Abstract
Human metapneumovirus (hMPV) is a recently discovered paramyxovirus that is known to cause respiratory tract infections in children and immunocompromised individuals. Given the difficulties of identifying hMPV by conventional culture, molecular techniques could improve the detection of this virus in clinical specimens. In this study, we developed a real-time reverse transcription-PCR (RT-PCR) assay designed to detect the four genetic lineages of hMPV. This assay and a commercial real-time nucleic acid sequence-based amplification (NASBA) assay (bioMérieux, Durham, NC) were used to determine the prevalence of hMPV in 114 immunosuppressed asymptomatic and symptomatic lung transplant recipients and 232 pediatric patients who were being evaluated for pertussis. hMPV was detected in 4.3% of the immunosuppressed lung transplant recipients and in 9.9% of children evaluated for pertussis. Both RT-PCR and NASBA assays were efficient in detection of hMPV infection in respiratory specimens. Even though hMPV was detected in a small number of the lung transplant recipients, it was still the most prevalent etiologic agent detected in patients with respiratory symptoms. In both of these diverse patient populations, hMPV infection was the most frequent viral respiratory tract infection identified. Given our findings, infection with hMPV infection should be determined as part of the differential diagnosis of respiratory illnesses.
Human metapneumovirus (hMPV) is a negative-sense, single-stranded RNA virus that was first described in 2001 as a novel paramyxovirus isolated from the respiratory tract of children in The Netherlands (23). Since its initial description, hMPV has been reported worldwide (8, 9, 14, 15, 17, 22, 23, 26, 28), particularly in children and immunocompromised adults (6, 18, 21). hMPV has two main genetic lineages, A and B, with two subtypes for each lineage (A1, A2, B1, and B2) (19, 21, 24). hMPV had gone unrecognized for many years because it displays very slow replication kinetics in vitro, does not replicate efficiently in continuous cell lines, and requires trypsin for growth in vitro (23). hMPV causes occasional upper respiratory tract infections, although lower respiratory tract infections can result in bronchiolitis, pneumonitis, and asthma exacerbation (7, 10, 23). Studies have closely associated a seasonal incidence of hMPV infections during late winter (January to April). In addition, 1.2 to 4.1% of asymptomatic individuals are positive for hMPV RNA by reverse transcription-PCR (RT-PCR), suggesting that inapparent infections are common (6, 23, 27).
Solid-organ transplant recipients, particularly lung transplant recipients, are susceptible to opportunistic respiratory infections that are mostly of unknown etiology. Among the potential posttransplant complications, obliterative bronchiolitis is the most significant. Respiratory viral infections have been postulated to be associated with the development of obliterative bronchiolitis, since immunosuppression leaves lung transplant recipients more susceptible to community-acquired infections (11). In this study, we have developed and compared a real-time RT-PCR assay targeting the nucleoprotein (N) gene and a nucleic acid sequence-based amplification (NASBA) assay targeting the matrix gene for detection of hMPV infection in respiratory specimens from lung transplant recipients and children who were being evaluated for pertussis to determine its prevalence in these two diverse patient populations.
MATERIALS AND METHODS
Sample collection.
Bronchoalveolar lavage (BAL) specimens were collected from adult lung transplant recipients. Bronchoscopies with bronchoalveolar lavage were performed at regular intervals according to University of Pittsburgh Medical Center transplantation protocols (1, 3, 6, 9, and 12 months posttransplant) and as indicated by symptomatic events such as fever, radiographic infiltrates, and decreased forced expiratory flow as determined by spirometry. One hundred microliters of BAL specimens was stored in lysis buffer (bioMérieux, Durham, NC) at −80°C in a total volume of 1 ml. Suspensions of nasopharyngeal secretions were obtained from a collection maintained by the Pediatric Molecular Microbiology Laboratory at Children's Hospital of Pittsburgh (PA). The secretions were collected with Dacron swabs and suspended in 500 μl of saline, and the suspensions were stored at −80°C as single-use aliquots (i.e., 100 μl) until needed (25). The swab specimens had been obtained as part of routine care of pediatric patients who were evaluated for pertussis between February and May 2005.
Nucleic acid extraction.
