Abstract
Pulsed-field gel electrophoresis (PFGE) of genomic macrorestriction fragments has been used by the Belgian Reference Laboratory for Staphylococci for national hospital surveys of methicillin-resistant Staphylococcus aureus since 1992. The sequencing of the polymorphic X region of the protein A gene (spa typing) offers significant advantages over PFGE in terms of speed, ease of interpretation, and exportability. To validate its potential use for national surveillance, we evaluated the robustness of spa typing compared with that of PFGE based on a collection of 217 S. aureus strains representative of the Belgian S. aureus epidemiology during the last 13 years. spa typing and PFGE both showed high discriminatory power (discriminatory indexes of 0.98 and 0.96, respectively) and achieved high concordance (95.9%) in type classification. Both methods also showed good concordance with multilocus sequence typing (MLST) (95.5%). However, we observed occasional “violations” of MLST clonal complex assignment by spa typing. Our results suggest that both PFGE and spa typing are reliable methods for long-term, nationwide epidemiological surveillance studies. We suggest that spa typing, which is a single-locus-based method, should preferably be used in combination with additional markers, such as staphylococcal cassette chromosome mec typing or resistance or virulence gene detection.
Staphylococcus aureus is a leading human pathogen, responsible for a wide range of diseases, from superficial skin infections to life-threatening conditions, such as endocarditis, pneumonia, and toxic shock syndrome (24). Strains of S. aureus resistant to methicillin (methicillin-resistant S. aureus [MRSA]) and to other β-lactams first emerged in 1961, shortly after the introduction of methicillin into clinical practice (21). Since then, MRSA has spread worldwide, causing outbreaks in the hospital setting (hospital-acquired [HA]-MRSA) as well as in the community (community-acquired [CA]-MRSA), becoming a major health issue (1).
During the last few decades, diverse typing methods, first phenotypic, then genotypic, have been used for monitoring and tracking MRSA spread. Among these, pulsed-field gel electrophoresis (PFGE) of genomic macrorestriction fragments has emerged as the gold standard method for MRSA epidemiology (36). PFGE proved to be a highly discriminatory and sensitive technique in microepidemiological (local or short term) and macroepidemiological (national, continental, or long term) surveys (2, 25, 28, 31, 37, 40). Nevertheless, some authors have argued that the stabilities of PFGE markers may be insufficient for the reliable application of PFGE to long-term or macroepidemiological studies (3). In addition, PFGE remains suboptimal regarding other aspects: it is a technically demanding and labor-intensive method, its interpretation leaves room for subjectivity (40), and interlaboratory result comparison remains difficult and dependent on strict adherence to standardized protocols and interpretation criteria (4, 11, 26).
In recent years, sequence-based methods, which provide fast, unambiguous, and exportable typing data, have been developed. Multilocus sequence typing (MLST) is a method based on the sequence polymorphism of ∼500-bp-long fragments of seven housekeeping genes (13). Initially designed for the study of bacterial population genetic structures, this technique has proved to be adequate for the long-term global epidemiology and study of the recent evolution of S. aureus (13, 14). A second technique, called staphylococcal cassette chromosome mec (SCCmec) typing, is based on the molecular characterization by multiplex PCR of the mobile genetic element carrying the methicillin resistance gene (mecA) (29). The combination of the MLST type and the SCCmec type, defined as the “clonal type,” is now used for the international nomenclature of MRSA clones (14). However, MLST typing remains too expensive and labor-intensive for its application to outbreak investigations and routine surveillance at a national level (14, 30).
The sequencing of the polymorphic X region of the protein A gene (spa), which contains a variable number of 24-bp-repeat regions flanked by well-conserved regions, is called spa typing (15). This single-locus-sequence-based typing method combines a number of technical advantages, such as rapidity, reproducibility, and portability (35, 36). Moreover, due to its repeat structure, the X region simultaneously indexes micro- and macrovariations, enabling the use of spa typing in both local and global epidemiological studies (22).
