Abstract
Molecular detection of Bordetella pertussis DNA is a sensitive and specific method for the rapid diagnosis of pertussis. In this study, a new molecular assay for the detection and differentiation of Bordetella spp. based on automated DNA extraction and real-time PCR was evaluated. The analytical sensitivity of the new assay was determined by Probit analysis of serial dilutions of both cloned PCR products IS481 and IS1001 and cell suspensions of B. pertussis, B. parapertussis, and B. bronchiseptica. The specificity was analyzed by testing a number of pathogens producing respiratory infections. Moreover, a total of 92 clinical samples were investigated. The results were compared to those obtained by an in-house assay based on manual DNA extraction, followed by real-time PCR and detection of IS481. The analytical sensitivity of the new assay for the detection of IS481 and IS1001 was determined to be 2.2 and 1.2 genome equivalents/μl, respectively. The analytical sensitivity for the detection of B. pertussis, B. parapertussis, and B. bronchiseptica was determined to be 1.6, 1.0, and 2.7 genome equivalents/μl, respectively. When clinical specimens were tested with the new assay, 46 of 92 were found to be positive for Bordetella DNA. With the in-house assay, 45 samples tested positive. The new molecular assay proved to be suitable for the rapid diagnosis of pertussis in the routine diagnostic laboratory.
Bordetella pertussis is the agent responsible for producing pertussis, also called whooping cough. B. parapertussis, B. bronchiseptica, and B. holmesii may also cause a pertussis-like respiratory disease but with a milder clinical course (7). Pertussis occurs worldwide, with an increased incidence among young unvaccinated infants. Neonates and elderly patients may show an atypical course of disease (2, 4). A rapid and reliable diagnostic method is thus essential for appropriate treatment and prophylaxis. Culture has been considered the gold standard for detection of B. pertussis but often lacks sensitivity, and a minimum of 4 days may be required to obtain results (1).
Today, PCR has been shown to be a sensitive and specific method for the rapid diagnosis of pertussis (3, 6, 11). In the Bordetella genome, there are repetitive insertion sequences present in a high copy number that may be useful for the amplification of different bacterial strains. Insertion sequences include IS481 in B. pertussis, B. holmesii, and, occasionally B. bronchiseptica, and IS1001 in B. parapertussis and, occasionally, B. bronchiseptica and B. holmesii. These sequences have been reported as target sequences in several PCR protocols (5, 10, 13, 15).
In the present study, the new artus Bordetella LC PCR Kit (QIAGEN Hamburg GmbH, Germany), which includes a multiplex PCR system consisting of four independent PCRs for qualitative detection and differentiation of B. pertussis, B. parapertussis, B. bronchiseptica, and the heterologous internal control (IC), was evaluated. After we tested these methods for analytical sensitivity and specificity, clinical specimens were investigated. The results were compared to those obtained by a qualitative in-house assay based on single amplification and detection of IS481.
MATERIALS AND METHODS
Study design.
In a first step, the analytical sensitivity and specificity of the new artus Bordetella LC PCR kit were investigated. Analytical sensitivity was determined by probit analysis of serial dilutions of cloned PCR products IS481 and IS1001 and cell suspensions of B. pertussis (strain DSM 5571; DSMZ GmbH, Braunschweig, Germany), B. parapertussis (strain DSM 13415; DSMZ), and B. bronchiseptica (strain DSM 13414; DSMZ). For determination of the lower limit of detection, dilution series (0.5 log in buffer AE; QIAGEN) for each of the strains were prepared. Each of the dilutions was analyzed on at least 2 days with at least 18 replicates per dilution. Specificity was tested using all known Bordetella species (B. pertussis DSM 5571, B. parapertussis DSM 13415, B. bronchiseptica DSM 13414, B. holmesii DSM 13416, B. avium DSM 11332, B. hinzii DSM 11333, B. trematum DSM 11334, and B. petrii DSM 12804; DSMZ), as well as a number of viral, bacterial, and fungal pathogens producing respiratory infections.
In a second step, a total of 92 clinical samples were investigated with the new molecular assay in an International Standards Organization (ISO9001, 2000)-certified routine clinical laboratory, the Molecular Diagnostics Laboratory, Institute of Hygiene. The results were compared to those obtained by a qualitative in-house assay published recently (9).
Samples.
A total of 92 nasopharyngeal swabs were investigated. All specimens were collected by using the Copan Venturi Transystem culture swab transport system (COPAN Italia Spa, Brescia, Italy) according to the manufacturer's instructions. Samples were derived from patients (45 females, 47 males; mean age, 8.7 years; age range, 0 to 86 years) with a clinical presentation compatible to B. pertussis infection.
