Abstract
Two recombinant phenotypic assays for human immunodeficiency virus (HIV) coreceptor usage and an HIV envelope genotypic predictor were employed on a set of clinically derived HIV type 1 (HIV-1) samples in order to evaluate the concordance between measures. Previously genotyped HIV-1 samples derived from antiretroviral-naïve individuals were tested for coreceptor usage using two independent phenotyping methods. Phenotypes were determined by validated recombinant assays that incorporate either an ∼2,500-bp (“Trofile” assay) or an ∼900-bp (TRT assay) fragment of the HIV envelope gp120. Population-based HIV envelope V3 loop sequences (∼105 bp) were derived by automated sequence analysis. Genotypic coreceptor predictions were performed using a support vector machine model trained on a separate genotype-Trofile phenotype data set. HIV coreceptor usage was obtained from both phenotypic assays for 74 samples, with an overall 85.1% concordance. There was no evidence of a difference in sensitivity between the two phenotypic assays. A bioinformatic algorithm based on a support vector machine using HIV V3 genotype data was able to achieve 86.5% and 79.7% concordance with the Trofile and TRT assays, respectively, approaching the degree of agreement between the two phenotype assays. In most cases, the phenotype assays and the bioinformatic approach gave similar results. However, in cases where there were differences in the tropism results, it was not clear which of the assays was “correct.” X4 (CXCR4-using) minority species in clinically derived samples likely complicate the interpretation of both phenotypic and genotypic assessments of HIV tropism.
Human immunodeficiency virus type 1 (HIV-1) can be characterized according to the host chemokine coreceptor used to gain entry into CD4-expressing cells. The phenotypic designations are R5 (for CCR5-using variants), X4 (for CXCR4-using variants), and R5/X4 (defined here either as a mixture of both R5 and X4 variants or as a dual-tropic variant able to enter cells using both CXCR4 and CCR5 coreceptors). Most population-based phenotype assays cannot readily distinguish between virus populations that are truly dual tropic and those that are comprised of mixtures of viruses with different coreceptor phenotypes (4). The fact that viral populations within an individual may contain heterogeneous mixtures composed of any combination of the three classes of virus complicates the accurate and reliable assessment of tropism in clinical samples.
Recently, with the development of coreceptor antagonists capable of blocking either the CCR5 (14, 18) or the CXCR4 (18, 20) coreceptor, the need for accurate HIV-phenotyping assays has increased. This is especially important in preliminary tests to screen patients for clinical trials of these agents and for monitoring the evolution of coreceptor usage under the pressure of CCR5 and/or CXCR4 antagonists (13).
Many, but not all, of the genetic determinants of coreceptor usage reside in the HIV envelope, in particular, the V3 loop (8, 10). Consequently, this region is highly useful for the training and development of computerized genotypic predictors of coreceptor usage, which have the potential to make the screening and testing of HIV-infected patients faster and more cost-effective. The simplest genotypic approach, known as the “11/25 rule” classifies a virus as X4 if positively charged amino acids (lysine or arginine) are present at positions 11 and/or 25 of the V3 loop (5, 7, 8). Evaluation of bioinformatics-based genotypic predictors, such as neural networks (17), position-specific scoring matrices (9), and support vector machines (SVMs), indicate that the coreceptor phenotypes may be accurately determined with genotypic predictors (19) when tested on clonally derived sequence data. However, both phenotypic assays and genotypic prediction methods are challenged when applied to heterogeneous sequence data obtained from actual clinical samples. If testing for HIV coreceptor usage becomes necessary for treatment with coreceptor inhibitors, it would be expected to be performed on clinically derived samples. Under these circumstances, a high level of reliability and repeatability should be expected.
Two previously validated recombinant phenotypic assays for HIV coreceptor use (the Trofile HIV Co-receptor Tropism Assay [formerly the PhenoSense HIV-Entry Assay] from Monogram Biosciences and the Tropism Recombinant Test [TRT], performed in the laboratory of Fabrizio Mammano at INSERM [to be available through Eurofins Viralliance Inc.]) were employed here, as well as genotypic analysis of the HIV V3 loop in 74 clinically derived HIV-1 samples in order to evaluate the concordance between phenotypic assays and to determine the ability of an SVM-based genotypic method to predict coreceptor usage.
