Abstract
The recently sequenced genome of Campylobacter jejuni RM1221 revealed the presence of three integrated bacteriophage-like elements. In this study, genes from the first element, a Mu-like bacteriophage, were amplified by PCR and used to probe pulsed-field gels of clinical C. jejuni strains obtained from a waterborne outbreak (Ontario, Canada, 2000). These highly similar strains differed only by their pulsed-field gel electrophoresis (PFGE) patterns due to an apparent insertion or deletion of a 40-kb fragment. Bacteriophage probes hybridized to these different bands in Southern blot analysis, indicating that homologues of bacteriophage genes were present in the outbreak strains. Investigation of the bacteriophage insertion sites in these isolates suggested that bacteriophage acquisition, loss, or transposition was responsible for the PFGE pattern variation. The bacteriophage gene sequences were similar, but not identical, in the outbreak strains and RM1221, indicating that differences may exist between the bacteriophages.
Campylobacter spp. are the most common cause of human bacterial diarrheal illness worldwide. While the precise number of cases is difficult to determine, it has been estimated that as much as 1% of the population in the United States and the United Kingdom is infected annually with the organism (27). Infections are commonly caused by the consumption of uncooked poultry, contaminated water, or raw milk. They have also been linked to the consumption of contaminated raw fruits and vegetables, as well as to the presence of pets (21). Most infections are sporadic events that tend to occur in the summer, while outbreaks, although infrequent, generally occur in early spring or late fall (23).
As infections occur, it is necessary to identify and differentiate the implicated isolates so that the sources of contamination, the routes of transmission, and clusters of infections can be identified (19). Methods commonly used to type Campylobacter spp. include serotyping of heat-labile or heat-stable antigens, ribotyping, amplified fragment length polymorphism analysis, randomly amplified polymorphic DNA typing, multiplex PCR, pulsed-field gel electrophoresis (PFGE), and nucleic acid sequencing (26). These methods can be recommended for different uses. For example, flaA (which encodes flagellin) restriction fragment length polymorphism (fla-RFLP) and flaA short variable region sequence (fla-SVR) typing are regarded as being best suited for routine surveillance of Campylobacter spp., while PFGE and multilocus sequence typing may be better suited for outbreak characterization and trace-back investigations (3).
PFGE has been extensively used to characterize isolates of Campylobacter jejuni and C. coli and is considered one of the most discriminatory methods available (5, 13, 15, 19). Due to its high degree of discrimination of human and animal isolates, it is not recommended for use for routine surveillance but, instead, is best used for differentiating outbreak-associated cases from unrelated background isolates during a possible outbreak (8, 9, 25). Hänninen et al. (8) found that changes in the PFGE patterns occurred after isolates were passed through chick intestines, while On (15) found that the PFGE patterns changed upon subculturing of the isolates. Variations in PFGE patterns are thought to be more frequently caused by recombination events than by point mutations in the restriction endonuclease sites (15). This phenomenon of genetic instability can be caused by natural transformation, recombination, or mobile elements such as transposons, plasmids, and lysogenic bacteriophages (26). Gibson et al. suggested that bacteriophages may affect the C. jejuni PFGE patterns, but this hypothesis has not been supported with scientific data (7).
The recently completed genomic sequence of chicken isolate RM1221 revealed the presence of three bacteriophage-like elements (6). The first element, Campylobacter Mu-like phage 1 (CMLP1), was determined to be a Mu-like bacteriophage, while the last two elements, CJIE2 and CJIE4, were identified as integrated elements predicted to contain bacteriophage-related genes. The Mu-like bacteriophage encodes genes related to bacteriophage function and transposase homologues but no known virulence determinants.