Isolation of viral nucleic acid from control material and patient specimens was done using the NucliSens Automated Extractor (bioMérieux, Durham, NC) according to the manufacturer's instructions. Briefly, 100 μl of sample was lysed in lysis buffer (bioMerieux, Durham, NC) for 30 min, following which a fixed volume and concentration of equine arteritis virus (EAV) was added as internal control for extraction and amplification in addition to diluted silica per the manufacturer's instructions. The solution was transferred into a closed system cartridge and placed onto the instrument for extraction. The procedure took approximately 1 h and the RNA was eluted in 50 μl of elution buffer (bioMerieux, Durham, NC), which was stored at −80°C in the same tube until use.
Virus and viral RNA.
Viral isolates of hMPV strains Can97-83 (strain A2) and Can98-75 (strain B2) stored in TRIzol buffer were kindly provided by Dean Erdman of the Centers for Disease Control and Prevention (Atlanta, GA). Can97-83 viral RNA was kindly provided by Ursula Buchholz at the National Institutes of Health (Bethesda, MD). Total RNA was extracted using the RNeasy kit (QIAGEN, Alameda, CA). Equine arteritis virus (EAV) was obtained from the American Type Culture Collection (VR-796; Rockville, MD) for preparation of an internal control for the hMPV RT-PCR assays. The virus was inoculated into a 75-cm2 flask of Vero cells near confluence. Infection resulted in detachment of the monolayer from the flask in less than 24 h. The virus was harvested from the supernatant following sonication and stored (500 μl:500 μl) in minimal essential medium with 10% fetal bovine serum at −80°C. A 1/50 dilution of the virus (106.25 50% tissue culture infectious doses/ml) was spiked into each specimen prior to extraction.
Real-time RT-PCR.
Reverse transcription and amplification of hMPV RNA was performed in a single tube using the in-house-developed assay targeting the nucleoprotein (N) gene. The primers Forward (5′-CATCAGGTAATATCCCACAAAATCAG-3′ and Reverse 5′-GTGAATATTAAGGCACCTACACATAATAARA-3′) and probe (5′-6-carboxyfluorescein-TCAGCACCAGACACAC- minor groove binding-3′) were designed using Primer Express software (Applied Biosystems). One-step RT qPCR MasterMix (Eurogentec, San Diego, CA) was used according to the manufacturer's instructions. This 25-μl reaction consisted of 2× reaction buffer, 8 mM MgCl2, 200× EuroScript RT (0.25 U/μl), RNase inhibitor (0.1 μl), 300 nM forward primer, 300 nM reverse primer, Taq DNA polymerase, 100 nM probe, template RNA, and sterile H2O. The amplification was carried out using an ABI PRISM 7000 (Applied Biosystems, Foster City, CA) with the following cycling conditions: RT step at 48°C for 30 min]; Taq activation step at 95°C for 10 min for cycle, followed by 45 cycles of a PCR step at 95°C for 15 s and 60°C for 1 min. Amplification of EAV served as an internal control in every patient specimen to determine the integrity of the sample controlling for both extraction and amplification procedures. The primers and assay conditions for EAV amplification were as previously described (1). RT-PCRs for hMPV and EAV were performed in separate wells.
Real-time NASBA.
Real-time NASBA was performed using NucliSens Basic kit reagents (bioMérieux) on an EasyQ Analyzer (bioMérieux, Durham, NC) for 120 min according to the manufacturer's instructions. hMPV-specific primer/beacon mix was provided by bioMérieux (Durham, NC) for virus amplification and detection. Each sample was tested in a 20-μl NASBA reaction mixture containing 5 μl of template, 5 μl of enzyme (1 enzyme sphere diluted in 55 μl of enzyme diluent), and 10 μl of reagent mastermix (1 reagent sphere, 80 μl of reagent diluent, 16 μl of 80 mM KCl, 10 μl of primer/beacon mix, and 14 μl of H2O). RNA was added to the reagent mastermix and incubated at 65°C for 2 min and 41°C for 2 min. Following the incubation, the enzyme was added and mixed by briefly flicking and spinning the tubes. The EasyQ analyzer was utilized for amplification and detection.