MATERIALS AND METHODS
Sample selection.
The samples represented a nonrandom selection of 93 baseline samples from the well-characterized HOMER cohort (11), a large cohort of antiretroviral-naïve individuals initiating triple-combination therapy in British Columbia. Baseline plasma samples from this cohort have been previously genotyped for the HIV envelope V3 loop sequence (1). Samples were chosen for analysis on the basis of the predicted phenotype using the 11/25 charge rule (5, 7, 8), a neural-network method (17), and results from a position-specific scoring matrix analysis (9) in order to overrepresent expected X4 variants to an approximate 50% prevalence. Note that the entire HOMER cohort was later phenotyped for HIV coreceptor usage; these results have been published (2).
Assay methods.
The two phenotypic assays (12, 21) use virus stocks pseudotyped with envelope sequences derived from patient plasma samples to infect cell lines engineered to express CCR5 or CXCR4. Some relevant differences in the phenotypic assays are highlighted in Table 1. The Trofile assay confirms coreceptor usage by verifying a decrease in relative light units (RLU) after the addition of a CCR5 or CXCR4 antagonist. The TRT assay did not perform this step at the time of these analyses. Note that the TRT assay was performed on PCR products amplified by our laboratory in British Columbia, Canada, and the Trofile assay was performed on the corresponding baseline plasma samples. More detailed information regarding the two phenotypic assays can be found in references 3, 4, and 12 (Trofile) and 4 and 21 (TRT).
TABLE 1.
Potentially relevant differences in phenotypic assays
| Parameter | Value
|
|
|---|---|---|
| Trofile | TRT | |
| Insert | Entire gp160, ∼2,500-bp RT-PCR product | ∼900-bp RT-PCR product spanning V1 to V3 |
| Vector | pCXAS-envelope expression vector plus HIV genomic-Luc vector | pNL43-ΔV |
| Construction | Restriction enzymes | Recombination |
| Producer cells | 293 | 293T |
| Target cells | U87-CD4-CCR5 and U87-CD4-CXCR4 | U373-CD4-CCR5 and U373-CD4-CXCR4 with HIV-1 long terminal repeat-lacZ cassette |
| Reporter gene | Luciferase | β-Galactosidase |
| Detection assay | RLU | OD using a colorimetric assay |
SVM prediction.
Population-based HIV envelope V3 loop sequences (∼105 bp) of corresponding baseline plasma samples were obtained using previously described standard automated sequence analysis (1). Genotypic predictions of HIV coreceptor usage were performed using an SVM analysis of the sequenced HIV V3 loop (http://www.geno2pheno.org).
Robust alignments of the envelope V3 region were obtained using the multiple-alignment software MUSCLE (6). SVMs were trained using all genotype-phenotype data from the HOMER cohort, determined using the Trofile assay, excluding the 74 samples compared here. Codon mixtures in sequence data are considered ambiguous with respect to sequence identity and were treated using an “X4-sensitive” strategy during training and a “combinatorial” strategy for prediction, as described previously (19). Briefly, in the combinatorial prediction strategy, nonambiguous V3 sequences are generated by random sampling from the ambiguous genotype, and a numeric score, indicating the probability of CXCR4 usage, is calculated for each of the nonambiguous sequences. The final score is taken as the 0.75 percentile of the individual scores. A cutoff of 0.4 on this score was chosen to predict X4 variants. This cutoff was determined by maximizing the area under the receiver operating characteristic (ROC) curve (AUC) for all matched genotype-phenotype samples in the HOMER cohort, excluding the 74 samples analyzed here.
RESULTS
Phenotypic-assay comparison.
