Abstract
Enteroaggregative Escherichia coli (EAEC) is an emerging enteric pathogen that causes acute and chronic diarrhea among children, human immunodeficiency virus-infected patients, and travelers to developing regions of the world. The pathogenesis of EAEC strains involves the production of biofilm. In this study, we determined the association between presence of putative EAEC virulence genes and biofilm formation in 57 EAEC isolates (as defined by HEp-2 adherence) from travelers with diarrhea and in 18 EAEC isolates from travelers without diarrhea. Twelve nondiarrheagenic E. coli isolates from healthy travelers were used as controls. Biofilm formation was measured by using a microtiter plate assay with the crystal violet staining method, and the presence of the putative EAEC virulence genes aap, aatA, aggR, astA, irp2, pet, set1A, and shf was determined by PCR. EAEC isolates were more likely to produce biofilm than nondiarrheagenic E. coli isolates (P = 0.027), and the production of biofilm was associated with the virulence genes aggR, set1A, aatA, and irp2, which were found in 16 (40%), 17 (43%), 10 (25%), and 27 (68%) of the biofilm producers versus only 4 (11%), 6 (6%), 2 (6%), and 15 (43%) in non-biofilm producers (P = 0.008 for aggR, P = 0.0004 for set1A, P = 0.029 for aatA, and P = 0.04 for irp2). Although the proportion of EAEC isolates producing biofilm in patients with diarrhea (51%) was similar to that in patients without diarrhea (61%), biofilm production was related to the carriage of aggR (P = 0.015), set1A (P = 0.001), and aatA (P = 0.025). Since aggR is a master regulator of EAEC, the presence of aap (P = 0.004), astA (P = 0.001), irp2 (P = 0.0006), pet (P = 0.002), and set1A (P = 0.014) in an aggR versus an aggR-lacking background was investigated and was also found to be associated with biofilm production. This study suggests that biofilm formation is a common phenomenon among EAEC isolates derived from travelers with or without diarrhea and that multiple genes associated with biofilm formation are regulated by aggR.
Bacterial diarrhea is one of the most common causes of morbidity and mortality among infants and children of developing nations (4, 21). Diarrheagenic Escherichia coli is the most commonly identified pathogen, and at least five distinct pathotypes have been characterized, including enterotoxigenic E. coli (ETEC), enteropathogenic E. coli, enterohemorrhagic E. coli, enteroinvasive E. coli, and enteroaggregative E. coli (EAEC). EAEC is an emerging food-borne pathotype that can cause acute and persistent diarrhea in children, human immunodeficiency virus-infected persons, and international travelers (1, 2, 15, 16, 24, 27). EAEC has been associated with diarrhea in both developing and industrialized countries (13, 24), including the United States (5, 24), and sometimes causes large outbreaks of gastrointestinal illness (14).
Aggregative adherence (AA) to the intestinal mucosa is the first step in EAEC pathogenesis (23). EAEC adheres to the small and large intestinal mucosal surface and stimulates mucus secretion, forming a thick aggregating biofilm (12, 21, 30, 32, 33). After adhesion, multiplication, and colonization on a surface (6), the bacteria surround themselves with exopolymeric substances and recruit more cells to form microcolonies interspersed with fluid-filled channels. Because of the restricted penetration of antimicrobials, decreased growth rate, and the expression of possible resistance genes, colonies in biofilm are not easily eradicated by bactericidal antibiotics (6) and may also be protected from attack by the intestinal immune system, resulting in prolonged infections.
The genetic determinants involved in biofilm production by EAEC have just begun to be understood (8, 18, 30, 34). Several virulence factors such as aggA, aggR, and aap (dispersin) are important in EAEC adherence to the intestinal mucosa and in stimulating biofilm formation (29, 30, 34). In addition, Sheikh et al. (30) have shown that fis and yafK, through the activation of aggR, are also important in biofilm formation. Several other genes, including aatA, pet (plasmid encoded toxins), ShET1 (Shigella enterotoxins), irp2 (yersiniabactin biosynthesis gene), and shf (cryptic open reading frame), are also thought to be involved in EAEC pathogenesis (7, 9, 11, 29, 31). The relationship of known or putative EAEC virulence genes and biofilm formation in isolates derived from subjects with travelers' diarrhea has not been studied.
MATERIALS AND METHODS
Study population and microbiology.