PFGE has been used by the Belgian Reference Laboratory for Staphylococci for both local outbreak investigations and national hospital surveys of MRSA since 1992 (6-9, 37, 39). spa typing offers significant advantages over PFGE in terms of speed, workflow capacity, ease of interpretation, and exportability. We performed this study to (i) validate the use of PFGE for long-term, nationwide epidemiology by the Reference Laboratory for Staphylococci, (ii) evaluate the robustness of spa typing and its concordance with PFGE and MLST, and (iii) establish, if possible, a “permutation nomenclature” from PFGE to spa typing to validate its potential use for the national surveillance of MRSA.
MATERIALS AND METHODS
Strain collection.
A group of 217 strains (93 MRSA and 124 methicillin-susceptible S. aureus [MSSA] strains) were selected from the Belgian Reference Laboratory for Staphylococci collection based on their PFGE profiles. The goal was to select strains with various degrees of genetic relatedness, collected all over the country during a 13-year period as part of national surveillance studies and outbreak investigations. Strains representative of all major epidemic Belgian HA PFGE types (6-9) as well as minor or sporadic PFGE types were selected from national surveillances in 1992, 1995, 1997, 2001, and 2003 (6-8, 12) and outbreaks (10). CA-MRSA strains (described in reference 5) as well as MSSA strains collected during a Belgian national survey in 2003 (M. Hallin, O. Denis, A. Deplano, R. De Mendonça, R. De Ryck, S. Rottiers, and M. J. Struelens, submitted for publication) were included, as they presented a large genotypic diversity.
DNA extraction.
DNA was extracted as described by Ünal et al. (41). Briefly, isolates cultured for 24 h on Columbia agar with 5% sheep blood were successively incubated with lysostaphin and proteinase K, boiled, and finally centrifuged. This lysate was used as a DNA template in all PCRs described below.
Identification and characterization of oxacillin resistance.
The presence of mecA and nuc genes was tested by PCR as previously described (17).
Molecular typing methods. (i) PFGE.
SmaI restriction fragments of genomic DNA were separated by PFGE with a CHEF mapper system (Bio-Rad Laboratories, Nazareth, Belgium) as previously described (12). Similarities among macrorestriction patterns were determined both by visual comparison and by computer matching with BioNumerics 4.0 software (Applied Maths, Ghent, Belgium). Bands in the size range between 36 and 700 kb were analyzed by the Dice similarity coefficient, with the position tolerance set to 0.8%, and dendrograms for similarity were built using the unweighted-pair group method using arithmetic averages. Patterns differing by less than seven fragments are considered to belong to the same PFGE group (represented by a capital letter), and those differing by zero to three fragments are considered to belong to the same PFGE type (represented by a number) (6).
(ii) spa typing.
spa typing was performed as described by Harmsen et al. (18), and spa types were determined with Ridom StaphType software version 1.4 (Ridom GmbH, Würzburg, Germany) and analyzed by the BURP algorithm, with the following default parameters: spa types shorter than five repeats were considered nongroupable, and spa types belonged to the same group (clonal complex [CC]) if the cost was less than or equal to six (according to the excision, duplication, substitution, and insertion/deletion model) (34).
(iii) MLST.
MLST was performed as described by Enright et al. (13) on all MRSA strains as well as on a subsample of 32 MSSA strains representative of every PFGE group. Sequence types (STs) were determined with the MLST database, accessible via http://www.mlst.net.
SCCmec typing.
SCCmec typing was performed by multiplex PCR as described by Oliveira and de Lencastre (29).
Typing system evaluation. (i) Typeability.
Typeability, defined as the proportion of strains that are assigned a type by a typing system, was calculated as described by Struelens (38).
(ii) DI.
Discriminatory index (DI), defined as the average probability that the typing system will assign a different type to two unrelated strains, was calculated as described by Hunter and Gaston (19) and the confidence intervals as described by Grundmann et al. (16).
(iii) Concordance.