Of the 93 samples tested, results were obtained for 78 (84.9%) samples with the Trofile coreceptor tropism assay and results were obtained for 90 (96.8%) samples with the TRT coreceptor usage assay. Note that no comparison can be made with respect to the number of results obtained by each assay, as the TRT assay was performed on PCR products amplified by our laboratory in British Columbia while the Trofile assay was performed on the corresponding baseline plasma samples. Results from the two phenotypic assays were obtained for 74 samples (79.6%) and were in general, but not complete, agreement.
Overall, 85.1% concordance was observed between the two assays. Of the 74 samples, 42 (56.8%) were identified as R5 by both assays and 21 (28.4%) were identified as R5/X4 by both assays. An additional eight (10.8%) were identified as R5 by the Trofile assay but as R5/X4 by the TRT assay, and the remaining three (4.1%) were identified as R5/X4 by the Trofile assay but as R5 by the TRT assay. The Trofile assay characterized one sample as being pure X4. This sample was characterized as being R5/X4 by the TRT assay but for the purposes of our analysis was classified as being R5/X4 by the Trofile assay and was therefore treated as concordant.
We then investigated the clinical parameters of samples with concordant and discordant results (Table 2) . A low CD4 cell count has been shown to be predictive of CXCR4-using HIV (22), and as expected, the samples identified as R5 in both assays showed significantly higher median CD4 cell counts (P < 0.01) than those samples identified as R5/X4 in both assays, although the plasma viral loads were not significantly different (P > 0.1). Median R5 RLU (Trofile assay) and R5 optical-density (OD) (TRT assay) values were consistently high in both assays, regardless of whether they were identified as R5 or R5/X4, implying a high efficiency of replication in the R5 cell lines. Discordant results were observed where R5/X4 was detected in the Trofile assay and not in the TRT assay, and vice versa (shown individually in Table 3). When samples were grouped as “discordant” and “concordant,” we observed no statistically significant differences between the viral loads or CD4 counts (P > 0.05). Also, when discordant groups were compared to each other, we observed no statistically significant differences between CD4 or viral loads (P ≫ 0.5; Wilcoxon rank sum test), although the small sample size of the discordant groups should be noted.
TABLE 2.
Summary of median baseline CD4 count and baseline viral load for samples categorized as either R5 or R5/X4 using the Trofile or TRT coreceptor usage assay
| TF-TRTa | Baseline no. of CD4 cells/mm3 (IQR)c | Log (VL base)b (IQR) | TF RLU
|
TRT
|
||
|---|---|---|---|---|---|---|
| R5 signal (log) (IQR) | X4 signal (log) (IQR) | R5 signal ODSample/ ODCtrl (IQR) | X4 signal ODSample/ODCtrl (IQR) | |||
| R5-R5 (n = 42) | 325 (217-457) | 5.0 (4.5-5.4) | 5.66 (5.3-5.9) | 1.87 (1.8-1.9) | 17.1 (15.3-18.6) | 1.1 (1.1-1.2) |
| R5/X4-R5/X4 (n = 21) | 100 (30-170) | 5.1 (4.6-5.6) | 5.27 (4.9-5.6) | 4.52 (3.6-5.2) | 18.4 (15.0-19.6) | 11.2 (2.2-19.2) |
| R5-R5/X4 (n = 8) | 165 (100-342) | 5.5 (5.2-5.7) | 5.49 (5.4-5.7) | 1.88 (1.8-2.0) | 19.3 (18.4-20.4) | 2.0 (1.6-3.3) |
| R5/X4-R5 (n = 3) | 100 (80-275) | 5.0 (4.8-5.6) | 5.69 (5.5-5.8) | 3.89 (3.2-3.9) | 14.4 (13.8-17.6) | 1.1 (1.1-1.2) |
TF, Trofile HIV coreceptor tropism assay; TRT, TRT coreceptor usage assay.
VL base, baseline viral load in log10 HIV RNA copies/ml.
IQR, interquartile range.
TABLE 3.