Stool samples were collected during the period from 1999 to 2004 from travelers from industrialized countries with diarrhea (n = 57) and without diarrhea (n = 30) during short-term stays in Mexico (57 isolates), India (10 isolates), and Guatemala (8 isolates) (10) and studied for the presence of enteric pathogens as previously described (19). A total of 75 EAEC isolates (57 from travelers with diarrhea and 18 from travelers without diarrhea) derived from fresh stool culture and stored in peptone stabs were used in the present study. Twelve E. coli isolates from subjects without diarrhea that did not demonstrate HEp-2 adherence and did not carry genes for ETEC heat-labile toxin (LT) and/or heat-stable toxin (ST) production were also used in the present study as nondiarrheagenic E. coli controls. The study was approved by the University of Texas Committee for the Protection of Human Subjects at the University of Texas Health Science Center at Houston, and written informed consent was obtained from each subject.
Detection of virulence factors by PCR.
E. coli isolates were picked from a single colony grown on Luria broth agar plates. EAEC virulence factors were identified by PCR using specific primers (Table 1) as described previously (7, 28), including the putative virulence genes irp2 of Yersinia species (28) and shET1 (set1A) of Shigella (7), which have been previously identified in some EAEC isolates.
TABLE 1.
Primers used in this study
| Genea | Putative function | Primer sequence | Amplicon size (bp) | Accession no. and/or reference | Starting PCR conditions |
|---|---|---|---|---|---|
| aggR* | Transcriptional activator | 5-CTAATTGTACAATCGATGTA-3′ | 308 | Z18751 (7) | 1 min at 94°C, 1 min at |
| for AAF/I and AAF/II | 5′-ATGAAGTAATTCTTGAAT-3′ | 42°C, 1 min at 72°C | |||
| aap* | Dispersin | 5′-ATGAAAAAAATTAAGTTTGTTATCTT-3′ | 351 | Z32523 | 1 min at 94°C, 1 min at |
| 5′-TTATTTAACCCATTCGGTTAGAGC-3′ | 52°C, 1 min at 72°C | ||||
| aatA* | Dispersin transporter | 5′-CTGGCGAAAGACTGTATCAT-3′ | 630 | X81423 | 1 min at 94°C, 1 min at |
| protein/CVD432 | 5′-CAATGTATAGAAATCCGCTGTT-3′ | 55°C, 1 min at 72°C | |||
| astA | EAST1 heat-stable toxin | 5′-ATGCCATCAACACAGTATAT-3′ | 110 | S81691 | 1 min at 94°C, 1 min at |
| 5′-GCGAGTGACGGCTTTGTAGT-3′ | 55°C, 1 min at 72°C | ||||
| pet | Plasmid encoded toxin | 5′-ACTGGCGGACTCATTGCTGT-3′ | 832 | AFO56581 | 1 min at 94°C, 1 min at |
| 5′-GCGTTTTTCCGTTCCCTATT-3′ | 55°C, 1 min at 72°C | ||||
| shf | Cryptic open reading | 5′-ACTTTCTCCCGAGACATTC-3′ | 613 | AF134403 (7) | 1 min at 94°C, 1 min at |
| frame | 5′-CTTTAGCGGGAGCATTCAT-3′ | 50°C, 1 min at 72°C | |||
| irp2† | Yersiniabactin | 5′-AAGGATTCGCTGTTACCGGAC-3′ | 264 | Schubert et al. (28) | 1 min at 94°C, 1 min at |
| biosynthesis | 5′-TCGTCGGGCAGCGTTTCTTCT-3′ | 55°C, 1 min at 72°C | |||
| set1A† | Shigella enterotoxin 1 | 5′-TCACGCTACCATCAAAGA-3′ | 309 | AF097644 | 1 min at 94°C, 1 min at |
| 5′-TATCCCCCTTTGGTGGTA-3′ | 55°C, 1 min at 72°C |
*, Plasmid-borne genes known to be regulated by AggR; †, chromosomal encoded gene.
Biofilm assay.