In May 2000, a waterborne outbreak of Escherichia coli O157:H7 and Campylobacter spp. occurred in Ontario, Canada. Isolates obtained from patients, farm animals, and the surrounding environment were characterized by a number of phenotypic and genotypic typing methods. Four of these isolates (isolates 00-2425, 00-2426, 00-2544, and 00-2856) were identified as being highly similar by serotyping, phage typing, fla-RFLP typing, and PFGE. Their PFGE patterns were indistinguishable, apart from a difference of a single band of approximately 40 kb in each pattern (4). Given that a 37-kb temperate bacteriophage was recently discovered in C. jejuni RM1221, we hypothesized that the PFGE heterogeneity may be due to the insertion or deletion of a bacteriophage element. In this study, PFGE blots from these isolates were hybridized with probes derived from CMLP1. The results indicated that the PFGE patterns differ by the presence or the absence of a bacteriophage homologous to CMLP1. Furthermore, differences among the bacteriophage insertion sites within the bacterial chromosomes were noted, which suggests that bacteriophage translocation might be responsible for generating some of the variability seen in the PFGE patterns of epidemiologically linked C. jejuni isolates.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
C. jejuni strains were obtained from a waterborne outbreak (Ontario, Canada, 2000) (3). All isolates used were characterized as outbreak strain 1 and were epidemiologically related (5). Isolates 00-2425, 00-2426, and 00-2544 were human case strains, while isolate 00-2856 was a bovine isolate. Further strain characteristics are listed in Table 1. Note that differences in the heat-labile serotype and phage type were considered to be of lower or undetermined significance, especially compared to the significance of the other characteristics assessed (5). Isolates were grown on Oxoid Mueller-Hinton agar containing 10% sheep blood and were incubated microaerophilically (10% CO2, 5% O2, and 85% N2) at 37°C for 36 to 48 h.
TABLE 1.
Characteristics of isolates used in this studya
| Isolate | HL serotype | Phage type | PFGE type
|
|
|---|---|---|---|---|
| SmaI | KpnI | |||
| 00-2425 | 125 | 33 | 2 | 2 |
| 00-2426 | 125 | 33 | 1 | 1 |
| 00-2544 | 125 | 35 | 4 | 1 |
| 00-2856 | 128 | 33 | 4 | 1 |
All isolates were biotype II; sequence type 21, based on Oxford multilocus sequence typing; flaA-SVR sequence type 36, based on the flagellin short variable region DNA sequence; fla-RFLP type 1; and heat-stable serotype O:2.
PFGE and nucleic acid transfer.
PFGE was performed with SmaI by the method of Ribot et al. (19) and then analyzed with Bionumerics software (version 4.5; Applied Maths, Kortrijk, Belgium). In preparation for Southern hybridization, the gels were immersed in depurination buffer (250 mM HCl) for 10 min at room temperature (RT), followed by immersion in denaturation buffer (1.5 M NaCl, 0.5 M NaOH) for 25 min at RT. The gels were rinsed in distilled water and then transferred to a Hybond-N+ membrane (Amersham Biosciences, Baie D'Urfe, PQ, Canada) by using a TurboBlotter apparatus (Schleicher & Schuell, Keene, NH). Transfer occurred in denaturation buffer over 2 h at RT. The gels were restained with ethidium bromide to verify that the transfer had occurred.
Southern blot probe production.
Probes were generated by PCR amplification of bacteriophage genes from either C. jejuni strain RM1221 or 00-2544 by using the primers and conditions listed in Table 2. Primers were designed on the basis of the bacteriophage genes present in RM1221 (GenBank accession number CP000025) by using either PrimerSelect software (DNASTAR Inc., Madison, WI) or Primer3 software (available free on the web at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Template DNA was prepared by using the Puregene genomic DNA purification kit (Gentra Systems, Minneapolis, MN), according to the manufacturer's recommendations. The 100-μl PCR mixtures consisted of 1× PCR buffer, 2 mM MgCl2, 0.5 μM each primer, 0.2 mM deoxynucleoside triphosphate mix, 200 to 1,000 ng of template DNA, and 5 U FastStart DNA polymerase (Roche, Laval, PQ, Canada). PCR was performed with a Perkin-Elmer 2400 thermocycler. The products were electrophoresed on 1.5% agarose gels and were then purified by using a QIAquick PCR purification kit (QIAGEN, Mississauga, ON, Canada), according to the manufacturer's instructions. The probe sequences were determined by the DNA Core Facility at the National Microbiology Laboratory prior to use in the hybridizations. Sequencing reactions were run by using BigDye Terminator 3.1 cycle sequencing kits (Applied Biosystems, Streetsville, ON, Canada), according to the manufacturer's instructions, and sequencing was performed with an ABI 3100 or 3730 DNA analyzer (Applied Biosystems).
TABLE 2.