Characteristics of samples with discordant results from the Trofile or TRT entry coreceptor assay
| TF-TRTa | Sample identifier | Baseline no. of CD4 cells/mm3 | Log (VL base)b | TRT
|
TF
|
Amino acid observed at codon:
|
|||
|---|---|---|---|---|---|---|---|---|---|
| R5 signal ODSample/ODCtrl | X4 signal ODSample/ODCtrl | RLU R5 signal (log) | RLU X4 signal (log) | 11 | 25 | ||||
| R5/X4-R5 | 1 | 450 | 4.6 | 14.4 | 1.1 | 5.31 | 3.89 | G | E/G |
| 2 | 60 | 5 | 21.3 | 1.3 | 5.88 | 2.48 | S | R | |
| 3 | 100 | 5.9 | 13.2 | 1.1 | 5.69 | 3.94 | G | R | |
| R5-R5/X4 | 4 | 150 | 5.5 | 20.0 | 1.6 | 5.71 | 1.86 | R/S | R |
| 5 | 320 | 4.2 | 21.9 | 2.2 | 5.40 | 1.89 | G | K | |
| 6 | 110 | 5.9 | 18.4 | 1.5 | 5.57 | 2.19 | G | A/E | |
| 7 | 410 | 5.9 | 19.1 | 1.5 | 6.22 | 1.99 | G | K | |
| 8 | 70 | 5.3 | 16.6 | 20.5 | 5.76 | 1.87 | G/S | D | |
| 9 | 20 | 5.5 | 18.3 | 1.8 | 3.80 | 1.73 | G | D | |
| 10 | 530 | 5.1 | 21.4 | 6.5 | 5.31 | 1.83 | G | E | |
| 11 | 180 | 5.7 | 19.4 | 16.4 | 5.38 | 2.25 | D/G | R | |
TF, Trofile HIV coreceptor tropism assay; TRT, TRT coreceptor usage assay.
VL base, baseline viral load in log10 HIV RNA copies/ml.
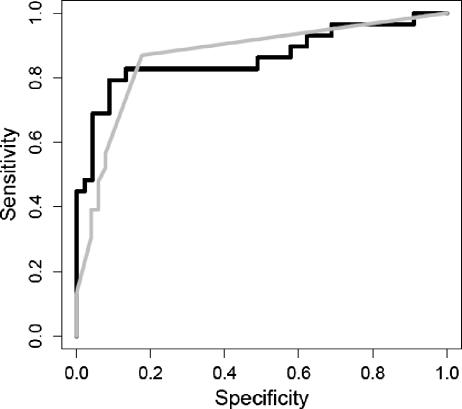
The median X4 signal measured in RLU (Trofile assay) or the OD of the sample divided by the OD of the control (ODSample/ODCtrl) (TRT assay) of the discordant samples was close to the lower quartile of the X4 signal for samples concordantly phenotyped as being R5/X4 by both assays. Therefore, a possible explanation for discordances is misclassification as a result of using OD or RLU “cutoff” values for categorizing samples as R5 or R5/X4. ROC curves examine the effects of all possible cutoff values (for X4 RLU [Trofile assay] or X4 OD [TRT assay]) on sensitivity (the ability to identify true positives) and specificity (the ability to identify false positives). Both assays appeared to use cutoff values that optimized their sensitivities and specificities by placing them in the upper left quadrant of the ROC graph (Fig. 1). In comparing the AUCs, the X4 RLU resulted in an AUC of 0.88 when the TRT assay was used as the gold standard, and the X4 ODs resulted in an AUC of 0.87 when the Trofile assay was used as the gold standard. There appeared to be no significant difference between the two assays when one assay was used as a reference and the cutoffs used for the other assay were varied, and vice versa (P > 0.1), suggesting that the use of different cutoff values in either assay would not improve the predictive performance of one phenotypic assay with respect to the other. Interestingly, 6 of the 11 discordant samples (54.5%) had positively charged amino acids at codons 11 and/or 25 (Table 3), implying a high probability that many of these discordant samples harbored X4 variants that gave only marginally detectable X4 RLU and/or OD.
FIG. 1.