Viral culture and immunofluorescent staining.
Two R-Mix shell vials containing a mixed monolayer of mink lung cells (strain Mv1Lu) and human adenocarcinoma cells (strain A549) were inoculated with 200 μl of clinical specimen according to the manufacturer's instructions (Diagnostic Hybrids, Athens, OH). Each vial was centrifuged at 700 × g for 60 min and incubated at 35°C. Screening of the R-Mix shell vial at 18 to 24 h postinoculation was accomplished with the first of the two vials. Coverslips were fixed with acetone and stained with a pool of respiratory virus fluorescent antibodies (adenovirus, influenza A and B viruses, respiratory syncytial virus [RSV], and parainfluenza viruses 1 to 3) (Bartels, Issaquah, WA) according to the manufacturer's instructions. Positive specimens were further identified with the second R-Mix shell vial by scraping and spotting 8-well slides. These slides were fixed with acetone again and stained with virus-specific monoclonal antibodies.
Construction of N-gene plasmid and RNA transcripts.
The full-length hMPV N gene was amplified by conventional two-step RT-PCR from the Can97-83 viral RNA. A gene-specific primer downstream of the N-gene region was designed for reverse transcription (5′-GCCTTGCGAGACATTATGATTTG-3′), and the forward (5′-GAAAATGTCTCTTCAAGGGATTCAC-3′) and the reverse primers (5′-GCCAATTTTGCCGCTTCAT-3′) were used to amplify the full-length N gene. The PCR product was gel purified using the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) according to the manufacturer's instructions and ligated into the pGEM-T Easy Vector (Promega, Madison, WI). Insertion of the correct sequences into the plasmid was confirmed by digesting the plasmid with PstI (Promega, Madison, WI) at 37°C for 3 h. In vitro transcription was performed under the control of the SP6 promoter using the Ribomax Large Scale RNA Production System (Promega, Madison, WI). The transcripts were DNase treated and RNA purified by phenol-chloroform extraction according to the manufacturer's instructions. The concentration of hMPV N-gene transcript was determined spectrophotometrically and used as a positive control for real-time RT-PCR during each run.
RESULTS
Sensitivity and specificity of molecular detection assays.
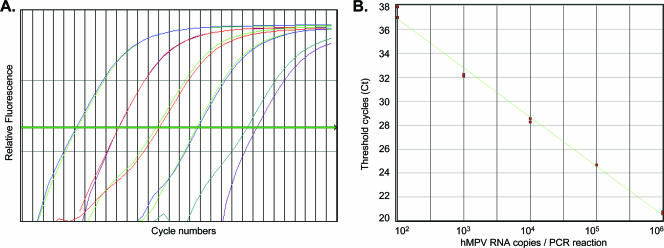
The sensitivity of the N-gene real-time RT-PCR assay using serially diluted transcripts was 100 RNA copies/RT-PCR (Fig. 1). Using these serially diluted transcripts as external standards, hMPV strains A (Can97-83) and B (Can98-75) were quantitated by real-time RT-PCR. Differences in sensitivities of amplification between RT-PCR and NASBA methods as well as between the two hMPV strains A and B were observed. The lower limits of detection of hMPV strains A and B by real-time RT-PCR was 50 and 100 copies/RT-PCR, respectively, and that by NASBA was 100 and 1,000 copies/reaction, respectively (data not shown). Both assays were highly specific for hMPV detection and failed to amplify RNA from other respiratory viruses such as influenza A and B viruses, respiratory syncytial virus, parainfluenza viruses 1 to 3, adenovirus, and sudden acute respiratory syndrome coronavirus.
FIG. 1.
Sensitivity of hMPV real-time RT-PCR. (A and B) Amplification plot and the external standard curve generated by serially diluted hMPV N-gene in vitro transcripts by real-time RT-PCR. The values shown in panel A represent hMPV RNA copies/RT-PCR. Based on copy numbers of the in vitro-transcribed RNA standards, the lower detection limit is 100 RNA copies/RT-PCR by real-time RT-PCR.
hMPV in adult lung transplant recipients.