Primer sequences and characteristicsa
| Primer name | Sequence (5′→3′) | Tm (°C) | Amplicon size (bp) | Position in RM1221 genome |
|---|---|---|---|---|
| Cje0215F | GCG AGT GAA GGC AAA AG | 46.0 | 351 | 207652-207668 |
| Cje0215R | TTC TCC ATA GCA AGT GAT AAA C | 46.0 | 208002-207981 | |
| Cje0251F | ATG GGG ATA AAT TTA GCA CTT G | 49.8 | 392 | 230266-230287 |
| Cje0251R | TAG GCC TTT AAC TTC ACT TTC AC | 49.5 | 230657-230636 | |
| Cje0270F | TTC ACC GCA AAG ATA AAA CTA A | 49.3 | 373 | 241603-241582 |
| Cje0270R | ACT ATA ATA TCA GCT GGG GAA CTA | 48.4 | 241231-241254 | |
| Cje0544F | AAT AGG GGA ATG CCA AAA A | 49.0 | 357 | 498853-498871 |
| Cje0544R | CTA CTA ATC TCA AAT ATC CTA CAT | 41.6 | 499209-499186 | |
| RMCj212F | CGA ATT TCG AGC TTT ACA GT | 54.9 | 206651-206670 | |
| RMCj275R | AAA ATG CGA ACA AGT TCA TT | 55.0 | 244602-244583 | |
| Cje0213F2 | TGA CTT TAA GGT TTG GAG GAT A | 55.7 | NA | |
| Cje0343F2 | ACT TTT TCT CAT TTT TGC ATA CC | 55.3 | NA | |
| Cje0344F | GCG ATC TAA TTC TTT ACC CAT A | 55.3 | NA | |
| Cje0344R | TTG ATA CCA ATT CTT CTT GTC A | 55.0 | 309513-309492 | |
| 344jk1 | TGA AAG TGA CTT CAA AGA AAA G | 54.1 | NA | |
| 522jk1 | TTA TTG CTT CTG CAT ATT CAT C | 54.8 | NA | |
| 522jk2 | ACG GCT TTA GTA TTA GCT TTT G | 55.0 | NA |
Primers were used to generate PCR amplicon probes for Southern hybridizations, to sequence the lambda library, or to investigate bacteriophage insertion sites. Primers were based on the bacteriophage genes present in RM1221 (GenBank accession number CP000025) or on the sequence obtained from isolate 00-2425. Tm, melting temperature of the primer; NA, not applicable (the sequence is not present in RM1221).
Southern hybridization and detection.
Probe preparation, hybridization, and detection were performed according to the instructions of the manufacturer of the AlkPhos direct labeling and CDP-Star detection system (Amersham Biosciences). One microliter of unlabeled denatured DNA was spotted on the membrane prior to cross-linking to serve as a positive hybridization control.
PCR.
PCR was performed to determine whether the same genes that flank CMLP1 in RM1221 flank the bacteriophage in the outbreak strains. Primers RMCj212F, complementary to a region in Cje0212, and Cje0215R amplified a 1,353-bp region upstream of CMLP1. A 3,371-bp amplicon was amplified from the region downstream of CMLP1 by primers directed to Cje0270 and Cje0275 (argC). Additional primer characteristics are listed in Table 2.
Creation and screening of genomic library.
A genomic library was prepared from partial Sau3A-I digests of C. jejuni 00-2425 by using a ZAP Express predigested kit (Stratagene, La Jolla, CA), according to the manufacturer's recommendations. The library was screened by Southern hybridizations with an enhanced chemiluminescence direct nucleic acid labeling and detection system (Amersham Biosciences) and probes specific for either Cje0215 or Cje0270. Clones containing DNA flanking the bacteriophage were sent to the DNA Core Facility for sequencing with vector-specific primers (primers T3 and T7), bacteriophage-specific primers (primers Cje0215R and Cje0270R), and additional primers designed for primer walking (Table 2). A BLAST search of the NCBI database was performed with the resultant sequences to identify the C. jejuni sequence. Specific primers based on the putative flanking sequence were designed (Table 2), and then PCR and sequencing were performed to confirm the flanking sequence identity.
Nucleotide sequence accession numbers.
The upstream and downstream insertion site sequences of the bacteriophage in C. jejuni isolate 00-2425 have been deposited in the GenBank database with accession numbers EF092315 and EF092316, respectively.
