Table 5).
TABLE 5.
Genetic differences between strains with identical PFGE patterns
| PFGE type (no. of isolates) | Variant locus | Predominant allele | Variant allele (no. of isolates) | No. of different nucleotides | No. of repeats involved | Distance to 3′ end (bp) | Genetic event | Evolution |
|---|---|---|---|---|---|---|---|---|
| A (15) | spa | 1 | 14 (2) | 24 | 1 | 316 | Deletion | Clonal |
| A16 (3) | clfB | 1 | 34 (2) | 18 | 1 | 226 | Deletion | Clonal |
| C (14) | clfA | 3 | 22 (2) | 1 | 1 | 224 | Point mutation | Clonal |
| C (14) | clfB | 3 | 21 (2) | 36 | 2 | 112 | Deletion | Clonal |
| C (14) | spa | 3 | 34 (2) | 24 | 1 | 390 | Deletion | Clonal |
| C10 (5) | spa | 5 | 19 (1) | 24 | 1 | 366 | Deletion | Clonal |
| C1 (18) | clfA | 3 | 16 (2) | 12 | 1 | 147 | Duplication | Clonal |
| C1 (18) | clfA | 3 | 22 (6) | 1 | 1 | 224 | Point mutation | Clonal |
| C1 (18) | spa | 3 | 34 (4) | 24 | 1 | 390 | Deletion | Clonal |
DISCUSSION
The data of the present study showed that the combination of partial 3′ DNA sequences of clfB and spa can serve as useful genetic markers for typing MRSA at the local epidemiological level. So far, PFGE has undoubtedly been the method of choice in epidemiological typing of MRSA. Over the last decade, a number of studies have tested and compared sequence-based typing schemes to analyze MRSA epidemiology with the goal of replacing PFGE. Of these, spa typing has received particular interest and has been suggested for epidemiological investigations. Here, we present evidence that spa typing alone based on the X-repeat region or almost the entire gene can clearly not match the resolution of PFGE in epidemiological investigations. On the basis of data obtained with clfB typing alone, it was proposed that the combination of spa and clfB for typing should lead to increased resolution in local epidemiological analyses (16). In this study, we show for the first time that the resolution obtained with the combination of partial 3′ spa and clfB sequences is indeed comparable to the performance of PFGE. Both markers are located outside the staphylococcal chromosome cassette (SCC mec), thus also allowing typing of methicillin-susceptible S. aureus.
Importance of clonally related MRSA.
The striking evidence for a difference in resolution between spa typing and PFGE was observed with the collection of clonally related MRSA isolates. Almost all previous studies that aimed to compare sequence-based methods to PFGE determined typing performance on the basis of collections of genetically diverse isolates from different major clones (12, 15, 16). Indeed, similar to these studies, when only the collection of genetically diverse isolates was considered, we observed no clear difference in resolution between PFGE and spa typing. Markers showed a fairly good performance in resolution (ID of >0.90), except for typing with fnbA.
It seems obvious that, if resolution is evaluated on isolates that were selected in order to represent the overall diversity of the species (e.g., obtained with MLST), most typing methods will show a great power in resolution. Therefore, we think that evaluation of performance in resolution should also be done with isolates that are genetically related. Our data are limited in the way that 48 related isolates from only one clone were considered. However, it is of note that most of the isolates of LTCs belonged to one of the four major clones encountered in our area, thereby increasing the proportion of related isolates within the overall 144 studied isolates. Also, this collection showed similar trends in levels of discrimination. Our evidence is supported by the only previous study that compared spa typing to PFGE at a local epidemiological level showing that the number of genotypes obtained with spa typing was lower than with PFGE (22).
Correlation between number of types and ID.
In general, a positive correlation between the number of types and ID was observed. However, when considering only the number of types or ID, significant information about the performance of a marker in epidemiological analyses might be missed. This was based on two observations. First, clfB systematically showed higher number of types than clfA or spa, even though ID values were comparable. Second, pairwise marker combinations with clfA resulted in a smaller number of types than for the combination of spa and clfB, whereas ID values were comparable. This could be explained with the calculation of ID which took into account the frequency of types, while unique genotypes (represented by only one isolate) did not affect the resulting ID value. For this reason, despite the higher number of unique genotypes observed in clfB compared to clfA (data not shown), there was no difference in ID value. In this study, we considered both the number of types and ID for the evaluation of performance. The combination of clfB and spa was found to be superior to combinations comprising clfA.