To test for biofilm formation, isolates grown overnight in Luria broth with 0.25% glucose at 37°C with agitation were diluted 1:100 in Dulbecco modified Eagle medium plus 0.45% glucose, and 200 μl of the bacterial suspension was inoculated into individual wells of sterile 96-well polystyrene microtiter plates. After incubation at 18 h at 37°C, a modified biofilm assay was carried out according to previously published methods (17), except that bacteria were fixed with 200 μl of Bouin fixative for 15 min at room temperature and rinsed once with phosphate-buffered saline. The fixed bacterial cells were then stained with 0.5% crystal violet for 15 min at room temperature and rinsed thoroughly with distilled water. After air drying, crystal violet was solubilized in 200 μl of ethanol-acetone (80:20 [vol/vol]) for 30 min, and the optical density at 570 nm (OD570) was measured by using a microplate reader (MultiskanSpectrum, Thermo Labsystems, Vantaa, Finland). Each assay was performed in quadruplicate and repeated on at least three different occasions. EAEC strain 042 served as a positive control (30) for the strong biofilm producer, and the nonpathogenic E. coli strains HS and E. coli DH5α served as negative controls for the non-biofilm producer.
Statistical analysis.
Statistical analyses were performed by using the Fisher exact test and chi-square test for categorical data, while the nonparametric Mann-Whitney test, as well as analysis of variance (GraphPad Prism 4) was used for continuous variables. The median OD570 and interquartile range values for clinical isolates were calculated by using GraphPad Prism 4 statistical software.
(This study was presented in part at the 106th General Meeting of the American Society for Microbiology, 21 to 25 May 2006, Orlando, FL.)
RESULTS
Biofilm formation by EAEC isolates.
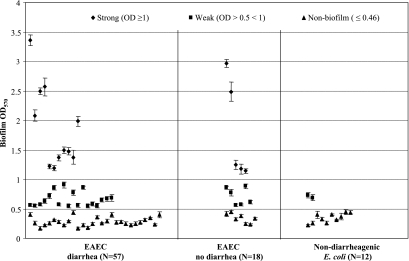
The biofilm mean OD570 values for the negative controls E. coli HS and E. coli DH5α were 0.377 ± 0.026 and 0.243 ± 0.029, respectively, whereas for the positive control EAEC 042 the mean OD570 value was 3.341 ± 0.074. Based on OD570 readings after incubation and crystal violet staining, E. coli isolates were deemed biofilm formers if the OD570 readings exceeded the mean plus two standard deviations of the E. coli HS and DH5α negative control strains (i.e., >0.46). We also arbitrarily classified samples as strong biofilm (OD570 ≥1), weak biofilm (OD570 > 0.46 < 1), and non-biofilm (OD570 ≤ 0.46) producers (Fig. 1) . For the entire group of isolates, the OD570 readings ranged from 0.16 to 3.36 U (Fig. 1). Forty (53%) of the isolates were classified as biofilm producers with a mean OD of 1.143 ± 0.744. Of the rest of the isolates, 35 (47%) were classified as non-biofilm producers with a mean OD of 0.292 ± 0.075. Based on the definition given above, 40 of the 75 (53%) EAEC isolates were biofilm producers; in contrast, only 2 of the 12 (17%) nondiarrheagenic E. coli isolates produced biofilm (P = 0.027; chi-squre test), and when they did it was of the weak phenotype (Fig. 1). Among the 75 EAEC isolates, 16 (21%) were strong biofilm (mean OD = 1.852 ± 0.722), 24 (32%) were weak (mean OD = 0.670 ± 0.127), and 35 (47%) were non-biofilm (mean OD = 0.292 ± 0.075) producers (Tables 2 and 3) . Among 75 EAEC isolates, 31 (54%) of the 57 isolates from Mexico, 5 (50%) of the 10 isolates from India, and 5 (62%) of the 8 isolates from Guatemala produced biofilm (the P value was not significant [NS]; Fisher's exact test).
FIG. 1.
Biofilm formation by EAEC isolates derived from travelers' with diarrhea or without diarrhea. Biofilm formation on polystyrene surface after 18 h was assessed by crystal violet staining. Each dot indicates the mean values with standard deviations. The biofilm assay was performed three times, with quadruplicates in each assay. Biofilm formation by nondiarrheagenic E. coli isolates was also tested. E coli isolates were deemed biofilm formers if the OD570 readings exceeded the mean plus two standard deviations of the E. coli strains HS and DH5α negative control for non-biofilm producers (i.e., >0.46).
TABLE 2.