RESULTS
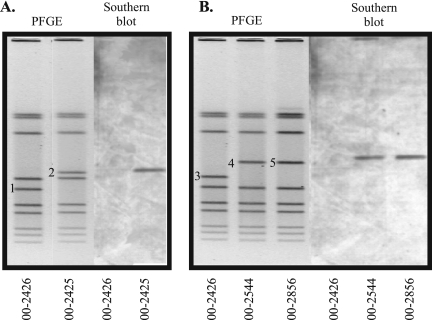
Southern hybridizations were performed to determine whether bacteriophages were responsible for the PFGE pattern variation seen with the outbreak isolates. Probes were amplified by PCR from genes present at the beginning, middle, and end of CMLP1 in RM1221. The targets for PCR included Cje0215, Cje0251, and Cje0270, which encode a putative bacteriophage repressor protein, a putative prophage MuSo1 F protein, and a putative bacteriophage DNA transposition protein A, respectively. One gene from the second bacteriophage-like element in RM1221, Cje0544 (which encodes a site-specific recombinase), was also amplified by PCR and used as a probe. These amplicons were used to probe a PFGE gel of four outbreak isolates (isolates 00-2425, 00-2426, 00-2544, and 00-2856). The results from the hybridizations indicated that bacteriophage genes were present in three isolates but absent from isolate 00-2426. Figure 1 shows the hybridization pattern obtained with the Cje0270-specific probe; identical patterns were generated with the probes for Cje0215 and Cje0251 (data not shown). The hybridization signals corresponded to the same bands that differed in each of the PFGE patterns rather than to the conserved bands (Fig. 1, bands 2, 4, and 5). The probe from the second bacteriophage-like element in RM1221 did not hybridize to DNA from any of the isolates. All probes hybridized to RM1221 in Southern blots (data not shown).
FIG. 1.
PFGE gel and Southern hybridization of highly related C. jejuni strains isolated from a waterborne outbreak. PFGE was performed with SmaI-digested DNA, and Southern hybridization was performed with a probe directed toward Cje0270, a gene that encodes a putative bacteriophage DNA transposition protein A in RM1221. The different bands in the PFGE gel are numbered 1 to 5. (A) Results of PFGE and Southern blot analysis for isolates 00-2426 and 00-2425; (B) results of PFGE and Southern blot analysis for isolates 00-2426, 00-2544, and 00-2856.
Additional Southern hybridizations performed with the outbreak strains indicated that homologues of Cje0221, Cje0226, Cje0232, and Cje0244 were also present in isolates 00-2425, 00-2544, and 00-2856 but were absent from isolate 00-2426 (data not shown). These genes encode a putative bacteriophage virion morphogenesis protein, a putative bacteriophage major tail tube protein, a putative bacteriophage tail protein, and a putative Mu-like prophage 1 protein, respectively. Thus, altogether, homologues of seven bacteriophage genes were shown to be present in three outbreak isolates and absent from a fourth. Given that these bacteriophage genes are present throughout CMLP1, these data suggested that homologues of the entire CMLP1 were present within the genomes of these isolates.
Since every section of chromosomal DNA is not necessarily visible on a PFGE gel, we sought to confirm that the bacteriophages were in fact present in the visible different PFGE bands (18). PFGE band sizes were therefore determined by using the “Metrics” function of the Bionumerics software. Bands 1 and 2 (Fig. 1) were calculated to be 156.99 kb and 196.09 kb, respectively, giving a difference in band size of 39.1 kb. Band 3 was determined to be 182.52 kb, while bands 4 and 5 were approximately 221.47 kb, thus giving a difference in band size of 38.95 kb. These differences in band sizes were consistent with the size of CMLP1 (which is approximately 37 kb). In addition, the sums of all the PFGE band sizes per strain were determined to be 1,792.58 kb, 1,754.81 kb, 1,793.34 kb, and 1,813.13 kb for isolates 00-2425, 00-2426, 00-2544, and 00-2856, respectively. These approximate genome sizes are consistent with the 1,777.83-kb size of the genome of previously sequenced strain C. jejuni NCTC 11168, suggesting that the visible bands represent the isolates' entire genome content and that the absence of a bacteriophage homologous to CMLP1 is the only detectable large-scale difference between isolate 00-2426 and the remaining three isolates (17).