Analysis of partial sequences.
Partial sequences comprising 500 bp were taken for analysis. For the majority of isolates, the 3′ partial sequences of spa comprised the entire X-repeat region with its small 24-bp repeat motif, while its 5′ partial sequence almost exclusively comprised the other 174-bp repeat motif. Partial sequences of fnbA showed a similar organization: its 3′ partial sequences were located within a region comprising most of the small 42-bp repeat motif, while its 5′ partial sequences were located in a region comprising the bigger 114-bp repeat motif. Because the 3′ and 5′ partial sequences of both markers each comprised a different repeat motif, this might explain the difference in resolution.
For both clfA and clfB, 3′ and 5′ partial sequences covered equally sized proportions of the same SD repeat region. Therefore, we expected, in contrast to our findings with fnbA and spa, no differences in resolution between 3′ and 5′ partial sequences. Surprisingly, similar to fnbA and spa, we observed an increased resolution of 3′ partial sequences compared to 5′ fragments. Yet, we have no explanation for such a difference in resolution within the same repeat-containing region.
Marker stability.
Epidemiological analyses at a local level aim at identifying transmission of isolates. This requires that markers used in these analyses are stable during the period of investigation, i.e., during several years. For the chosen 3′ partial sequence markers of clfB and spa, we observed no genetic changes in pairs of isolates collected from the same patients at intervals of time of several months to 4 years. These results are in concordance with two previous studies on clfB and spa, which did not find any genetic changes during the period of investigation of 21 months and 5 years, respectively (10, 16). A third, more detailed, study of the in vivo stability of the X region showed that the first genetic changes appeared after an average period of 5.8 years (70 months) (14). The overall congruence between previous findings and our findings led us to conclude that both markers are sufficiently stable for epidemiological investigations at a local level.
For all markers, when the entire repeat regions were considered, we observed more genetic changes than for partial sequences, indicating that entire repeat regions were less stable. Yet, if compared to PFGE, this stability was still superior. In addition, while the number of genetic changes in sequence-based markers increased with the period of time, there was no such correlation observable for PFGE. Thus, with regards to marker stability, there is a clear advantage of sequence-based markers over PFGE for use in epidemiological investigations.
A limitation of our approach resides in the fact that the second isolate might not be a direct descendant of the first isolate (e.g., strain replacement). This was likely not the case in our collection, because all pairs of isolates have either identical or related PFGE patterns, confirming their clonal relatedness. In addition, we would have expected that replacement of clones occurred independently of time. The pairs identified here were obtained from all pairs of isolates from the surveillance of MRSA over a 15-year period of time, and they were the most distant couples in time. They represent the most likely cases where marker changes can be observed during long-term carriage. The analysis of additional couples of isolates that are less distant in time would probably not significantly change the current outcome.
Stability of markers can also be assessed by the analysis of isolates from well-defined outbreaks. In a previous study, it was shown that, considering identical periods of time, LTC isolates showed a greater number of related PFGE patterns than outbreak isolates (3). We therefore propose, for local epidemiological investigations using these sequence-based markers, that a chain of transmission should be suspected only when indistinguishable isolates are found.
Genetic relatedness.
Whereas during local epidemiological investigations, the DLST markers are highly stable, they probably undergo changes during long-term epidemiological investigations (e.g., the international surveillance of clones). In these cases, related genotypes should also be considered. Such relatedness could be established using phylogenetic tools. However, for repeat regions of genes, these methods fail to work, as deletion or duplication of a repeat will not be considered a single genetic event. We overcame this problem by considering repeat regions that are more conserved (DLST-R). Grouping of isolates with this approach did indeed show some congruence with PFGE and MLST data. However, several exceptions were observed, which led us to reconsider the definition of clones and their biological or epidemiological relevance.