Concordance between methods was calculated as described by Robinson et al. (33). Briefly, all possible pairs of strains are examined, and their types are classified as “identical” or “different” by the two methods and then cross-classified in a two-by-two table. Concordance, expressed in percentages, corresponds to the proportion of pairs for which the two methods are in agreement.
RESULTS
PFGE and spa typing: discriminatory power and concordance.
The PFGE patterns of the 217 strains belonged to 52 groups and 85 types. Sixty-one types belonging to 33 groups were represented by a single strain.
Ninety-five spa types were found (62 were represented by a single strain). Eighty-nine of them were distributed by the BURP algorithm in 13 groups (CCs) and 10 singletons (groups represented by a single type), while 6 spa types shorter than five repeats were excluded from the clustering (Table 1).
TABLE 1.
Discriminatory power and concordance of PFGE and spa typing based on a collection of 217 S. aureus strains
| Method | Classification level | Typeability (%) | Value for classification level
|
DI (95% CI) | Concordance (%) with:
|
||
|---|---|---|---|---|---|---|---|
| No. | Most frequent (%) | PFGE type | PFGE group | ||||
| PFGE | Type | 100 | 83 | 28 | 0.961 (0.949-0.973) | ||
| Group | 100 | 52 | 28 | 0.935 (0.922-0.948) | |||
| spa typing | Type | 100 | 95 | 18 | 0.976 (0.969-983) | 95.9 | 94.7 |
| CC | 97a | 23 | 43 | 0.892 (0.874-0.911) | 92.1 | 93.4 | |
Six spa types, shorter than five repeats, were excluded.
The two methods showed both a high discriminatory power (DIs above 0.95 for both methods) and excellent concordance (95.9%) (Table 1).
Discrepancies between the two methods were mainly due to strains classified as singletons by one method but presenting the same type as epidemic strains by the other method. For example, a large majority of strains that we considered sporadic based on their PFGE patterns (having a difference of more than six fragments compared to any other strain in the database) belonged to spa CC002 or spa CC008, harboring the same spa type as epidemic strains (Fig. 1). Moreover, strains belonging to several distinct epidemic PFGE types were also undistinguishable by spa type: PFGE type A20 and A21 strains were mainly t008 (6 of 8 A20 strains and 3 of 5 A21 strains), while PFGE type G10 strains (all 3) and certain C3 strains (11 of 22) shared spa type t002.
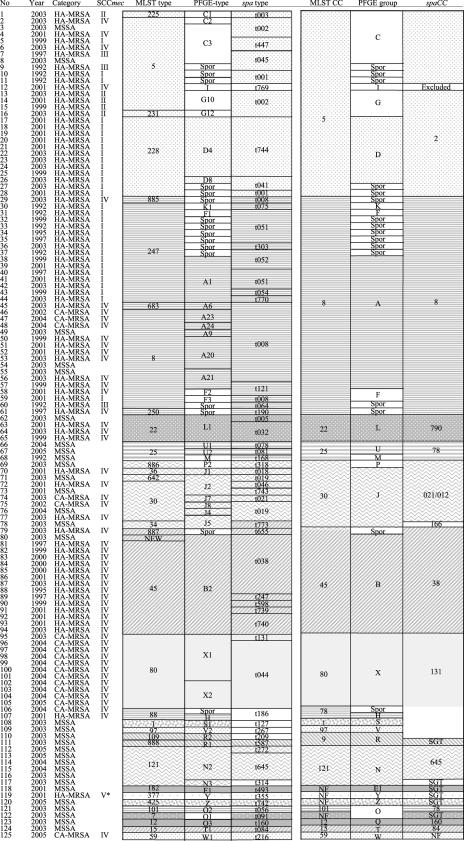
FIG. 1.
Years of isolation and typing results for 125 S. aureus strains obtained by SCCmec typing, MLST, spa typing, and PFGE. Clustering results are indicated as follows: congruent groups or CCs obtained by MLST, spa typing, and PFGE are represented by shaded blocks, and discordant results are represented in separated, colorless boxes. SGT, singleton; Spor, sporadic; NF, no founder assigned; NEW, MLST profile containing one new allele, with no ST assigned yet; *, not typeable by the Oliveira and de Lencastre method (29).