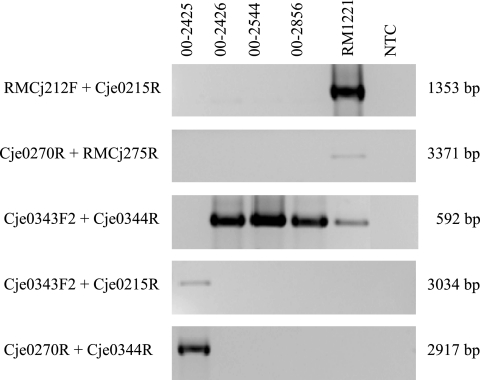
As the bacteriophages present in the outbreak isolates showed homology to CMLP1, a PCR screen was performed to determine if they are also in the same location within the chromosome. Primers directed toward Cje0212, a chromosomal homologue of a pathogenicity protein, and Cje0215, successfully amplified the region upstream of CMLP1 in RM1221 but failed to produce an amplicon from isolates 00-2425, 00-2426, 00-2544, and 00-2856 (Fig. 2). Likewise, the downstream region was successfully amplified only in RM1221 with primers directed to bacteriophage gene Cje0270 and chromosomal gene Cje0275, which encodes N-acetyl-gamma-glutamylphosphate reductase. No amplicons were obtained when the outbreak isolates were tested with these PCR primers (Fig. 2). These results indicated that either the flanking genes were divergent or the bacteriophages in the outbreak strains were in a different genomic location(s).
FIG. 2.
PCR analyses of bacteriophage insertion sites in C. jejuni outbreak isolates. The isolates tested, the primers used, and the amplicon sizes are listed above, to the left, and to the right of the agarose gel images, respectively. Additional primer characteristics are described in Table 2. All amplicons were electrophoresed on 1.5% agarose gels at 100 V. NTC, no-template control.
To establish if the last possibility was true, a genomic library of isolate 00-2425 was created in a lambda phage vector. The library was screened by Southern hybridization with probes directed toward either Cje0215 or Cje0270. Vectors containing sequences that flanked the bacteriophage were sequenced by using vector-specific and bacteriophage-specific primers. The resulting sequences indicated that the bacteriophage in isolate 00-2425 was inserted between Cje0343 (panB, which encodes 3-methyl-2-oxobutanoate hydroxymethyltransferase) and Cje0344 (which encodes a beta-lactamase). The bacteriophage was positioned in the opposite orientation within the genome compared with the orientation of CMLP1 in RM1221, with the beginning of the bacteriophage located upstream of Cje0344 and the end of the bacteriophage located downstream of Cje0343. The sequencing results also revealed that Cje0213 and Cje0215 were present and separated by almost 1,800 bp of DNA and that there were no regions homologous to Cje0214 of CMLP1. The last complete bacteriophage gene present was Cje0270. Cje0271, Cje0272, and Cje0273 were absent; and only 32 bp upstream of Cje0274 and 18 bp within this gene were present.
The location of the insertion site was confirmed by PCR with a genomic DNA template and primers directed to the bacteriophage and the flanking sequence. PCR with primers Cje0215R and Cje0344R, as well as primers Cje0270R and Cje0343F2, successfully produced amplicons from isolate 00-2425 but failed with the template from isolate 00-2426 (Fig. 2). Cje0343F2 and Cje0344R, however, produced a 592-bp amplicon from 00-2426 but not with 00-2425, adding support to the possibility that Cje0343 and Cje0344 are adjacent in the former isolate but not in the latter one.
The location of the bacteriophages in isolates 00-2544 and 00-2856 has yet to be determined, although the PCR screening assay used to confirm the presence of the insertion site in isolate 00-2425 failed to produce any amplicons in these strains, thus suggesting the bacteriophages are present in another location within the chromosomes of these isolates (Fig. 2).
DISCUSSION
Bacteriophages are thought to be major contributors to genomic diversification (14). They are widespread and have been shown to be present in most bacteria, including E. coli, Salmonella enterica, and Vibrio cholerae (24). Their effect on PFGE patterns has been established for bacteria such as Staphylococcus epidermidis, Shigella sonnei, and E. coli (1, 10-12, 22). In these organisms, bacteriophage lysogeny resulted in diverse changes in PFGE banding patterns, which varied from minor changes, as in the case of staphylococci, to major alterations in which up to seven bands differed, as in the case of E. coli K-12. Consequently, these studies demonstrate that bacteriophage acquisition, loss, or transposition can affect the bacterial chromosome structure and, in turn, molecular typing results.