Markers of any typing system will over time accumulate mutations and thus are subject to change. The types of differences and in what period of time they occur depend on the nature of markers. Isolates that likely have evolved from each other could be grouped in a clone. The cutoff to define a clone has always arbitrarily been chosen (e.g., six [or less] band differences for PFGE, >80% similarity with other typing techniques). As observed in our study, such grouping of isolates depends on the markers used. This suggests that these groupings have no biological or epidemiological significance. In contrast, at a higher level, the evolutionary significance of clonal complexes defined by MLST data of S. aureus has been established. Further research is needed to describe the infrastructure of these clonal complexes.
Microevolutionary changes of markers.
Our study suggests that the occurrence of either point mutations or loss or gain of repeats (repeat mutations) depends on the studied marker. Analysis of both LTC isolates and isolates from groups showing indistinguishable PFGE patterns suggested that only point mutations occurred in clfA, only repeat mutations (duplication and deletion of repeats) occurred in clfB, and both point mutations and repeat mutations occurred in spa. To confound these trends, more work needs to be done with a greater number of isolates. If true, this could mean that depending on the gene, evolution could be the result of different patterns of mutations (point mutations and/or repeat mutations).
We found that genetic differences between highly related genotypes were the result of clonal changes and indicated clonal microevolution. No evidence for replacement of one or several repeats by foreign repeats could be suspected. This is in line with the results of previous population genetic analyses studying S. aureus evolution, which showed a predominant clonal structure of even highly diverse genes (7, 17, 19). It is noteworthy that in the present study, conclusions have been limited to isolates with single locus differences, while other isolates of the same PFGE group contained differences in at least two loci and were not considered.
A simple DLST scheme.
So far, S. aureus typing methods based on repeat-containing regions were performed by amplifying and sequencing the entire region, then determining each repeat type, and finally, identifying the genotype (15, 16). We showed that the majority of genotypic variation can be captured with an arbitrary approach by considering only 500 bp starting from the 3′ end of a marker. By defining a given marker length, we deliberately discarded information about repeat organization and considered alleles only at the level of nucleotides. As a result of this decision, no special search algorithms and assembly procedures were needed. In addition, this way we ensured that genetic differences contributed with equal importance to allele differences and thus to discriminatory power. Simple comparison with an international or local database comprising presently available allele sequences could serve for epidemiological interrogation.
Single-strand sequencing strategy.
Sequencing both strands is the currently accepted standard for reliable sequence analyses. However, a recent study reported that by single-strand sequencing of a set of housekeeping MLST genes, only 0.2% of 2,795 alleles were wrongly identified (20). This low error rate should be taken into account but will probably not affect conclusions of local epidemiological investigations. Therefore, we consider that technical errors associated with single-strand sequencing are outweighed by the increase in speed and the reduction in costs associated with analysis.
Sequencing technology has constantly been improved, and state-of-the-art techniques allow reliable sequence reads of up to 650 bp in only one strand. Sequence analysis software allocates quality values to peaks in electrophoretic patterns, which can be used for standardization and evaluation of reliability. As our method requires only 500 bp of clfB and spa, we therefore propose that single-strand sequencing is adequate for typing.
The main reason to choose partial sequences has been to reduce time, cost, and labor associated with typing. Indeed, by reducing the work amount involved in typing an isolate to one DNA extraction, two PCRs, and two sequencing reactions, including the analysis with a sequencer, the associated costs could be reduced to $12 on the basis of the presented protocols. As indicated in Materials and Methods, all procedures were performed in a 96-well format, which resulted in an average time investment of 2.5 days (18 h of work) for 96 samples. All procedures could easily be incorporated in a robotized work flow, and technical procedures could be further improved, thereby reducing the turnaround time.
Other typing methods have recently been developed, including multilocus variable-number tandem repeat analysis- and single-nucleotide polymorphism-based genotyping assays (9, 23). Both methods are characterized by low cost and time investments combined with great typeability and little ambiguity in data sets. Yet, no sequencing is required. To evaluate whether they could be used as alternatives to PFGE, comparison of their performances, especially the discriminatory power using collections of related isolates and the stability of the markers as suggested in this study, will be needed.
Conclusions.