A total of 234 BAL specimens from 114 lung transplant recipients were tested for hMPV infection by both real-time RT-PCR and real-time NASBA. At the time of collection, 156 of these BAL specimens were from asymptomatic lung transplant recipients and 78 were from symptomatic patients. Samples were collected spanning a 1-year period from May 2004 to April 2005. The patients' ages ranged from 22 to 75 years old, with an average age of 51.7. The cohort was comprised of 41.2% males and 58.8% females.
All samples included in the study were extracted and amplified without inhibition, as evidenced by positive EAV cyclic threshold values (data not shown). Out of 114 patients, 5 (4.3%) specimens from different patients were positive for hMPV by either RT-PCR or NASBA, all occurring during the late winter season between February and April 2005 (Fig. 2A). Two of these specimens were positive by both assays (Table 1). Of these five specimens positive for hMPV, three were positive by RT-PCR (1.2%), while NASBA detected four (1.7%) hMPV. Of the five hMPV-positive specimens, only one (patient 1130) was asymptomatic at the time of sample collection, which was obtained during a routine follow-up, while the other four (patients 1119, 1124, 1396, and 1422) had respiratory symptoms and underwent bronchoscopies. There were no significant changes in forced expiratory volume in 1 s in lung transplant recipients following hMPV infection. In addition to hMPV, two patients, 1124 and 1119, were positive for cytomegalovirus pp65 antigenemia, while patient 1396 was also positive for Candida albicans, Streptococcus viridans, Neisseria sp., and Propionibacterium sp. infections from BAL specimens. Patient 1119 was rejecting the graft at the time of hMPV infection.
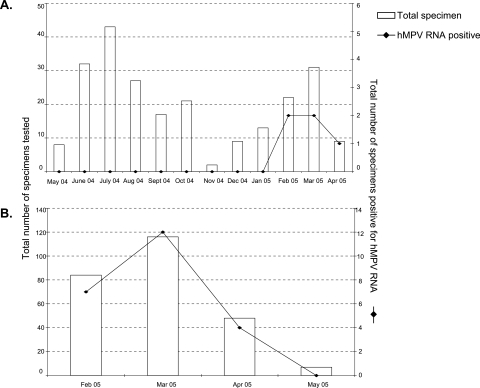
FIG. 2.
Seasonal variation of hMPV prevalence. (A) Graphical representation of the total number of respiratory specimens received from lung transplant recipients during May 2004 to April 2005 and the number of samples positive for hMPV RNA. (B) Graphical representation of the total number of respiratory specimens received from pediatric patients during February to May 2005 and the number of hMPV-positive samples detected during each month.
TABLE 1.
Associated clinical data from specimens positive for hMPV RNAa
| Study identity no. | Test result
|
Respiratory symptoms present | Type of lung transplant | Rejection | Baseline FEV1c (PFEV1) | FEV1b | |
|---|---|---|---|---|---|---|---|
| TaqMan RT-PCR | NASBA | ||||||
| 1119 | Positive | Negative | Yes | Heart/lung | Minimal ACRd | 2.62 | 1.18 |
| 1124 | Positive | Positive | Yes | Double | No | ND | 2.78 |
| 1130 | Positive | Positive | No | Left single | No | 1.43 | 1.5 |
| 1396 | Negative | Positive | Yes | Heart/lung | No | 1.53 | 1.63 |
| 1422 | Negative | Positive | Yes | Double | No | ND | 1.23 |
The clinical data correspond to the time at which the specimens were collected for hMPV RNA detection.
FEV1, forced expiratory volume in 1 s.
Baseline FEV1 (PFEV1), preceding FEV1 values prior to the onset of symptoms within a month. ND, not done.
ACR, acute rejection.
An immunofluorescence assay was used to determine the prevalence of other respiratory viruses in the lung transplant recipients. In this patient population, one specimen was positive for adenovirus (0.008%) while none of the specimens tested positive for influenza A, influenza B, RSV, or paramyxoviruses 1 to 3 (data not shown). Respiratory specimens taken throughout the entire year from these highly immunosuppressed patients showed that hMPV infection was the etiologic agent during the months (February to April) when hMPV is prevalent in the community (Fig. 2A).
hMPV in children evaluated for pertussis.