Temperate bacteriophages were identified in C. fetus (originally characterized as Vibrio fetus) during the late 1960s and 1970s; however, their effect on Campylobacter typing methods was never addressed (2, 20). In this study, we have identified the presence of bacteriophage genes in the genomes of three of four highly related C. jejuni strains isolated from a waterborne outbreak. Southern hybridization experiments indicated that a total of seven CMLP1 bacteriophage gene homologues (Cje0215, Cje0221, Cje0226, Cje0232, Cje0244, Cje0251, and Cje0270) were present in isolates 00-2425, 00-2544, and 00-2856 but not in isolate 00-2426. A probe from the second bacteriophage-like element in RM1221 did not hybridize to DNA from any of the isolates, suggesting that a homologue of CJIE2 was absent from these outbreak isolates. These findings indicated that a bacteriophage with extensive sequence homology to CMLP1 was present in the outbreak isolates. Interestingly, hybridizations of a PFGE gel of the isolates revealed that the bacteriophage-specific probes bound to the same bands that differed by the approximate size of CMLP1. This supports the hypothesis that the PFGE pattern variation seen between isolates 00-2425 and 00-2426 was due to the insertion/deletion of a temperate bacteriophage, while the variation seen between isolates 00-2425, 00-2544, and 00-2856 was due to bacteriophage translocation.
The lack of hybridization of bacteriophage probes to isolate 00-2426 was further supported by comparative genomic hybridizations of isolates 00-2425 and 00-2426, which indicated the presence of a bacteriophage with homology to CMLP1 in the former isolate but not in the latter isolate (data not shown). The absence of bacteriophage genes in highly related isolate 00-2426 suggests either the acquisition of bacteriophages in the first three strains or the loss of bacteriophage in the last strain.
To investigate the possibility of bacteriophage translocation, the locations of the bacteriophage insertion sites in isolates 00-2425, 00-2544, and 00-2856 were investigated. PCR with RM1221-specific primers demonstrated that the bacteriophages in these isolates were inserted in a different genomic location(s) than CMLP1 in RM1221. The bacteriophage in 00-2425 was determined to be between genes Cje0343 and Cje0344, with its orientation opposite that of CMLP1 in RM1221. The bacteriophages of the two remaining isolates were not present in the same chromosomal location as either of the bacteriophages in 00-2425 or RM1221. These results were not unexpected, given that isolates 00-2544 and 00-2856 have different PFGE patterns and that Southern blots of PFGE gels indicate that the bacteriophage was present in a different band. The results are also consistent with those of Parker et al., who determined that CMLP1 is differentially located in a number of C. jejuni isolates (16). Altogether, these data suggest that CMLP1 inserts randomly into the chromosome and may have the potential to translocate.
Sequencing of the isolate 00-2425 bacteriophage insertion site indicated that there were some differences in gene content near the ends of the bacteriophages, as one gene at the beginning and four genes at the end were absent compared to the sequence of CMLP1. As well, partial nucleotide sequence determinations of these bacteriophages indicated that although they were nearly identical, there was significant sequence divergence in selected regions when the sequences of these bacteriophages were compared with the sequence of CMLP1 (unpublished data). Therefore, these observations suggest that there may be differences not only in bacteriophage insertion sites but also in gene content. Whether the CMLP1 homologues in these four isolates also show sequence divergence among themselves will be the subject of future investigations.
This study has demonstrated that the different PFGE patterns of four epidemiologically linked C. jejuni strains was due to the presence, absence, or translocation of a temperate bacteriophage(s). This indicates that the mechanisms contributing to PFGE diversity in C. jejuni isolates are not limited to natural transformation and suggests that temperate bacteriophages may play an important role in Campylobacter biology.
Acknowledgments
We are indebted to the DNA Core Facility at the National Microbiology Laboratory for primer synthesis and sequencing; Dobryan Tracz for CGH analysis; as well as Katarina Strank, Jason Allen, Cynthia Misfeldt, and Lorelee Tschetter of Emerging Bacterial Pathogens, National Microbiology Laboratory, for PFGE electrophoresis and analysis.