Automated sample preparation protocol.
For the new molecular assay, preparation of bacterial DNA started with dipping the swab into a mixture of 380 μl of MagNA pure bacteria lysis buffer (Roche) and 20 μl of proteinase K solution (20 g/liter). The tip of the swab was cut, and the suspension (including the swab tip) was incubated at 65°C for 10 min and at 95°C for another 10 min. After removal of the swab, 400 μl of the suspension were transferred into the MagNA pure compact sample tube. Automated DNA extraction was done on the MagNA pure compact instrument (Roche) by using the MagNA pure compact nucleic acid isolation kit I (Roche). Prior to the start of DNA extraction, the instrument adds the heterologous IC automatically. For extraction of B. pertussis DNA, the DNA bacterium purification protocol was chosen. The elution volume was set to be 50 μl.
Manual sample preparation protocol.
For the in-house assay, manual isolation of B. pertussis DNA was done as described recently (9).
Amplification and detection by the new molecular assay.
Real-time PCR was performed on the LightCycler 2.0 CE/IVD (LC) instrument (Roche). All samples were tested with the artus Bordetella LC PCR kit (QIAGEN). This assay includes a multiplex PCR system consisting of four independent PCRs for the qualitative detection of B. pertussis, B. parapertussis, and B. bronchiseptica and a heterologous IC in a single LC glass capillary. With the first PCR, an 81-bp sequence specific for the insertion sequence IS481 present in high copy numbers in B. pertussis, B. holmesii, and, occasionally, B. bronchiseptica is amplified. The second PCR provides specific amplification of a 104-bp sequence specific for the insertion sequence IS1001 present in high copy numbers in B. parapertussis and occasionally B. bronchiseptica and B. holmesii. Both of the amplification systems use the same fluorescence dyes and are thus detected in the same LC channel. With the third PCR, a 135-bp sequence specific for the filamentous hemagglutinin gene, present as a single-copy gene in B. pertussis, B. parapertussis, and B. bronchiseptica is amplified. This specific amplification product allows the differentiation of B. pertussis, B. parapertussis, and B. bronchiseptica by melting-curve analysis. B. holmesii is not detected with this system. In addition, the assay includes amplification of a heterologous IC (a specially designed artificial DNA product) with a fourth PCR.
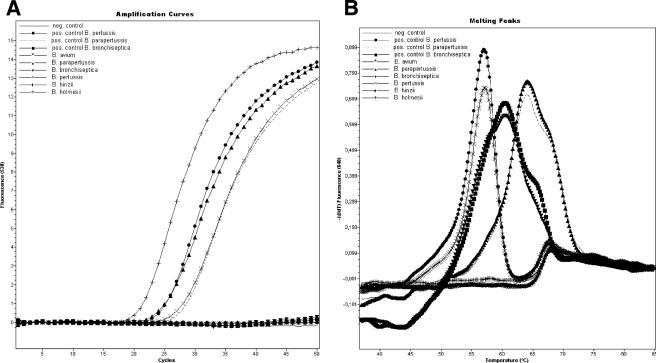
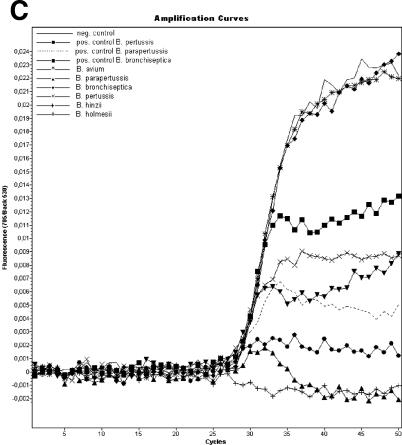
For real-time PCR, the PCR master mixture contained 13 μl of Bordetella LC Master and 2 μl of Bordetella LC Mg-Sol. A 5-μl aliquot of extracted sample was added to 15 μl of PCR master mixture in each LC glass capillary. After this, LC capillaries were sealed, inserted into the specially designed LC Carousel (Roche), and centrifuged at 3,000 rpm for 15 s. The cycling protocol was as follows: 1 cycle of 95°C for 10 min, followed by 50 cycles consisting of denaturation for 1 s at 95°C, annealing for 20 s at 55°C, and elongation for 15 s at 72°C. After the final cycle, the melting curve was started with the initial temperature set at 95°C for 1 s, followed by cooling to 35°C for 15 s; the thermal chamber temperature was then slowly (0.1°C/s) increased to 85°C, and the fluorescence was measured continuously. Fluorescence curves were analyzed with the LC software (LightCycler SW 4.05.415). The LC software provides three different fluorescence channels for final estimation of results obtained by the new molecular assay. Channel 530 was selected for both the IS481 and IS1001 amplification products, channel 640 was used for the amplification product specific for the filamentous hemagglutinin gene, and channel 705/Back530 was selected for the IC. Melting-curve analysis in channel 640 allows a differentiation between B. pertussis, B. parapertussis, and B. bronchiseptica, with melting peaks of 57, 64, and 60°C, respectively. Strains of B. bronchiseptica that do not include the IS481 and the IS1001 sequences in their genome are detected by the presence of the pathogenic factor gene only. Detection of B. holmesii is not provided by melting-curve analysis.