Receiver operating characteristic curve of the Trofile HIV coreceptor assay versus the TRT coreceptor assay. The gray line uses the Trofile coreceptor results as the reference while varying the cutoff for the X4 ODSample/X4 ODCtrl data obtained from the TRT assay. The black line uses the TRT coreceptor results as the reference while varying the cutoff for the X4 RLU data obtained from the Trofile assay.
Genotypic prediction.
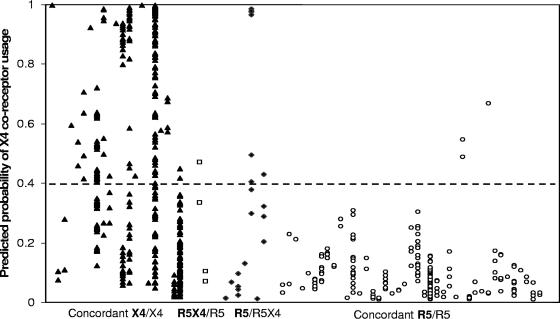
Using the HIV envelope V3 sequences, genotypic predictions of coreceptor usage were made using an SVM trained on separate V3 genotype-phenotype (Trofile assay) data, as described above, excluding the 74 samples tested here. In the data set of 74 clinically derived isolates, amino acid mixtures at ≥1 codon of the HIV V3 loop were observed in 56 (75.7%) sequences. According to the “combinatorial” prediction strategy, mixture-containing sequences were expanded into all possible permutations (median, 2 permutations per isolate; range, 1 to 128), giving rise to multiple values for each sample, each falling between 0 (a low likelihood of being X4) and 1 (a high likelihood of being X4) (Fig. 2).
FIG. 2.
SVM-predicted probabilities of CXCR4 coreceptor usage. Each position on the x axis represents a sample from a single patient. Sequences containing amino acid mixtures were expanded into all possible permutations per sample, and each permutation was assigned a score based on its probability of harboring X4-containing virus (where 0 is low probability and 1 is high probability). Sample permutations were tested and scored individually, and the probability of being X4 is indicated on the y axis. Each individual sample permutation is represented by a single symbol, generating a series of vertically aligned symbols for each clinically derived sample. The samples were grouped along the x axis, labeled according to the results obtained from both phenotypic assays as “R5” or “R5/X4,” and ordered by “Trofile assay/TRT assay.” The solid triangles represent samples phenotyped as R5/X4 by both assays, open squares represent samples phenotyped as R5/X4 by the Trofile assay and as R5 by the TRT assay, solid diamonds represent samples phenotyped as R5 by the Trofile assay and as R5/X4 by the TRT assay, and open circles represent samples phenotyped as R5 by both assays. The dashed horizontal line represents the SVM cutoff, above which samples are classified as R5/X4.
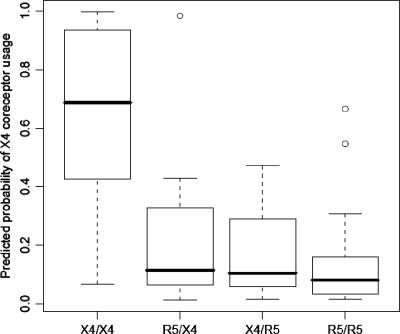
The samples were grouped according to the results obtained from both phenotypic coreceptor assays (concordant R5/R5, concordant R5X4/R5X4, discordant R5X4/R5, and discordant R5/R5X4, where the Trofile result is indicated first and in boldface). When comparing groups, the maximum X4 score per sample was used. With regard to the Trofile assay, 16 of the 24 samples phenotyped as R5/X4 were predicted to be R5/X4 by the SVM, resulting in a sensitivity of 67%. A total of 48 of the 50 samples phenotyped as R5 were predicted to be R5 by the SVM, yielding a specificity of 96%. Similarly, for the TRT assay, 16 of the 28 samples phenotyped as R5/X4 were predicted to be R5/X4 by the SVM, yielding a sensitivity of 55%, and 43 of the 45 samples phenotyped as R4 were predicted to be R5, yielding a specificity of 96%. In comparison, the “11/25” rule had a sensitivity and a specificity for predicting X4 usage in the Trofile assay of 54% and 44%, respectively, and 52% and 42% for the TRT assay. The median X4 scores of the samples classified as R5/X4 by both phenotypic coreceptor assays was 0.68. This score was significantly higher (Fig. 3) than the median 0.08 X4 score of the samples classified as R5 by both phenotypic coreceptor