Concordance with MLST.
To asses the performance of PFGE and spa typing compared to that of MLST, we worked on a 125-strain subset (93 MRSA and 32 MSSA strains) representative of all the PFGE groups present in the 217-strain collection. MLST showed a DI of 0.93, identifying 33 different STs, of which 23 were represented by a single strain. The 33 STs belonged to 20 CCs.
The performances of PFGE and spa typing in terms of concordance with MLST were equally good (95.5 and 95.4%, respectively).
PFGE and MLST.
MLST CC22, CC9, CC80, and CC121 corresponded to one PFGE group each (Fig. 1). CC30, CC45, CC78, and CC25 each contained two distinct PFGE groups. Conversely, two strains belonging to distinct CCs (ST101 and ST7) fell into the same PFGE group.
A more complex picture was observed with strains of CC5 and CC8 lineages: PFGE subdivided them into 9 and 12 distinct groups, respectively (Fig. 1). Most of those groups corresponded to sporadic PFGE patterns but also included PFGE groups C, G, and D (containing epidemic HA-MRSA PFGE types C3, G10, and D4), all of which belonged to CC5 and some of which (PFGE types C3 and G10) belonged to the same ST (ST5). In the same way, strains of PFGE types A20 and A21 (considered two distinct epidemic HA-MRSA clones) both belonged to ST 8.
spa typing and MLST.
As for PFGE, each MLST CC corresponded to one spa CC, as was the case for CC5, CC8, CC45, CC22, and CC25 (Fig. 1). Exceptions included CC30 and CC121, both of which corresponded to one spa CC and one spa singleton.
All strains belonging to CC80 shared the same spa CC (CC131). However, spa CC131 also included strains belonging to four other MLST CCs (two ST88, one ST97, one ST1, and one ST109 strain) (Table 2) .
TABLE 2.
Typing characteristics of 16 strains belonging to spa CC131
| No. of strains | Category | spa profile | spa type | MLST profile | ST | CC | SCCmec type | PFGE type |
|---|---|---|---|---|---|---|---|---|
| 1 | CA-MRSA | 072312----------343334 | t131 | 1-3-1-14-11-51-10 | ST80 | CC80 | IV | X1 |
| 10 | CA-MRSA | 072312--------34343334 | t044 | 1-3-1-14-11-51-10 | ST80 | CC80 | IV | X1/X2 |
| 2 | CA-MRSA, HA-MRSA | 07--122117131334343334 | t186 | 22-1-14-23-12-4-31 | ST88 | CC78 | IV | Sporadic/H |
| 1 | MSSA | 0723--2116------343313 | t127 | 1-1-1-1-1-1-1 | ST1 | CC1 | S1 | |
| 1 | MSSA | 0723122117----34343334 | t267 | 3-1-1-1-1-5-3 | ST97 | CC97 | V2 | |
| 1 | MSSA | 071612--23----------34 | t209 | 3-27-1-1-1-1-10 | ST109 | CC9 | R2 |
As for PFGE types, several spa types corresponded to the same ST. Conversely, in CC5 and CC8 as well as in CC30, spa typing was not able to distinguish STs. Strains belonging to distinct STs which harbored the exact same spa type were as follows: t008 was associated with ST683, ST8, and a new ST (a ST885; t002 with ST5 and ST231; t001 with ST5 and ST228; t038 with ST45 and a new ST (a single-locus variant of ST45); and t019 with ST30 and ST642. However, such a violation of MLST ST “assignment” also happened occasionally for PFGE typing: PFGE J5 was associated with ST642 and ST30, J2 with ST30 and ST34, and B2 with ST45 and a new ST (a single-locus variant of ST45).