Occurrence of biofilm formation (BF+) and presence of various genes in E. coli isolates from U.S. travelers to Mexico, India, and Guatemala in EAEC isolates (n = 75)
| Genea | Biofilm formation [no. of isolates (%)]b
|
Pc | |||
|---|---|---|---|---|---|
| Strong (n = 16) | Weak (n = 24) | Non-BF (n = 35) | All BF+ (n = 40) | ||
| aggR* | 8 (50) | 8 (33) | 4 (11) | 16 (40) | 0.008 |
| set1A† | 8 (50) | 9 (38) | 2 (6) | 17 (43) | 0.0004 |
| aatA* | 6 (38) | 4 (17) | 2 (6) | 10 (25) | 0.029 |
| irp2† | 9 (56) | 18 (75) | 15 (43) | 27 (68) | 0.038 |
| aap* | 6 (38) | 7 (29) | 7 (20) | 13 (33) | 0.298 |
| pet | 8 (50) | 8 (33) | 8 (23) | 16 (40) | 0.14 |
| shf | 3 (19) | 7 (29) | 14 (40) | 10 (25) | 0.217 |
| astA | 12 (75) | 14 (58) | 25 (71) | 26 (65) | 0.624 |
*, plasmid-borne genes known to be regulated by AggR; †, chromosomal encoded gene.
Strong, OD570 ≥ 1; weak, OD570 > 0.46 < 1; Non-BF, OD570 ≤ 0.46.
P values represent biofilm formers versus non-biofilm formers (Fisher's exact test).
TABLE 3.
Occurrence of biofilm formation (BF+) and presence of various genes in E. coli isolates from U.S. travelers to Mexico, India, and Guatemala in non-EAEC isolates (n = 12)
| Genea | Biofilm formation [no. of isolates (%)]b
|
Pc | |||
|---|---|---|---|---|---|
| Strong (n = 0) | Weak (n = 2) | Non-BF (n = 10) | All BF+ (n = 2) | ||
| aggR* | 0 | 0 | 0 | 0 | 1.000 |
| set1A† | 0 | 0 | 4 (40) | 0 | 0.515 |
| aatA* | 0 | 0 | 0 | 0 | 1.000 |
| irp2† | 0 | 1 (50) | 8 (80) | 1 (50) | 0.455 |
| aap* | 0 | 0 | 0 | 0 | 1.000 |
| pet | 0 | 0 | 3 (30) | 0 | 1.000 |
| shf | 0 | 1 (50) | 2 (30) | 1 (50) | 0.455 |
| astA | 0 | 1 (50) | 4 (40) | 1 (50) | 1.000 |
*, plasmid-borne genes known to be regulated by AggR; †, chromosomal encoded gene.
Strong, OD570 ≥ 1; weak, OD570 > 0.46 < 1; Non-BF, OD570 ≤ 0.46.
P values represent biofilm formers versus non-biofilm formers (Fisher's exact test).
Association between the presence of putative EAEC virulence genes and biofilm formation.
We observed an association between the presence of aggR and biofilm formation. Of the 75 EAEC isolates, 16 (40%) of 40 the biofilm producers contained aggR versus only 4 of 35 (11%) of the non-biofilm producers (P = 0.008; Fisher's exact test) (Tables 2 and 3). Among these isolates, aggR was detected in 8 of 16 (50%) of the strong biofilm producers and in 8 of 24 (33%) of the weak biofilm producers. We also identified an association with the presence of the set1A gene and biofilm production, which was present in 17 of 40 (43%) of biofilm producers versus 2 of 35 (6%) of non-biofilm producers (P = 0.0004; Fisher's exact test) (Tables 2 and 3). Of 40 biofilm-positive isolates, 10 (25%) carried the aatA gene compared to only 2 of 35 (6%) biofilm-negative isolates (P = 0.029; Fisher's exact test) (Tables 2 and 3).