assays (P ≪ 0.001). It also exceeded the median X4 scores for the samples classified as R5 by the Trofile assay and as R5/X4 by the TRT assay (0.11; P < 0.01), and vice versa (0.10; P < 0.01). There was no significant difference between the X4 scores of the two groups with discordant phenotypic coreceptor classifications (P = 0.99). Two outliers in the R5/R5 category and one outlier in the R5/R5X4 category can be identified in Fig. 3. These outliers had a very high probability of harboring X4, as determined by the genotypic predictor, but were categorized as R5 according to one or both of the phenotypic coreceptor assays. Of note, samples classified as R5/X4 virus by both phenotypic assays were more heterogeneous (i.e., contained more amino acid mixtures in the V3 loop) than samples classified exclusively as R5 (median for R5/X4 samples, 1.5 amino acid mixtures/sequence; interquartile range for R5/X4 samples, 0.75 to 4.25; median for R5 samples, 1.0 amino acid mixtures/sequence; interquartile range for R5 samples, 1.0 to 2.0).
FIG. 3.
Box plot summary of the predicted probability of CXCR4 coreceptor usage. Mixtures were separated into all possible permutations per sample. The samples were grouped and labeled according to the results obtained from both phenotypic assays as “R5” or “R5/X4” and ordered by “Trofile assay/TRT assay.” The maximum score observed for each isolate was used to create the box plot. The boxes represent the interquartile range, and the solid line inside each box represents the median of the maxima of the group of samples. The box plot whiskers represent the range of the data in each category, and outliers are represented as open circles.
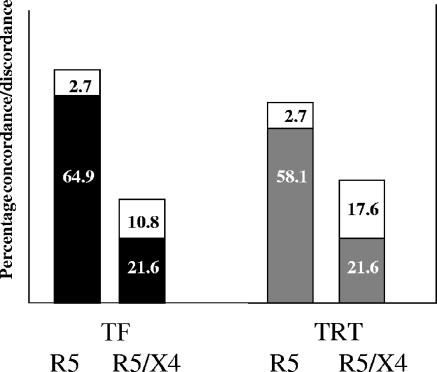
The results of the two phenotypic coreceptor assays were compared to the V3 sequence-based genotypic SVM predictions for the 74 patient-derived samples (19). The Trofile assay and SVM genotypic predictor resulted in concordance for 64 (86.5%) samples, whereas the TRT assay and SVM resulted in concordance for 59 (79.7%) samples (Fig. 4). Both the SVM and the Trofile coreceptor assay had a higher prevalence of classifying samples as R5 than the TRT assay.
FIG. 4.
Concordances between phenotypic coreceptor assays and an SVM bioinformatic predictor. The black bars represent the concordances between the Trofile coreceptor assay (TF) and SVM, and the gray bars represent concordances between the TRT coreceptor assay and SVM. Percent discordances are represented by the white bars.
DISCUSSION
This study compared two well-characterized recombinant phenotypic coreceptor assays on a set of clinically derived plasma samples selected to contain roughly equal numbers of R5- and X4-containing HIV-1 isolates. The overall concordance of 85.1% between the assays indicates that although they are in good agreement with each other, there remains considerable disagreement. Discordant samples appear to show no significant difference between viral loads or CD4 counts, although concordant R5/R5 samples had significantly higher CD4 counts than samples identified by any assay as harboring X4 variants and concordant X4/X4 samples had much higher X4 signals as measured by RLU (Trofile assay) or OD (TRT assay). The SVM and Trofile coreceptor assay had higher concordance and a greater prevalence of classifying samples as R5 than the TRT assay. This may be a result of the SVM being trained on sequence data phenotyped by the Trofile coreceptor assay. The small number of discordances (n = 11) makes the determination of associations in observed clinical or assay parameters between the two discordant groups difficult. A more thorough analysis of discordances would require a larger sample size.