In summary, despite concordance percentages similar to these seen with MLST, PFGE classification in groups and types rarely violated the MLST assignment of CCs and STs, while this type of violation was much more frequent for the spa classification into types and CCs. To quantify this problem, we examined the concordance tables for each method with MLST and compared the percentages of discordant pairs of strains that belonged to distinct STs or CCs by MLST but were clumped into the same type, group, or CC by either PFGE or spa typing (Table 3). Up to 12% of the discrepancies between spa type and MLST ST and 61% of those between spa CC and MLST CC fell into this category, compared with only 5% of the discrepancies between PFGE type and MLST ST and 0.2% of those between PFGE group and MLST CC.
TABLE 3.
Concordance of spa typing and PFGE with MLST and clonal type based on a 125-strain subset
| Classification level | No. of strains | Concordance (%) with:
|
% Violation of assignment tob:
|
|||
|---|---|---|---|---|---|---|
| MLST ST | MLST CC | Clonal type | MLST ST | MLST CC | ||
| spa type | 125 | 95.4 | 12 | 0 | ||
| spa CC | 124a | 98.6 | 61 | |||
| spa type plus SCCmec type | 93 | 94.8 | ||||
| PFGE type | 125 | 95.5 | 5 | 0 | ||
| PFGE group | 125 | 91.5 | 0 | |||
| PFGE type plus SCCmec type | 93 | 96.1 | ||||
One spa type, shorter than five repeats, was excluded.
Calculated as the proportion of discordant pairs of strains in the two-by-two concordance table that were considered different by MLST but similar by the other typing method.
SCCmec typing and clonal type assignment.
Given the clustering of several PFGE types and groups into common MLST STs and the discrepancies observed between spa types and PFGE types as described above, the SCCmec types of MRSA strains were determined to examine whether the combination of SCCmec with the different typing methods could resolve conflicting assignments.
The most frequent SCCmec type among the 93 MRSA strains was type IV (n = 53), followed by type I (n = 30). Following the method described by Oliveira and de Lencastre (29), six strains were of SCCmec type II, three strains were of SCCmec type III, and one was nontypeable (SCCmec type V) (20) (Fig. 1). The combination of SCCmec typing with the three methods allowed an increase of concordance from 95.5% between PFGE type and MLST ST to 96.1% between PFGE type plus SCCmec type and clonal type (Table 3) and from 95.3% between PFGE and spa typing to 95.6% between PFGE plus SCCmec typing and spa typing plus SCCmec typing. This minor improvement in intermethod agreement for PFGE was due to the fact that PFGE G10/ST5/spa CC002 strains harbored SCCmec type II, while PFGE C3/ST5/spa CC002 strains harbored SCCmec I, III, or IV. However, SCCmec typing did not help to resolve the conflicts within CC8 and CC30 lineages, since all ST30/spa CC021/CC012 strains harbored SCCmec type IV, all ST247/spa CC008 strains (epidemic-PFGE A1 and PFGE-sporadic types) harbored SCCmec type I, and all except one ST8/spa CC008 strain (PFGE-epidemic A20 and A21 as well as PFGE-sporadic types) harbored SCCmec type IV (Fig. 1).
DISCUSSION
PFGE has been used by the Belgian National Reference Laboratory for Staphylococci since 1992 for both local and national investigations (6-9, 37). It is considered the “gold standard” for typing S. aureus strains (36). However, spa typing possesses significant advantages over PFGE in terms of speed, ease of interpretation, and exportability (35, 36). The goal of this study was to evaluate the robustness of spa typing and its concordance with PFGE and MLST based on a strain collection representative of the Belgian S. aureus population.
Ideally, the test population for evaluating a typing systems should (i) be of large size (n > 100), (ii) reflect as much as possible the diversity expected in the microbial population to which the typing system will be applied, (iii) be constituted by strains that are epidemiologically unrelated, and (iv) not be selected on the basis of type characteristics (38). The strain collection described here fulfils most of these criteria, as it was large and diverse but was, however, selected on the basis of PFGE results to specifically cover the range of patterns identified over a decade in a particular region. Regarding the epidemiological relatedness of the strains studied, we selected strains coming from different locations if from the same year and from distinct years if from the same location.