Of 40 biofilm producers, 27 (68%) carried irp2 versus 15 (43%) of 35 non-biofilm producers (P = 0.038; Fisher's exact test) (Tables 2 and 3). No significant relationship between the presence of other putative virulence factors, including aap, astA, pet,and shf, and the biofilm phenotype were identified. Thirteen (13 of 40 or 32%) biofilm-producing EAEC isolates carried four genes (aggR, set1A, aatA, and/or aap) simultaneously in contrast to none (0 of 35) of the non-biofilm-producing EAEC isolates (P = 0.0001; Fisher's exact test). Of these 13 biofilm-producing isolates, 10, 2, and 1 isolate were derived from Mexico, India, and Guatemala, respectively (P = NS; Fisher's exact test). Of interest, 10 of the 12 nondiarrheagenic E. coli isolates had at least one or more of the genes studied (set1A, irp2, pet, astA, and shf) (Tables 2 and 3); however, they demonstrated weak or no biofilm formation.
The median biofilm OD value was significantly higher in biofilm producers carrying aggR (median OD of 0.72 versus 0.40), set1A (median OD of 0.86 versus 0.40), and aatA (median OD of 1.376 versus 0.44) than in isolates lacking these genes (P = 0.001 for aggR; P = 0.0003 for set1A; and P = 0.01 for aatA; Mann-Whitney test). In contrast, no significant association between biofilm and carriage of the aap, astA, pet, and shf genes were observed.
Association of biofilm and virulence genes with diarrhea.
As shown in the Tables 4 and 5, the proportion of EAEC isolates producing biofilm from patients with diarrhea (29 of 57 [51%]) was similar to that of subjects without travelers' diarrhea (11 of 18 [61%]; P = NS; Fisher's exact test), and biofilm production by EAEC isolates from travelers' diarrhea was related to the carriage of virulence genes, particularly aggR, set1A, and aatA (P = 0.015 for aggR, P = 0.001 for set1A, and P = 0.025 for aatA; Fisher's exact test).
TABLE 4.
Incidence of biofilm (BF+) formation and the presence of various genes among EAEC isolates from patients with diarrhea (n = 57)
| Genea | Biofilm formation [no. of isolates (%)]
|
Pb | |
|---|---|---|---|
| BF+ (n = 29) | BF− (n = 28) | ||
| aggR* | 12 (41) | 3 (11) | 0.015 |
| set1A† | 14 (48) | 2 (7) | 0.001 |
| aatA* | 8 (27) | 1 (4) | 0.025 |
| irp2† | 19 (65) | 11 (39) | 0.065 |
| aap† | 9 (31) | 5 (18) | 0.360 |
| pet | 12 (41) | 5 (18) | 0.082 |
| shf | 9 (31) | 11 (39) | 0.585 |
| astA | 23 (79) | 22 (79) | 1.000 |
*, plasmid-borne genes known to be regulated by AggR; †, chromosomal encoded gene.
P values represent biofilm formers versus non-biofilm formers (Fisher's exact test).
TABLE 5.
Incidence of biofilm (BF+) formation and the presence of various genes among EAEC isolates from patients without diarrhea (n = 18)
| Genea | Biofilm formation [no. of isolates (%)]
|
Pb | |
|---|---|---|---|
| BF+ (n = 11) | BF− (n = 7) | ||
| aggR* | 4 (36) | 1 (14) | 0.596 |
| set1A† | 3 (27) | 0 | 0.245 |
| aatA* | 2 (18) | 1 (14) | 1.000 |
| irp2† | 8 (73) | 4 (57) | 0.627 |
| aap† | 4 (36) | 2 (29) | 1.000 |
| pet | 4 (36) | 3 (43) | 1.000 |
| shf | 1 (9) | 3 (43) | 0.245 |
| astA | 3 (27) | 3 (43) | 0.627 |
*, plasmid-borne genes known to be regulated by AggR; †, chromosomal encoded gene.
P values represent biofilm formers versus non-biofilm formers (Fisher's exact test).
Role of AggR regulator and biofilm formation.
Since AggR is a master regulator in the pAA plasmid, the relatedness of aggR to other genes was studied. Twelve biofilm producers had both aggR and set1A genes (eight strong, six weak), and none of non-biofilm producers had both these genes together. However, 20 biofilm producers (5 strong, 15 weak) and 34 non-biofilm producers did not possess both genes (P < 0.0001; Fisher's exact test). The presence of aap, astA, irp2, pet, and set1A virulence genes in the aggR background for biofilm production showed that the presence of these genes was associated with biofilm as long as aggR was also present. The pooled median biofilm ODs of set1A+/aggR+ versus set1A+/aggR− (P = 0.014), aap+/aggR+ versus aap+/aggR− (P = 0.004), astA+/aggR+ versus astA+/aggR− (P = 0.0019), irp2+/aggR+ versus irp2+/aggR− (P = 0.0006), and pet+/aggR+ versus pet+/aggR− (P = 0.0028) were statistically significant (Mann-Whitney test) (Fig. 2).