Some potentially relevant differences between the two assays may account for many of the observed differences between coreceptor phenotypes observed here. These differences include the size of the viral RNA fragment excised and amplified, the cells used to express the coreceptors, and the reporter gene. While the Trofile HIV assay uses the entire gp160 gene fragment, approximately 2,500 base pairs, the TRT assay uses a 900-bp-long viral fragment that spans the V1-V3 region, including 150-bp-long extensions on each side to allow homologous recombination during transfection (21). One hypothesis that may require further investigation is whether amplification of a larger DNA fragment leads to a greater loss of minority species during PCR amplification. Other potential reasons for discordance between assay results have been reviewed by Coakley et al. (4), including the interaction between the viral vector Gag proteins and the patient-derived envelope proteins, which may affect pseudotyping efficiency and the usage of reporter cell lines. The use of different target cell lines could be another source of discrepancy between the assays, as the tropism assays made use of human malignant glioma cell lines (U87 and U373) engineered to express CD4 and various chemokine coreceptors on their surfaces. Previous studies have shown that coreceptor utilization may be influenced by the level of receptor expression and the ratio of the receptors to each other (16). However, results from at least one study suggest that coreceptor levels do not impact the ability to measure coreceptor usage (15).
Clinical samples contain a heterogeneous viral population. Studies of the detection of minority species within interindividual virus populations have shown that the limit of detection is in the range of 10 to 20% for the phenotypic coreceptor assays (4) while the limit of detection of conventional genotypic assays (consisting of reverse transcription (RT)-PCR, followed by population [“bulk”] sequencing) is approximately 25%. This is largely because low-level signals cannot be detected by conventional automated DNA sequencing, a factor that limits both the phenotypic coreceptor assays and genotypic predictors (SVM).
Developing a reliable genotypic predictor of coreceptor usage requires a well-validated phenotypic coreceptor assay to use as a reference. Although the “11/25” rule for determining tropism, which categorizes variants as syncytium inducing if they display positively charged residues at codons 11 and/or 25 within the V3 loop and non-syncytium inducing if they do not, is reasonably specific, it lacks sensitivity (<60%) for predicting coreceptor usage (9). SVM analysis of clinically derived sequences appears to correlate well with the results of phenotypic coreceptor assays and has higher sensitivity and specificity than other bioinformatic models for determining coreceptor usage from clonal data (19). However, the inability to consistently detect minority species in clinically derived sequence data presents difficulties both in using genotypic predictors and in training these predictors with sequence data phenotyped using assays that are also unable to consistently detect minority species.
In summary, the two phenotypic coreceptor assays tested here generated largely concordant results. However, in cases where there were differences in the assay results, it was not clear which of the assays was “correct.” The discordances did not show significant trends that could be attributed to differences between the two phenotypic coreceptor assays or to patient characteristics. It is likely that the presence of heterogeneous viral populations in clinically derived isolates posed difficulties of detection for both phenotypic and genotypic/bioinformatic approaches for determining coreceptor use. For the purposes of determining coreceptor usage in a clinical setting, an understanding of the limitations of the test used is essential. Although genotypic predictive models have the potential to reduce the cost and time for determining the coreceptor usage of patient-derived isolates, validation of genotypic methods against a verified reference would be required before they can be adopted for clinical use.
Footnotes
Published ahead of print on 22 November 2006.