PFGE is known to be a highly discriminatory and valuable technique for outbreak investigation (31, 37, 40). However, it has been argued that the stability of PFGE may be insufficient for its reliable application to long-term epidemiological studies: high degrees of genetic variation leading to multiple PFGE profiles have been observed for pandemic clones with “long” evolutionary histories. This was attributed to the heterogeneous selective pressure exerted in different hospital ecosystems (1). This has also been attributed to the fact that large chromosome fragments undergo faster variations than smaller ones (3), leading to a progressive drift of PFGE patterns over time. In this study, we showed the excellent concordance between PFGE type and MLST ST and between PFGE type plus SCCmec type and clonal type based on a nationwide strain collection covering a 13-year period. Our results not only validate the use of PFGE for long-term, nationwide epidemiological studies but also provide a good basis for a nomenclature permutation table between MLST ST (with or without SCCmec type) and PFGE type (with or without SCCmec type). However, PFGE is a labor-intensive method and requires technical skills, and despite the efforts made to standardize protocols and interpretation criteria, interlaboratory result comparison remains difficult (11, 26).
spa typing is a single-polymorphic-locus-sequence-based typing method. This technique possesses a significant number of practical advantages over PFGE. It is a rapid, reproducible, and easy-to-perform technique, providing unambiguous and exportable data (35, 36). In addition, an extensive study has demonstrated that it is applicable to both local and global epidemiological studies (22). Using a large national collection, spa typing was confirmed in the present study to be as discriminatory and concordant with MLST as PFGE. However, we observed occasional “violations” of MLST ST assignment by spa typing with strains belonging to the same CC but to different STs which harbored the same spa type. Furthermore, strains belonging to distant MLST CCs (CC80, CC78, CC1, CC9, and CC97) had very similar spa profiles which clustered in a unique spa CC (spa CC131). We suspect that this phenomenon could be due to intergenomic recombination involving the spa locus. Although this type of event is presumed to be rare (22), it should have occurred, in this case, at least four times from one lineage to other lineages. Such a phenomenon was recently described by Robinson and Enright for another lineage (MLST ST239). The authors described a large chromosomal replacement, encompassing the spa locus, from ST30 to ST8 which generated ST239 (32). They speculated that this type of large replacement may be lineage specific. However, our findings suggest that this type of event may be more extensive than previously appreciated. Therefore, the extent and range of genetic lineages of S. aureus involved in large horizontal transfer in this chromosome region warrant further investigation.
spa typing was also highly concordant with PFGE. The majority of discrepancies observed between the two methods were encountered in MLST CC8 and CC5 and were of two different kinds. First, the large majority of MRSA strains which were classified as sporadic based on their PFGE profiles harbored the same spa type as epidemic MRSA belonging to spa CC008 (MLST CC8) and spa CC002 (MLST CC5). This could be due to the fact that PFGE may be in this case too discriminating as a result of the patterns of these strains drifting over time, as discussed above. This could also reflect genetic events such as the insertion/deletion of different mobile genetic elements that may be, to some extent, involved in the epidemic behavior of S. aureus and lead to different PFGE patterns while spa and MLST types remain indistinguishable.
The clustering of several epidemic HA-MRSA PFGE types in common spa CCs and spa types (as well as in common MLST ST) also deserves comment. This was the case for the clustering of G10 and C3 PFGE types into spa CC002/MLST ST5 and for the clustering of A20 and A21 PFGE types into spa CC008/MLST ST8. Again, one can speculate that all these discrepancies are due to the fact that PFGE may be too discriminating. However, these epidemic clones are known to possess, besides their specific PFGE profiles, several other distinctive features that are of potential epidemiological relevance. As described previously (7, 9), PFGE G10 and C3 strains have completely different susceptibility profiles for macrolides and tetracyclines: G10 strains usually harbor the tetM and ermC genes (conferring a constitutive resistance to tetracycline/minocycline and an inducible resistance to macrolides/lincosamides/streptogramines [MLS], respectively), while C3 isolates are susceptible to those drugs. G10 strains also harbor SCCmec type II, while C3 strains harbor SCCmec types I, III, and IV. Furthermore, G10 strains frequently carry the TSST-1 gene (7). In the same way, A20 strains posses the ermA gene (conferring a constitutive resistance to MLS), while A21 strains either are susceptible to MLS or harbor the ermC gene. All these observations suggest, as mentioned by others (8, 25), a parallel evolution of strains belonging to the same successful lineage but possessing different resistance and/or virulence genes acquired by insertion of large mobile elements, like staphylococcal pathogenicity island 1 for TSST-1 and transposon Tn544 for ermA (23, 27). The same mechanisms could explain the differences observed in their PFGE patterns.