A total of 232 nasopharyngeal samples, each from a different pediatric patient undergoing clinical evaluation for pertussis, were tested for hMPV infection by real-time RT-PCR and NASBA. This group of patients had a median age of 3 years, and 53.5% were males. Both assays together detected hMPV RNA in a total of 23 (9.9%) specimens. Twenty-two (9.4%) specimens were positive for hMPV RNA by real-time RT-PCR, and 15 (6.4%) were positive by NASBA. Of the 22 RT-PCR positive specimens, 8 were negative by NASBA, whereas RT-PCR failed to detect only 1 of the 15 NASBA-positive samples (Table 2).
TABLE 2.
Comparison of real-time RT-PCR and NASBA assays for detection of hMPV RNA in pediatric patientsa
| Real-time RT-PCR test result | No. NASBA positive | No. NASBA negative | Total |
|---|---|---|---|
| Positive | 14 | 8 | 22 (9.4%) |
| Negative | 1 | 209 | 210 |
| Total | 15 (6.4%) | 217 | 232 |
The numbers in parentheses represent percentages of positives in the total population tested.
Other pathogens detected in this group during the same time period included Bordetella pertussis, which was detected by PCR from 17 specimens (7.3%); RSV, which was detected by R-Mix shell vial culture and immunofluorescent staining from 5 specimens (2.1%); and Mycoplasma pneumoniae, which was detected from 1 (0.4%) respiratory specimen. Though tested on different respiratory specimen types, none of the hMPV-positive patients were coinfected with any of the other pathogens. The hMPV positivity peaked during the month of March and disappeared by May (Fig. 2B).
DISCUSSION
Since its discovery in 2001, hMPV has been found primarily in children, the elderly, and immunosuppressed individuals. Diagnosis of hMPV is difficult by the conventional culture techniques because of slow growth and subtle cytopathic effect. Real-time RT-PCR has been described previously for rapid detection of hMPV RNA (5, 16). In this study, we have developed a real-time RT-PCR assay that is designed to detect the N gene of all known hMPV lineages. hMPV has two main genetic lineages, A and B, with two subtypes within each lineage (A1, A2, B1, and B2). The genetic homology at the nucleotide level within each lineage is 94%, and between each lineage it is 84% (3). Can97-83 and Can98-75, representative of strains A2 and B2, respectively, were available for validation; however, the primers and probe for the real-time RT-PCR assay were designed to detect all four known genetic lineages based on available sequence data. In addition to the real-time RT-PCR, we also used a real-time NASBA assay available for hMPV from bioMerieux. Amplification of hMPV genotype B had a lower sensitivity than genotype A by both of the molecular methods of detection. This was probably due to the differences in the nucleotide homology between genotypes A and B (2, 4) in clinical isolates. NASBA was able to detect two additional specimens that were negative by RT-PCR in the transplant group, whereas in the pediatric group, RT-PCR detected eight hMPV RNA-positive specimens that NASBA failed to detect. These discrepancies between the two tests could be due to their different target regions in the hMPV genome or to a low concentration of hMPV RNA.
Since it has been suggested that hMPV can cause respiratory infection in immunocompromised individuals, we examined its incidence in lung transplant recipients. Out of 114 patients tested at different time intervals, hMPV RNA was detected in 5 patients (4.3%) by either RT-PCR or NASBA. The onset of hMPV infection varied from less than 1 month to about 1 year posttransplant. Out of the 156 specimens collected from asymptomatic patients, only 1 (0.6%) tested positive for hMPV, while 4 (5.1%) were positive from symptomatic patients. The infection was not persistent and resolved without major complications. However, compared to other respiratory infections, hMPV was a predominant infection in this cohort of transplant recipients. Recent studies by Gerna et al. (7) and Larcher et al. (13) have found a high prevalence of hMPV in symptomatic lung transplant recipients. However, Kumar et al. (11) observed hMPV as the etiologic agent in only 1 of 50 patients with respiratory tract infections from lung transplant recipients.