Amplification and detection by the in-house assay.
For the in-house assay, amplification and detection of Bordetella DNA was done as described recently (9).
RESULTS
The analytical sensitivity of the artus Bordetella LC PCR kit for the detection of IS481 and IS1001 was determined to be 2.2 and 1.2 genome equivalents/μl, respectively. The analytical sensitivity for the detection of the filamentous hemagglutinin gene, which allows the discrimination between B. pertussis, B. parapertussis, and B. bronchiseptica by melting-curve analysis, was determined to be 1.6, 1.0, and 2.7 genome equivalents/μl for B. pertussis, B. parapertussis, and B. bronchiseptica, respectively (Table 1) . Specificity testing of all known Bordetella spp. and of pathogens producing respiratory infections gave results as expected (Table 2). The results obtained from strains of B. pertussis, B. parapertussis, B. bronchiseptica, B. avium, B. hinzii, and B. holmesii are shown in Fig. 1.
TABLE 1.
Evaluation of analytical sensitivity
| Target | Sensitivity (genome equivalents/μl)
|
|
|---|---|---|
| Probit (P ≤ 0.05) | Confidence interval | |
| Insertion sequences | ||
| IS481 | 2.2 | 1.3-4.9 |
| IS1001 | 1.2 | 0.7-2.6 |
| Filamentous hemagglutinin genes | ||
| B. pertussis | 1.6 | 1.0-3.3 |
| B. parapertussis | 1.0 | 0.6-2.9 |
| B. bronchiseptica | 2.7 | 1.5-7.6 |
TABLE 2.
Results of cross-reactivity testing
| Pathogena | Presence (+) or absence (−) as determined by:
|
||
|---|---|---|---|
| Channel 530 IS481/IS1001 | Channel 640 melting curve | Channel 705/Back530 internal control | |
| Bordetella pertussis (DSM 5517) | + | + | + |
| Bordetella parapertussis (DSM 13415) | + | + | + |
| Bordetella bronchiseptica (DSM 13414) | − | + | + |
| Bordetella avium (DSM 11332) | − | − | + |
| Bordetella hinzii (DSM 11333) | − | − | + |
| Bordetella holmesii (DSM 13416) | + | − | + |
| Bordetella petrii (DSM 12804) | − | − | + |
| Bordetella trematum (DSM 11334) | − | − | + |
| Haemophilus influenzae | − | − | + |
| Haemophilus parainfluenzae | − | − | + |
| Staphylococcus aureus | − | − | + |
| Staphylococcus epidermidis | − | − | + |
| Neisseria meningitidis | − | − | + |
| Neisseria gonorrhoeae | − | − | + |
| Streptococcus pyogenes | − | − | + |
| Streptococcus pneumoniae | − | − | + |
| Pseudomonas aeruginosa | − | − | + |
| Escherichia coli | − | − | + |
| Corynebacterium diphtheriae | − | − | + |
| Klebsiella pneumoniae | − | − | + |
| Aeromonas hydrophila | − | − | + |
| Legionella pneumophila | − | − | + |
| Mycoplasma pneumophila | − | − | + |
| Mycobacterium tuberculosis | − | − | + |
| Chlamydia trachomatis | − | − | + |
| Helicobacter pylori | − | − | + |
| Candida albicans | − | − | + |
| Acinetobacter sp. | − | − | + |
| HSV-1 | − | − | + |
| HSV-2 | − | − | + |
| HIV-1 | − | − | + |
| CMV-Toledo | − | − | + |
| Enterovirus | − | − | + |
| WNV | − | − | + |
| HTLV-1 | − | − | + |
| HTLV-2 | − | − | + |
| HHV-6A | − | − | + |
| HHV-6B | − | − | + |
| HHV-7 | − | − | + |
| HHV-8 | − | − | + |
| EBV | − | − | + |
HSV-1, herpes simplex virus type 1, etc.; CMV-Toledo, cytomegalovirus-Toledo virus; WNV, West Nile virus; HTLV-1, human T-cell leukemia virus type 1, etc.; HHV-6A, human herpesvirus type 6A, etc.; EBV, Epstein-Barr virus.