REFERENCES
- 1.Brumme, Z. L., W. W. Y. Dong, B. Yip, B. Wynhoven, N. G. Hoffman, R. Swanstrom, M. A. Jensen, J. I. Mullins, R. S. Hogg, J. S. G. Montaner, and P. R. Harrigan. 2004. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial antiretroviral therapy. AIDS 18:F1-F9. [DOI] [PubMed] [Google Scholar]
- 2.Brumme, Z. L., J. Goodrich, H. B. Mayer, C. J. Brumme, B. M. Henrick, B. Wynhoven, J. J. Asselin, P. K. Cheung, R. S. Hogg, J. S. G. Montaner, and P. R. Harrigan. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naïve individuals. J. Infect. Dis. 192:466-474. [DOI] [PubMed] [Google Scholar]
- 3.Capon, D., and C. J. Petropoulos. November. 1998. Compositions and methods for determining anti-viral drug susceptibility and resistance and anti-viral drug screening. U.S. patent 5,837,464.
- 4.Coakley, E., C. J. Petropoulos, and J. M. Whitcomb. 2005. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 18:9-15. [DOI] [PubMed] [Google Scholar]
- 5.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodificiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation of the third variable domain of the human immunodificiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, M. A., A. B. van't Wout, D. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Predicting HIV-1 co-receptor usage using sequence analysis. AIDS Rev. 5:104-112. [PubMed] [Google Scholar]
- 10.Hoffman, N. G., F. Seilier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogg, R. S., B. Yip, K. J. Chan, E. Wood, K. J. Craib, M. V. O'Shaughnessy, and J. S. Montaner. 2001. Rates of disease progression by baseline CD4 cell count and viral load after initiating triple-drug therapy. JAMA 286:2568-2577. [DOI] [PubMed] [Google Scholar]
- 12.Huang, W., T. Wrin, J. Yap, S. Fransen, J. Beauchaine, M. Reddy, E. E. Paxinos, N. T. Parkin, J. M. Whitcomb, and C. J. Petropoulos. 2001. A rapid, multifunctional HIV-1 entry assay for measuring drug susceptibility, coreceptor tropism, and antibody neutralization, abstr. 1968. Progr. Abstracts 41st Annu. Intersci. Conf. Antimicrob. Agents Chemother.
- 13.Kuhmann, S. E., and J. P. Moore. 2005. The HIV-1 phenotypic variants: deadly and deadlier. J. Viral Entry 1:4-16. [Google Scholar]
- 14.Maeda, K., H. Nakata, H. Ogata, Y. Koh, T. Miyakawa, and H. Mitsuya. 2004. The current status of, and challenges in, the development of CCR5 inhibitors as therapeutics for HIV-1 infection. Curr. Opin. Pharmacol. 4:447-452. [DOI] [PubMed] [Google Scholar]
- 15.Princen, K., S. Hatse, K. Vermeire, G. J. Bridger, R. T. Skerlj, E. De Clercq, and D. Schols. 2003. The antiviral activity of the CXCR4 antagonist AMD 3100 is independent of the cytokine-induced CXCR4/HIV coreceptor expression level. AIDS Res. Hum. Retrovir. 19:1135-1139. [DOI] [PubMed] [Google Scholar]
- 16.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 18.Seibert, C., and T. P. Sakmar. 2004. Small-molecule antagonists of CCR5 and CXCR4: a promising new class of anti-HIV-1 drugs. Curr. Pharm. Des. 10:2041-2062. [DOI] [PubMed] [Google Scholar]
- 19.Sing, T., R. Harrigan, N. Beerenwinkel, M. Daumer, R. Kaiser, A. Low, P. K. Cheung, and T. Lengauer. 2005. Immunologic markers improve genotypic prediction of HIV-1 coreceptor usage, abstr. 30. Abstr. 1st Int. Workshop Targeting HIV Entry, Bethesda, MD.
- 20.Tanaka, Y., K. Okuma, R. Tanaka, S. Kumakura, A. Shimoyamada, K. Hirose, M. Yanaka, T. Murakami, and N. Yamamoto. 2006. Development of novel orally bioavailable CXCR4 antagonists, KRH-3955 and KRH-3140: binding specificity, pharmacokinetics and anti-HIV-1 activity in vivo and in vitro, abstract 49LB. Abstr. 13th Conf. Retrovir. Opportunistic Infect., Denver, CO.
- 21.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2000. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]