As expected and in agreement with previous studies (12, 20, 28), a higher prevalence of hMPV infection was observed in symptomatic children. Surprisingly, hMPV was the predominant infection in this cohort, followed by Bordetella pertussis and RSV. Also, coinfection of hMPV infection was not observed with other respiratory pathogens. Our study further confirms previous data suggesting a higher prevalence of hMPV infection during the months of February to April (6, 22).
In conclusion, using two molecular biology-based assays we have examined the prevalence of hMPV infection in two diverse patient populations. In lung transplant recipients, 4.3% of patients showed evidence of hMPV infection, most of whom were symptomatic at the time of specimen collection. In the pediatric population undergoing evaluation for pertussis, however, a high prevalence (9.9%) of hMPV infection was observed. These data and the seasonal predominance of hMPV infection suggest that it is not an opportunistic pathogen that selectively infects immunosuppressed patients. Further studies to determine the prevalence of hMPV infection in immunosuppressed individuals are needed.
Acknowledgments
Portions of this study were used in fulfillment of the requirements for an M.S. degree by R.D. in the University of Pittsburgh Graduate School of Public Health.
We thank BioMerieux for partly supporting this project by supplying hMPV primers and molecular beacon and the reagents required for NASBA testing.
Footnotes
Published ahead of print on 25 October 2006.
REFERENCES
- 1.Balasuriya, U. B., C. M. Leutenegger, J. B. Topol, W. H. McCollum, P. J. Timoney, and N. J. MacLachlan. 2002. Detection of equine arteritis virus by real-time TaqMan reverse transcription-PCR assay. J. Virol. Methods 101:21-28. [DOI] [PubMed] [Google Scholar]
- 2.Bastien, N., S. Normand, T. Taylor, D. Ward, T. C. Peret, G. Boivin, L. J. Anderson, and Y. Li. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 93:51-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biacchesi, S., M. H. Skiadopoulos, G. Boivin, C. T. Hanson, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Boivin, G., I. Mackay, T. P. Sloots, S. Madhi, F. Freymuth, D. Wolf, Y. Shemer-Avni, H. Ludewick, G. C. Gray, and E. LeBlanc. 2004. Global genetic diversity of human metapneumovirus fusion gene. Emerg. Infect. Dis. 10:1154-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouscambert-Duchamp, M., B. Lina, A. Trompette, H. Moret, J. Motte, and L. Andreoletti. 2005. Detection of human metapneumovirus RNA sequences in nasopharyngeal aspirates of young French children with acute bronchiolitis by real-time reverse transcriptase PCR and phylogenetic analysis. J. Clin. Microbiol. 43:1411-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falsey, A. R., D. Erdman, L. J. Anderson, and E. E. Walsh. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785-790. [DOI] [PubMed] [Google Scholar]
- 7.Gerna, G., P. Vitulo, F. Rovida, D. Lilleri, C. Pellegrini, T. Oggionni, G. Campanini, F. Baldanti, and M. G. Revello. 2006. Impact of human metapneumovirus and human cytomegalovirus versus other respiratory viruses on the lower respiratory tract infections of lung transplant recipients. J. Med. Virol. 78:408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray, G. C., A. W. Capuano, S. F. Setterquist, J. L. Sanchez, J. S. Neville, J. Olson, M. G. Lebeck, T. McCarthy, Y. Abed, and G. Boivin. 2006. Human metapneumovirus, Peru. Emerg. Infect. Dis. 12:347-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaida, A., N. Iritani, H. Kubo, M. Shiomi, U. Kohdera, and T. Murakami. 2006. Seasonal distribution and phylogenetic analysis of human metapneumovirus among children in Osaka City, Japan. J. Clin. Virol. 35:394-399. [DOI] [PubMed] [Google Scholar]