By using a local epidemiological collection of isolates, we showed clear differences in discriminatory power between PFGE, clfA, clfB, fnbA, and spa. None of the markers alone could match the resolution of PFGE, but comparable values were obtained by combining the 500-bp 3′ partial sequences of spa and clfB repeat regions. We showed that these two markers were stable over a period of time similar to those of local epidemiological investigations. The combination of high typeability, reproducibility, and discriminatory power, together with low cost, ease of use, and unambiguous definition of types, renders these two markers candidates of choice for epidemiological analyses.
Acknowledgments
We thank H. de Lencastre, M. Aires de Sousa, K. W. Larssen, W. Wannet, M. Struelens, J. Etienne, and F. Vandenesch for providing us with strains used in the collection of genetically diverse MRSA strains. Furthermore, we thank Aline Wenger at the Institute of Microbiology in Lausanne for strains used in the collection of related MRSA strains.
This work was funded by the Swiss National Foundation for Research (grant 3200B0-101044) and Roche Research Foundation (grant 116-2006).
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, E. Spalburg, B. Strommenger, M. J. Struelens, F. C. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanc, D. S. 2004. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infection, Genetics and Evolution. 6th Int. Meet. Microb. Epidemiolog. Markers 4:193-197. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, D. S., M. J. Struelens, A. Deplano, R. De Ryck, P. M. Hauser, C. Petignat, and P. Francioli. 2001. Epidemiological validation of pulsed-field gel electrophoresis patterns for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 39:3442-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elphinstone, M. S., G. N. Hinten, M. J. Anderson, and C. J. Nock. 2003. An inexpensive and high-throughput procedure to extract and purify total genomic DNA for population studies. Mol. Ecol. Notes 3:317-320. [Google Scholar]
- 5.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. J. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 9.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. van Leeuwen, A. van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frenay, H. M. E., A. E. Bunschoten, L. M. Schouls, W. J. van Leeuwen, C. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 11.Gomes, A. R., S. Vinga, M. Zavolan, and H. de Lencastre. 2005. Analysis of the genetic variability of virulence-related loci in epidemic clones of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 49:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmsen, D., H. Claus, and U. Vogel. 2005. DNA sequence-based tandem repeat analysis of the clfB gene is less discriminatory than spa typing for methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 294:525-528. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, P. R. 1990. Reproducibility and indexes of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahl, B. C., A. Mellmann, S. Deiwick, G. Peters, and D. Harmsen. 2005. Variation of the polymorphic region X of the protein A gene during persistent airway infection of cystic fibrosis patients reflects two independent mechanisms of genetic change in Staphylococcus aureus. J. Clin. Microbiol. 43:502-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. A. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koreen, L., S. V. Ramaswamy, S. Naidich, I. V. Koreen, G. R. Graff, E. A. Graviss, and B. N. Kreiswirth. 2005. Comparative sequencing of the serine-aspartate repeat-encoding region of the clumping factor B gene (clfB) for resolution within clonal groups of Staphylococcus aureus. J. Clin. Microbiol. 43:3985-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn, G., P. Francioli, and D. S. Blanc. 2006. Evidence for clonal evolution among highly polymorphic genes in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 188:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 30:2058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt, S., B. Pichon, R. George, and J. Green. 2006. A bioinformatics pipeline for high-throughput microbial multilocus sequence typing (MLST) analyses. Clin. Microbiol. Infect. 12:1144-1146. [DOI] [PubMed] [Google Scholar]
- 21.fRobinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens, A. J., F. Huygens, J. Inman-Bamber, E. P. Price, G. R. Nimmo, J. Schooneveldt, W. Munckhof, and P. M. Giffard. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43-51. [DOI] [PubMed] [Google Scholar]
- 24.Struelens, M. J., Y. De Gheldre, and A. Deplano. 1998. Comparative and library epidemiological typing systems: outbreak investigations versus surveillance systems. Infect. Control Hosp. Epidemiol. 19:565-569. [DOI] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vauterin, L., and P. Vauterin. 2006. Integrated databasing and analysis, p. 141-217. In E. Stackebrandt (ed.), Molecular identification, systematics, and population structure of prokaryotes. Springer-Verlag, Berlin, Germany.