Footnotes
Published ahead of print on 29 November 2006.
REFERENCES
- 1.Bielaszewska, M., R. Prager, W. Zhang, A. W. Friedrich, A. Mellmann, H. Tschape, and H. Karch. 2006. Chromosomal dynamism in progeny of outbreak-related sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 72:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokkenheuser, V. D., N. J. Richardson, J. H. Bryner, D. J. Roux, A. B. Schutte, H. J. Koornhof, I. Freiman, and E. Hartman. 1979. Detection of enteric campylobacteriosis in children. J. Clin. Microbiol. 9:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, C. G., L. Bryden, W. R. Cuff, P. L. Johnson, F. Jamieson, B. Ciebin, and G. Wang. 2005. Use of the Oxford multilocus sequence typing protocol and sequencing of the flagellin short variable region to characterize isolates from a large outbreak of waterborne Campylobacter sp. strains in Walkerton, Ontario, Canada. J. Clin. Microbiol. 43:2080-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, C. G., L. Price, R. Ahmed, D. L. Woodward, P. L. Melito, F. G. Rodgers, F. Jamieson, B. Ciebin, A. Li, and A. Ellis. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson, J., E. Lorenz, and R. J. Owen. 1997. Lineages within Campylobacter jejuni defined by numerical analysis of pulsed-field gel electrophoretic DNA profiles. J. Med. Microbiol. 46:157-163. [DOI] [PubMed] [Google Scholar]
- 8. Hänninen, M. L., M. Hakkinen, and H. Rautelin. 1999. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:2272-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hänninen, M. L., S. Pajarre, M. L. Klossner, and H. Rautelin. 1998. Typing of human Campylobacter jejuni isolates in Finland by pulsed-field gel electrophoresis. J. Clin. Microbiol. 36:1787-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iguchi, A., R. Osawa, J. Kawano, A. Shimizu, J. Terajima, and H. Watanabe. 2003. Effects of lysogeny of Shiga toxin 2-encoding bacteriophages on pulsed-field gel electrophoresis fragment pattern of Escherichia coli K-12. Curr. Microbiol. 46:224-227. [DOI] [PubMed] [Google Scholar]
- 11.Lina, B., M. Bes, F. Vandenesch, T. Greenland, J. Etienne, and J. Fleurette. 1993. Role of bacteriophages in genomic variability of related coagulase-negative staphylococci. FEMS Microbiol. Lett. 109:273-277. [DOI] [PubMed] [Google Scholar]
- 12.Murase, T., S. Yamai, and H. Watanabe. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48-50. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen, E. M., J. Engberg, V. Fussing, L. Petersen, C. H. Brogren, and S. L. W. On. 2000. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J. Clin. Microbiol. 38:3800-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 15.On, S. L. W. 1998. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol. Lett. 165:341-346. [DOI] [PubMed] [Google Scholar]
- 16.Parker, C. T., B. Quinones, W. G. Miller, S. T. Horn, and R. E. Mandrell. 2006. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 44:4125-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 18.Perkins, J. D., J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1992. SfiI genomic cleavage map of Escherichia coli K-12 strain MG1655. Nucleic Acids Res. 20:1129-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie, A. E., J. H. Bryner, and J. W. Foley. 1983. Role of DNA and bacteriophage in Campylobacter auto-agglutination. J. Med. Microbiol. 16:333-340. [DOI] [PubMed] [Google Scholar]
- 21.Stern, N. J. 1992. Reservoirs of Campylobacter jejuni and approaches for intervention in poultry, p. 49-60. In I. Nachamkin, M. J. Blaser, and L. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, DC.
- 22.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 9-19. In I. Nachamkin, M. J. Blaser, and L. Tompkins (ed.), Campylobacter jejuni: current status and future trends. American Society for Microbiology, Washington, DC.
- 24.Waldor, M. K. 1998. Bacteriophage biology and bacterial virulence. Trends Microbiol. 6:295-297. [DOI] [PubMed] [Google Scholar]
- 25.Wassenaar, T. M., B. Geilhausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 2000. The increasing incidence of human campylobacteriosis: report and proceedings of a WHO consultation of experts. World Health Organization, Copenhagen, Denmark.