FIG. 1.
Detection of B. avium, B. bronchiseptica, B. hinzii, B. holmesii, B. parapertussis, and B. pertussis with the new artus Bordetella LC PCR kit. (A) Plots for the IS481 and IS1001 amplification products in channel 530. (B) Melting curves of amplification products specific for the filamentous hemagglutinin gene in channel 640. (C) Plots for the heterologous IC in channel 705/Back530.
When clinical specimens were tested with the new assay, 46 of 92 samples were found to be positive for Bordetella DNA. Differentiation by melting-curve analysis in channel 640 yielded B. pertussis in 37 samples, B. parapertussis in 1 sample, and B. bronchiseptica in another sample. In the remaining seven samples, Bordetella DNA was detected just by analysis of fluorescence curves in channel 530. The heterologous IC was consistently detected in all samples except those that contained high target concentration. In these samples, amplification of the IC was suppressed because of competitive inhibition (14). When clinical specimens were tested with the in-house assay, 45 of 92 samples tested positive for the presence of IS481. The discrepant sample tested positive for B. parapertussis DNA with the new assay. All runs included no-template controls, and no contamination was observed during the entire study.
For the extraction of eight samples on the MagNA Pure Compact instrument (Roche), 60 min were required. This included a 5-min setup time for the instrument and 25 min for the pipetting steps prior to automated DNA extraction. After centrifugation, real-time PCR required another 55 min.
DISCUSSION
A rapid and reliable diagnostic method is essential for the appropriate treatment and prophylaxis of pertussis. Culture has been considered the gold standard for the detection of B. pertussis but often lacks sensitivity, and a minimum of 4 days may be required to obtain results (1). Several PCR approaches have been reported to be more sensitive and specific than culture for the diagnosis of pertussis (5, 10, 13, 15). These assays only provide detection of IS481 and IS1001 without differentiation of Bordetella spp.
In the present study, a new molecular assay for the detection and differentiation of Bordetella spp. based on automated DNA extraction and real-time PCR useful for the routine diagnostic laboratory was evaluated and compared to an in-house assay published recently. This new assay includes a multiplex PCR system consisting of four independent PCRs for the qualitative detection of insertion sequence IS481, insertion sequence IS1001, the filamentous hemagglutinin gene, and the heterologous IC in a single LC glass capillary. Differentiation between B. pertussis, B. parapertussis, and B. bronchiseptica is provided by melting-curve analysis of the amplification product specific for the filamentous hemagglutinin gene, which is present as a single-copy gene in the bacterium. FHA is a surface-exposed and secreted protein in Bordetella species that has been proposed to function as an adhesion and an immunomodulator, indicating a critical role for FHA in host specificity (8).
IS481 and IS1001 are repetitive insertion sequences that may occur in a high copy number. Comparative analysis of the genomes of B. pertussis and B. parapertussis revealed that the B. pertussis genome might have up to 238 copies of IS481 and that the B. parapertussis genome might have up to 22 copies of IS1001 (12). Therefore, IS481-based PCR systems are expected to be about 2 logs more sensitive in the detection of B. pertussis than single-copy-target-based PCR systems. This explains why 46 of the samples gave a positive PCR result for the IS481/IS1001 target, indicating the presence of a human pathogenic Bordetella species, but only 39 of these samples gave a melting curve from the filamentous hemagglutinin gene-specific amplification product in channel 640, allowing the discrimination between B. pertussis, B. parapertussis, and B. bronchiseptica.
When clinical specimens were tested with the new assay, an additional sample was found to be positive for Bordetella DNA. Differentiation based on melting-peak analysis resulted in the identification of B. parapertussis. The negative result obtained by the in-house assay can be explained by absence of IS481 in B. parapertussis (15).