FIG. 2.
Biofilm formation by EAEC isolates based on the presence of various genes in aggR background. Biofilm formation on a polystyrene surface after 18 h was assessed by crystal violet staining. Values are based on pooled median biofilm OD570 values shown by the box-and-whiskers plot which represents an upper quartile, median, and lower quartile.
DISCUSSION
The objective of this study was to determine the occurrence of various EAEC virulence factors and of biofilm formation among clinical isolates of EAEC collected from travelers from industrialized countries to Mexico, India, and Guatemala during short-terms stays. Of 75 EAEC isolates, 45 (53%) produced biofilm on an abiotic surface, which is lower than the rates reported by others (34) and may be due to our more stringent definition of biofilm formation or to differences in study design. In contrast to the findings of a previous study (34) wherein isolates were initially screened for biofilm and then characterized for aggregative phenotype, we initially screened for the aggregative phenotype and then characterized the isolates for biofilm formation. The frequency of biofilm formation by EAEC versus nondiarrheagenic E. coli isolates was significant, confirming that biofilm formation is more common in EAEC isolates than in nondiarrheagenic E. coli isolates (30, 34).
Isolates carrying aggR alone or in combination with other virulence genes (so-called typical EAEC) were strongly associated with biofilm formation. aggR is an EAEC master regulator formerly described as a transcriptional activator for AAF/I expression (19, 20) and later proved also to be required for AAF/II expression (9). Although other investigators have reported that no correlation exists between aggR and biofilm formation (26, 30), our findings are consistent with the work of Wakimoto et al. (34). In a related study, EAEC adhesins were found to be allelic in nature, and biofilm formation was shared by all members of the AAF family among Indian isolates (3). Taken together, these results suggest the sequence of factors AggR → AAF allele → biofilm production though other pathways can produce the same phenotype.
We also investigated the relationship of dispersin and its transporter in biofilm formation. Dispersin is a 10-kDa secreted protein coded by the aap gene that coats the bacterial surface and was selected for study since this protein promotes the dispersal of EAEC on the intestinal mucosa (29). Biofilm formation also correlated with the presence of the aatA gene that codes for a transporter protein that is a homolog of E. coli outer membrane protein, TolC (22). This was also of interest to us because the transporter facilitates the export of dispersin across the outer membrane in EAEC (29) and is the target for probe CVD 432, which is widely used to identify EAEC by molecular methods.
A novel finding in our study was the fact that the chromosomally located genes, set1A (Shigella enterotoxin 1) and irp2 (yersiniabactin biosynthesis gene) were also associated with EAEC biofilm production. set1A encodes an oligomeric enterotoxin, ShET1 (7, 11) and it contributes to secretory diarrhea caused by EAEC and Shigella infections. EAEC strains carrying set1A have been associated with diarrhea in Brazilian children and adult travelers from Spain (25). irp2 in turn encodes a protein involved in yersiniabactin expression and is designated iron-repressible high-molecular-weight protein 2 (Irp2) (7, 28). irp2 represents part of an unstable pathogenicity island that was acquired as a result of a horizontal transfer. Since set1A is located on the same island as a cluster of AggR-regulated genes, the relationship may be coincidental since it travels together with other putative virulence genes involved in biofilm formation.
In the present study, the proportion of EAEC isolates that were biofilm producers was similar in travelers with diarrhea and in travelers with asymptomatic colonization (2) and may explain in part the ability of EAEC to cause chronic colonization (1).
Since aggR is a master regulator that controls both plasmid and chromosomally encoded genes in EAEC, the biofilm formation ability of EAEC isolates carrying aap, astA, irp2, pet, and set1A with or without an aggR background was examined. EAEC isolates possessing the aap, astA, irp2, pet, and set1A genes in an aggR background produced more biofilm than those with an aggR-lacking background. This suggests that aggR regulates other genes needed for biofilm formation in EAEC. Recently, Nataro and Dudley have shown that AggR not only controls the plasmid-borne AAF fimbriae but also chromosomal genes located on an island located at 94 min of the EAEC chromosome (E. Dudley and J. Nataro, unpublished results). Further, it seems that additional genes are also under the control of aggR such as aatA (the dispersin transporter) and aap (dispersin). Sheikh et al. (30) have proved that the Fis protein contributes to biofilm production through AAF/II biogenesis, apparently by the activation of aggR expression.