In conclusion, the excellent concordance we found between PFGE and MLST in our collection validates the use of PFGE by the Belgian Reference Laboratory for Staphylococci for long-term, nationwide epidemiological surveillance studies. We also have shown that spa typing can be used for the same purpose but with some caution. Since spa typing is a single-locus-based method, we suggest that additional markers, such as SCCmec type and resistance or virulence genes, should be determined, especially in the rapidly evolving CC8 and CC5 lineages. Furthermore, the occasional “violation” of MLST CC assignment by spa typing, which is presumably due to recombination events involving the spa locus, underlines the need for the confirmation of the MLST profiles of a subset of representative strains.
Acknowledgments
This work was partially supported by a grant from the Fonds Erasme, Université Libre de Bruxelles, Brussels, Belgium, to Marie Hallin.
We gratefully thank Sylvianne Rottiers, Sébastien Crèvecoeur, and Aram Afsar for their skilled technical assistance and continuous support.
Footnotes
Published ahead of print on 8 November 2006.
REFERENCES
- 1.Aires de Sousa, M., and H. de Lencastre. 2004. Bridges from hospitals to the laboratory: genetic portraits of methicillin-resistant Staphylococcus aureus clones. FEMS Immunol. Med. Microbiol. 40:101-111. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., I. S. Sanches, M. L. Ferro, M. J. Vaz, Z. Saraiva, T. Tendeiro, J. Serra, and H. de Lencastre. 1998. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J. Clin. Microbiol. 36:2590-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc, D. S., P. Francioli, and P. M. Hauser. 2002. Poor value of pulsed-field gel electrophoresis to investigate long-term scale epidemiology of methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2:145-148. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 5.Denis, O., A. Deplano, H. De Beenhouwer, M. Hallin, G. Huysmans, M. G. Garrino, Y. Glupczynski, X. Malaviolle, A. Vergison, and M. J. Struelens. 2005. Polyclonal emergence and importation of community-acquired methicillin-resistant Staphylococcus aureus strains harbouring Panton-Valentine leucocidin genes in Belgium. J. Antimicrob. Chemother. 56:1103-1106. [DOI] [PubMed] [Google Scholar]
- 6.Denis, O., A. Deplano, R. De Ryck, C. Nonhoff, and M. J. Struelens. 2003. Emergence and spread of gentamicin-susceptible strains of methicillin-resistant Staphylococcus aureus in Belgian hospitals. Microb. Drug Resist. 9:61-71. [DOI] [PubMed] [Google Scholar]
- 7.Denis, O., A. Deplano, C. Nonhoff, R. De Ryck, R. de Mendonca, S. Rottiers, R. Vanhoof, and M. J. Struelens. 2004. National surveillance of methicillin-resistant Staphylococcus aureus in Belgian hospitals indicates rapid diversification of epidemic clones. Antimicrob. Agents Chemother. 48:3625-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis, O., A. Deplano, C. Nonhoff, M. Hallin, R. De Ryck, R. Vanhoof, R. De mendonça, and M. Struelens. 2006. In vitro activities of ceftobiprole, tigecycline, daptomycin, and 19 other antimicrobials against methicillin-resistant Staphylococcus aureus strains from a national survey of Belgian hospitals. Antimicrob. Agents Chemother. 50:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, O., J. Magdalena, A. Deplano, C. Nonhoff, E. Hendrickx, and M. J. Struelens. 2002. Molecular epidemiology of resistance to macrolides-lincosamides-streptogramins in methicillin-resistant Staphylococcus aureus (MRSA) causing bloodstream infections in patients admitted to Belgian hospitals. J. Antimicrob. Chemother. 50:755-757. [DOI] [PubMed] [Google Scholar]