In conclusion, the new molecular assay includes all of the features required for molecular detection and differentiation of Bordetella spp. DNA. It proved to be suitable for the routine diagnostic laboratory, allowing a rapid and safe diagnosis of pertussis.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Dragsted, D. M., B. Dohn, J. Madsen, and J. S. Jensen. 2004. Comparison of culture and PCR for detection of Bordetella pertussis and parapertussis under routine laboratory conditions. J. Med. Microbiol. 53:749-754. [DOI] [PubMed] [Google Scholar]
- 2.Edwards, K., and D. M. Freeman. 2006. Adolescent and adult pertussis: disease burden and prevention. Curr. Opin. Pediatr. 18:77-80. [DOI] [PubMed] [Google Scholar]
- 3.Fredericks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-486. [DOI] [PubMed] [Google Scholar]
- 4.Galanis, E., A. S. King, P. Varughese, S. A. Halperin, and IMPACT investigators. 2006. Changing epidemiology and emerging risk groups for pertussis. CMAJ 14:451-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glare, E. M., J. C. Paton, R. R. Premier, A. J. Lawrence, and I. T. Nisbet. 1990. Analysis of a repetitive DNA sequence from Bordetella pertussis and its application to the diagnosis of pertussis using the polymerase chain reaction. J. Clin. Microbiol. 128:1982-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He, Q., G. Schmidt-Schlapfer, M. Just, H. C. Matter, S. Nikkari, M. K. Viljanen, and J. Mertsola. 1996. Impact of polymerase chain reaction on clinical pertussis research: Finnish and Swiss experiences. J. Infect. Dis. 174:1288-1295. [DOI] [PubMed] [Google Scholar]
- 7.Heininger, U., K. Stehr, S. Schmitt-Grohe, C. Lorenz, R. Rost, P. D. Christenson, M. Uberall, and J. D. Cherry. 1994. Clinical characteristics of illness caused by Bordetella parapertussis compared with illness caused by Bordetella pertussis. Pediatr. Infect. Dis. J. 13:306-309. [DOI] [PubMed] [Google Scholar]
- 8.Inatsuka, C. S., S. M. Julio, and P. A. Cotter. 2005. Bordetella filamentous hemagglutinin plays a critical role in immunomodulation, suggesting a mechanism for host specificity. Proc. Natl. Acad. Sci. USA 102:18578-18583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koidl, C., M. Bozic, J. Berg, M. Stöcher, G. Mühlbauer, E. Marth, and H. H. Kessler. 2005. Addition of a homologous internal control to a real-time PCR assay for detection of Bordetella pertussis. Clin. Chem. 51:2404-2406. [DOI] [PubMed] [Google Scholar]
- 10.Kosters, K., U. Reischl, J. Schmetz, M. Riffelmann, and C. H. Wirsing von Konig. 2002. Real-time LightCycler PCR for detection and discrimination of Bordetella pertussis and Bordetella parapertussis. J. Clin. Microbiol. 40:1719-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffelholz, M. J., C. J. Thompson, K. S. Long, and M. J. Gilchrist. 1999. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J. Clin. Microbiol. 37:2872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkhill, J., M. Sebaihia, A. Preston, L. D. Murphy, N. Thomson, D. E. Harris, M. T. Holden, C. M. Churcher, S. D. Bentley, K. L. Mungall, A. M. Cerdeno-Tarraga, L. Temple, K. James, B. Harris, M. A. Quail, M. Achtman, R. Atkin, S. Baker, D. Basham, N. Bason, I. Cherevach, T. Chillingworth, M. Collins, A. Cronin, P. Davis, J. Doggett, T. Feltwell, A. Goble, N. Hamlin, H. Hauser, S. Holroyd, K. Jagels, S. Leather, S. Moule, H. Norberczak, S. O′Neil, D. Ormond, C. Price, E. Rabbinowitsch, S. Rutter, M. Sanders, D. Saunders, K. Seeger, S. Sharp, M. Simmonds, J. Skelton, R. Squares, K. Stevens, L. Unwin, S. Whitehead, B. G. Barrell, and D. J. Maskell. 2003. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis, and Bordetella bronchiseptica. Nat. Genet. 35:32-40. [DOI] [PubMed] [Google Scholar]
- 13.Reischl, U., N. Lehn, G. N. Sanden, and M. J. Loeffelholz. 2001. Real-Time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J. Clin. Microbiol. 39:1963-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stöcher, M., and J. Berg. 2002. Normalized quantification of human cytomegalovirus DNA by competitive real-time PCR on the LightCycler instrument. J. Clin. Microbiol. 40:4547-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Templeton, K. E., S. A. Scheltinga, A. van der Zee, B. M. Diederen, A. M. van Kruijssen, H. Goossens, E. Kuijper, and E. C. Claas. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J. Clin. Microbiol. 41:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]