Nine HEp-2 adherent isolates produced biofilm but did not carry either aggR, or aatA, or irp2, or set1A genes, indicating that there are additional factors involved in biofilm production in EAEC. It has recently been shown that atypical EAEC strain C1096 harboring Incl1 plasmid encoding a type IV pilus can contribute to biofilm formation (8). Additional work is needed to characterize HEp-2-adherent, biofilm-forming EAEC strains that are atypical (negative for aggR).
In summary, the present study suggests that biofilm formation is a common phenomenon among EAEC isolates derived from travelers with or without diarrhea. In vitro biofilm production from EAEC isolates is associated with aggR, and its regulated genes aatA, irp2, and set1A. Multiple EAEC genes located on the plasmid, as well as on its chromosome, are associated with biofilm formation.
Acknowledgments
This study was supported by the NIH NIAID RO1 A1 054948.
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Adachi, J. A., C. D. Ericsson, Z. D. Jiang, M. W. DuPont, S. R. Pallegar, and H. L. DuPont. 2002. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J. Infect. Dis. 185:1681-1683. [DOI] [PubMed] [Google Scholar]
- 2.Adachi, J. A., Z. D. Jiang, J. J. Mathewson, M. P. Verenkar, S. Thompson, F. Martinez-Sandoval, R. Steffen, C. D. Ericsson, and H. L. DuPont. 2001. Enteroaggregative Escherichia coli as a major etiologic agent in traveler's diarrhea in 3 regions of the world. Clin. Infect. Dis. 32:1706-1709. [DOI] [PubMed] [Google Scholar]
- 3.Bhardwaj, R., S. Majumdar, N. K. Ganguly, N. Taneja, S. Dutta, T. Ramamurthy, and A. Chakraborti. 2006. Characterization of adhesin variants in Indian isolates of enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 258:274-283. [DOI] [PubMed] [Google Scholar]
- 4.Clarke, S. C. 2001. Diarrhoeagenic Escherichia coli: an emerging problem? Diagn. Microbiol. Infect. Dis. 41:93-98. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, M. B., J. P. Nataro, D. I. Bernstein, J. Hawkins, N. Roberts, and M. A. Staat. 2005. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J. Pediatr. 146:54-61. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley, E. G., C. Abe, J. M. Ghigo, P. Latour-Lambert, J. C. Hormazabal, and J. P. Nataro. 2006. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect. Immun. 74:2102-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elias, W. P., Jr., J. R. Czeczulin, I. R. Henderson, L. R. Trabulsi, and J. P. Nataro. 1999. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 181:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg, D. E., Z. D. Jiang, R. Steffen, M. P. Verenker, and H. L. DuPont. 2002. Markers of inflammation in bacterial diarrhea among travelers, with a focus on enteroaggregative Escherichia coli pathogenicity. J. Infect. Dis. 185:944-949. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, I. R., S. Hicks, F. Navarro-Garcia, W. P. Elias, A. D. Philips, and J. P. Nataro. 1999. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect. Immun. 67:5338-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks, S., D. C. Candy, and A. D. Phillips. 1996. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect. Immun. 64:4751-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppertz, H. I., S. Rutkowski, S. Aleksic, and H. Karch. 1997. Acute and chronic diarrhoea and abdominal colic associated with enteroaggregative Escherichia coli in young children living in western Europe. Lancet 349:1660-1662. [DOI] [PubMed] [Google Scholar]
- 14.Itoh, Y., I. Nagano, M. Kunishima, and T. Ezaki. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 35:2546-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, Z. D., B. Lowe, M. P. Verenkar, D. Ashley, R. Steffen, N. Tornieporth, F. von Sonnenburg, P. Waiyaki, and H. L. DuPont. 2002. Prevalence of enteric pathogens among international travelers with diarrhea acquired in Kenya (Mombasa), India (Goa), or Jamaica (Montego Bay). J. Infect. Dis. 185:497-502. [DOI] [PubMed] [Google Scholar]