- 10.Deplano, A., R. de Mendonca, R. De Ryck, and M. J. Struelens. 2006. External quality assessment of molecular typing of Staphylococcus aureus isolates by a network of laboratories. J. Clin. Microbiol. 44:3236-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frenay, H. M., A. E. Bunschoten, L. M. Schouls, W. J. van Leeuwen, C. M. Vandenbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 16.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallin, M., N. Maes, B. Byl, F. Jacobs, Y. De Gheldre, and M. J. Struelens. 2003. Clinical impact of a PCR assay for identification of Staphylococcus aureus and determination of methicillin resistance directly from blood cultures. J. Clin. Microbiol. 41:3942-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jevons, M. P. 1961. ‘Celbenin’-resistant Staphylococci. Br. Med. J. 1:124-125. [Google Scholar]
- 22.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsay, J. A., and M. T. Holden. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378-385. [DOI] [PubMed] [Google Scholar]
- 24.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 25.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murchan, S., M. E. Kaufmann, A. Deplano, R. De Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick, R. P., P. Schlievert, and A. Ruzin. 2001. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 3:585-594. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira, D., I. Santos-Sanches, R. Mato, M. Tamayo, G. Ribeiro, D. Costa, and H. de Lencastre. 1998. Virtually all methicillin-resistant clones of Staphylococcus aureus (MRSA) infections in the largest Portuguese teaching hospital are caused by two internationally spread multiresistant strains: the ‘Iberian’ and the ‘Brazilian’ clones of MRSA. Clin. Microbiol. Infect. 4:373-384. [DOI] [PubMed] [Google Scholar]
- 29.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peacock, S. J., G. D. de Silva, A. Justice, A. Cowland, C. E. Moore, C. G. Winearls, and N. P. Day. 2002. Comparison of multilocus sequence typing and pulsed-field gel electrophoresis as tools for typing Staphylococcus aureus isolates in a microepidemiological setting. J. Clin. Microbiol. 40:3764-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts, R. B., H. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, et al. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, D. A., S. K. Hollingshead, J. M. Musser, A. J. Parkinson, D. E. Briles, and M. J. Crain. 1998. The IS1167 insertion sequence is a phylogenetically informative marker among isolates of serotype 6B Streptococcus pneumoniae. J. Mol. Evol. 47:222-229. [DOI] [PubMed] [Google Scholar]
- 34.Sammeth, M., T. Weniger, D. Harmsen, and J. Stoye. 2005. Alignment of tandem repeats with excision, duplication, substitution and indels (EDSI), p. 276-290. In R. Casadio and G. Myers (ed.), Lecture notes in computer science. Springer, Berlin, Germany.
- 35.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strommenger, B., C. Kettlitz, T. Weniger, D. Harmsen, A. W. Friedrich, and W. Witte. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struelens, M. J., R. Bax, A. Deplano, W. G. Quint, and A. van Belkum. 1993. Concordant clonal delineation of methicillin-resistant Staphylococcus aureus by macrorestriction analysis and polymerase chain reaction genome fingerprinting. J. Clin. Microbiol. 31:1964-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 39.Struelens, M. J., O. Ronveaux, B. Jans, R. Mertens, et al. 1996. Methicillin-resistant Staphylococcus aureus epidemiology and control in Belgian hospitals, 1991 to 1995. Infect. Control Hosp. Epidemiol. 17:503-508. [DOI] [PubMed] [Google Scholar]
- 40.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hebert, B. Hill, and R. Hollis. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ünal, S., J. Hoskins, J. E. Flokowitsch, C. Y. Wu, D. A. Preston, and P. L. Skatrud. 1992. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J. Clin. Microbiol. 30:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]