- 10.Kim, Y. K., and H. J. Lee. 2005. Human metapneumovirus-associated lower respiratory tract infections in Korean infants and young children. Pediatr. Infect. Dis. J. 24:1111-1112. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, D., D. Erdman, S. Keshavjee, T. Peret, R. Tellier, D. Hadjiliadis, G. Johnson, M. Ayers, D. Siegal, and A. Humar. 2005. Clinical impact of community-acquired respiratory viruses on bronchiolitis obliterans after lung transplant. Am. J. Transplant. 5:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuypers, J., N. Wright, L. Corey, and R. Morrow. 2005. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J. Clin. Virol. 33:299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larcher, C., C. Geltner, H. Fischer, D. Nachbaur, L. C. Muller, and H. P. Huemer. 2005. Human metapneumovirus infection in lung transplant recipients: clinical presentation and epidemiology. J. Heart Lung Transplant. 24:1891-1901. [DOI] [PubMed] [Google Scholar]
- 14.Luchsinger, V., C. Escobar, and L. F. Avendano. 2005. Detection of human metapneumovirus in children hospitalized for acute lower respiratory infection in Santiago, Chile. Rev. Med. Chile 133:1059-1064. [DOI] [PubMed] [Google Scholar]
- 15.Ludewick, H. P., Y. Abed, N. N. van G. Boivin, K. P. Klugman, and S. A. Madhi. 2005. Human metapneumovirus genetic variability, South Africa. Emerg. Infect. Dis. 11:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maertzdorf, J., C. K. Wang, J. B. Brown, J. D. Quinto, M. Chu, G. M. de, B. G. van den Hoogen, R. Spaete, A. D. Osterhaus, and R. A. Fouchier. 2004. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 42:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maggi, F., M. Pifferi, M. Vatteroni, C. Fornai, E. Tempestini, S. Anzilotti, L. Lanini, E. Andreoli, V. Ragazzo, M. Pistello, S. Specter, and M. Bendinelli. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahalingam, S., J. Schwarze, A. Zaid, M. Nissen, T. Sloots, S. Tauro, J. Storer, R. Alvarez, and R. A. Tripp. 2006. Perspective on the host response to human metapneumovirus infection: what can we learn from respiratory syncytial virus infections? Microbes Infect. 8:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peret, T. C., G. Boivin, Y. Li, M. Couillard, C. Humphrey, A. D. Osterhaus, D. D. Erdman, and L. J. Anderson. 2002. Characterization of human metapneumoviruses isolated from patients in North America. J. Infect. Dis. 185:1660-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Principi, N., S. Esposito, and S. Bosis. 2004. Human metapneumovirus and lower respiratory tract disease in children. N. Engl. J. Med. 350:1788-1790. [PubMed] [Google Scholar]
- 21.Prins, J. M., and K. C. Wolthers. 2004. Human metapneumovirus: a new pathogen in children and adults. Neth. J. Med. 62:177-179. [PubMed] [Google Scholar]
- 22.Rao, B. L., S. S. Gandhe, S. D. Pawar, V. A. Arankalle, S. C. Shah, and A. A. Kinikar. 2004. First detection of human metapneumovirus in children with acute respiratory infection in India: a preliminary report. J. Clin. Microbiol. 42:5961-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Hoogen, B. G., J. C. de Jong, J. Groen, T. Kuiken, G. R. de, R. A. Fouchier, and A. D. Osterhaus. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Hoogen, B. G., S. Herfst, L. Sprong, P. A. Cane, E. Forleo-Neto, R. L. de Swart, A. D. Osterhaus, and R. A. Fouchier. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadowsky, R. M., R. H. Michaels, T. Libert, L. A. Kingsley, and G. D. Ehrlich. 1996. Multiplex PCR-based assay for detection of Bordetella pertussis in nasopharyngeal swab specimens. J. Clin. Microbiol. 34:2645-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werno, A. M., T. P. Anderson, L. C. Jennings, P. M. Jackson, and D. R. Murdoch. 2004. Human metapneumovirus in children with bronchiolitis or pneumonia in New Zealand. J. Paediatr. Child Health 40:549-551. [DOI] [PubMed] [Google Scholar]
- 27.Williams, J. V., P. A. Harris, S. J. Tollefson, L. L. Halburnt-Rush, J. M. Pingsterhaus, K. M. Edwards, P. F. Wright, and J. E. Crowe, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf, D. G., Z. Zakay-Rones, A. Fadeela, D. Greenberg, and R. Dagan. 2003. High seroprevalence of human metapneumovirus among young children in Israel. J. Infect. Dis. 188:1865-1867. [DOI] [PubMed] [Google Scholar]