- 16.Mathewson, J. J., P. C. Johnson, H. L. DuPont, D. R. Morgan, S. A. Thornton, L. V. Wood, and C. D. Ericsson. 1985. A newly recognized cause of travelers' diarrhea: enteroadherent Escherichia coli. J. Infect. Dis. 151:471-475. [DOI] [PubMed] [Google Scholar]
- 17.Mohamed, J. A., W. Huang, S. R. Nallapareddy, F. Teng, and B. E. Murray. 2004. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. Infect. Immun. 72:3658-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira, C. G., S. M. Carneiro, J. P. Nataro, L. R. Trabulsi, and W. P. Elias. 2003. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 226:79-85. [DOI] [PubMed] [Google Scholar]
- 19.Nataro, J. P., Y. Deng, D. R. Maneval, A. L. German, W. C. Martin, and M. M. Levine. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nataro, J. P., J. B. Kaper, R. Robins-Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6:829-831. [DOI] [PubMed] [Google Scholar]
- 21.Nataro, J. P., T. Steiner, and R. L. Guerrant. 1998. Enteroaggregative Escherichia coli. Emerg. Infect. Dis. 4:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishi, J., J. Sheikh, K. Mizuguchi, B. Luisi, V. Burland, A. Boutin, D. J. Rose, F. R. Blattner, and J. P. Nataro. 2003. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 278:45680-45689. [DOI] [PubMed] [Google Scholar]
- 23.Okeke, I. N., A. Lamikanra, J. Czeczulin, F. Dubovsky, J. B. Kaper, and J. P. Nataro. 2000. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J. Infect. Dis. 181:252-260. [DOI] [PubMed] [Google Scholar]
- 24.Okeke, I. N., and J. P. Nataro. 2001. Enteroaggregative Escherichia coli. Lancet Infect. Dis. 1:304-313. [DOI] [PubMed] [Google Scholar]
- 25.Piva, I. C., A. L. Pereira, L. R. Ferraz, R. S. Silva, A. C. Vieira, J. E. Blanco, M. Blanco, J. Blanco, and L. G. Giugliano. 2003. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia, Brazil. J. Clin. Microbiol. 41:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarantuya, J., J. Nishi, N. Wakimoto, S. Erdene, J. P. Nataro, J. Sheikh, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, K. Miyata, and Y. Kawano. 2004. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J. Clin. Microbiol. 42:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scaletsky, I. C., S. H. Fabbricotti, S. O. Silva, M. B. Morais, and U. FagundesNeto. 2002. HEp-2-adherent Escherichia coli strains associated with acute infantile diarrhea, Sao Paulo, Brazil. Emerg. Infect. Dis. 8:855-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert, S., A. Rakin, H. Karch, E. Carniel, and J. Heesemann. 1998. Prevalence of the “high-pathogenicity island” of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infect. Immun. 66:480-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheikh, J., J. R. Czeczulin, S. Harrington, S. Hicks, I. R. Henderson, C. Le Bouguenec, P. Gounon, A. Phillips, and J. P. Nataro. 2002. A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Investig. 110:1329-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheikh, J., S. Hicks, M. Dall'Agnol, A. D. Phillips, and J. P. Nataro. 2001. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41:983-997. [DOI] [PubMed] [Google Scholar]
- 31.Suzart, S., B. E. Guth, M. Z. Pedroso, U. M. Okafor, and T. A. Gomes. 2001. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with or without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 201:163-168. [DOI] [PubMed] [Google Scholar]
- 32.Tzipori, S., J. Montanaro, R. M. Robins-Browne, P. Vial, R. Gibson, and M. M. Levine. 1992. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect. Immun. 60:5302-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vila, J., A. Gene, M. Vargas, J. Gascon, C. Latorre, and M. T. Jimenez de Anta. 1998. A case-control study of diarrhea in children caused by Escherichia coli producing heat-stable enterotoxin (EAST-1). J. Med. Microbiol. 47:889-891. [DOI] [PubMed] [Google Scholar]
- 34.Wakimoto, N., J. Nishi, J. Sheikh, J. P. Nataro, J. Sarantuya, M. Iwashita, K. Manago, K. Tokuda, M. Yoshinaga, and Y. Kawano. 2004. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am. J. Trop. Med. Hyg. 71:687-690. [PubMed] [Google Scholar]